Abstract
The Golgi-associated, γ-adaptin homologous, ADP-ribosylation factor (ARF)-interacting proteins (GGAs) are adaptors that sort receptors from the trans-Golgi network into the endosomal/lysosomal pathway. The GGAs and TOM1 (GAT) domains of the GGAs are responsible for their ARF-dependent localization. The 2.4-Å crystal structure of the GAT domain of human GGA1 reveals a three-helix bundle, with a long N-terminal helical extension that is not conserved in GAT domains that do not bind ARF. The ARF binding site is located in the N-terminal extension and is separate from the core three-helix bundle. An unanticipated structural similarity to the N-terminal domain of syntaxin 1a was discovered, comprising the entire three-helix bundle. A conserved binding site on helices 2 and 3 of the GAT domain three-helix bundle is predicted to interact with coiled-coil-containing proteins. We propose that the GAT domain is descended from the same ancestor as the syntaxin 1a N-terminal domain, and that both protein families share a common function in binding coiled-coil domain proteins.
The Golgi-associated, γ-adaptin homologous, ADP-ribosylation factor (ARF)-interacting proteins (GGAs) are a family of multimodular trafficking adaptors that sort receptors from the trans-Golgi network into the endosomal/lysosomal pathway (1–6). The GGA proteins consist of four modules: the Vps27, HRS, STAM (VHS); GGAs and TOM1 (GAT); hinge; and γ-adaptin ear (GAE) domains. The VHS domain at the N terminus is responsible for selecting receptor cargo by interacting with specific acidic cluster/dileucine signals in the cytoplasmic tails of the receptors (7–10). The GAT domain binds to the GTP-bound form of ARF1 and ARF3, which are responsible for targeting the GGAs to the trans-Golgi network (1, 3, 11). These isoforms of the ARF small G protein cycle between GTP and GDP associated states in a regulated manner, and thereby control the localization of their effectors (12). The hinge region contains a clathrin-binding sequence and is responsible for clathrin association (8, 11, 13, 14). The hinge region of GGA1 and GGA3 also contains an autoinhibitory sequence that, when phosphorylated by casein kinase II, blocks receptor binding to the VHS domain (15). The GAE domain is responsible for binding accessory proteins, including Rabaptin-5 and γ-synergin (2, 5, 8, 16).
The role of the GGAs in trafficking key lysosomal enzyme receptors, including the cation-dependent and cation-independent mannose 6-phosphate receptors, has led to intense interest in the structural characterization of the GGAs. The structures of the GGA1 and GGA3 VHS domain bound to signal peptides from the mannose 6-phosphate receptors have been determined (17–19). The structures of the related GAE domains of γ-adaptin have also been determined (20, 21), although the structures of GGA GAE domains have not. The GAT domain is predicted to be highly helical in structure on the basis of sequence on sequence analysis, but no 3D structure of a GAT domain has been reported.
GAT domains occur in TOM1 and TOM1-like proteins, in addition to the GGAs. There are 25 identified GAT domain proteins from various species in the Pfam database (22). The isolated GAT domains of GGAs bind to ARF1-GTP with ≈5 μM affinity, and the combined VHS-GAT domain binds with ≈10-fold higher affinity (23). TOM1 and TOM1-like are little characterized, but the mouse ortholog of TOM1-like has been dubbed “Srcasm” (24) for its interaction with the Src family tyrosine kinase Fyn. TOM1 and TOM1-like have the presence of an N-terminal VHS domain in common with the GGAs. However, the TOM1 protein binds neither Arf-GTP nor acidic-cluster/dileucine containing signal sequences (7, 11). A putative outlier GAT domain has been proposed in the yeast cytoskeletal/endocytic adaptor Lsb5p (25). A region of Lsb5p that falls within a putative GAT domain was recently found to interact with the endocytic adaptor protein Sla1p (25), and the possibility of an interaction between Lsb5p and ARF has been raised (25).
We undertook to determine the structure of a GAT domain from one of the GGAs. We subcloned and expressed the GAT domains of human GGAs 1–3 and attempted to crystallize each alone and in complex with ARF1-guanosine 5′-[γ-thio]triphosphate (GTPγS). We obtained suitable crystals of the GGA1 GAT domain, and we now report the 2.4-Å crystal structure of the GAT domain of human GGA1.
Methods
Protein Expression and Purification.
The GAT domain of GGA1 was subcloned into the pHIS-parallel2 vector (26). Overexpression of the His6-tagged protein in Escherichia coli Rosetta (DE3) cells (Novagen) was induced with isopropyl-β-D-thiogalactoside at 20°C. The protein was purified by using a Ni2+-nitrilotriacetate (NTA) column, the His6 tag was removed with TEV protease, and the protein was subjected to a second purification with Ni2+-NTA resin (Qiagen, Valencia, CA). The cleaved protein contains the vector-derived sequence GAMGS followed by the GGA1 residues 166–302. After dialysis in 50 mM Tris⋅HCl, pH 8/500 mM NaCl/1 mM DTT, the GAT domain was soluble at a concentration of 2 mg/ml. The protein yield was 7 mg/liter bacterial culture. The protein was stored at −80°C in dialysis buffer supplemented with 10% glycerol. The procedures for the cloning, expression, and purification of Δ1–17ARF1Q71L were essentially the same. After purification of the His6-tagged protein with Ni2+-NTA resin as described above, the cleaved protein was loaded onto a Superdex 75 gel filtration column equilibrated with 50 mM Tris⋅HCl, pH 9/150 mM NaCl/1 mM DTT by using a FPLC system (Amersham Pharmacia). The protein was stored at −80°C in elution buffer with 10% glycerol at a concentration of 19 mg/ml. To obtain a complex of these proteins, Δ1–17ARF1Q71L was activated by using GTPγS in a procedure modified from ref. 27 and subsequently incubated with the GAT domain. The complex was isolated by using a Superdex 75 gel filtration column equilibrated with 25 mM Hepes, pH 7.4/100 mM NaCl/1 mM DTT/1 mM EDTA/1 mM MgCl2. The proteins eluted on the gel-filtration column at a volume consistent with a heterodimeric complex of 1:1 stoichiometry. The complex was soluble at a concentration of 20 mg/ml and was stored at −80°C in elution buffer with 10% glycerol. Selenomethionyl (SeMet) GGA1 GAT domain and Δ1–17ARF1Q71L, respectively, were expressed in E. coli B834 cells in defined media. The purification procedures for the SeMet proteins were the same as above.
Crystallization, X-Ray Diffraction Data Collection, and Structure Determination.
Proteins were dialyzed into 20 mM Hepes, pH 8.2/50 mM NaCl/10 mM DTT for crystallization. Optimal crystallization conditions were found by using the Hampton Research (Riverside, CA) Crystal Screen I. A hanging droplet consisting of 2 μl of the protein solution (8 mg/ml) mixed with an equal volume of reservoir solution containing 0.1 M sodium citrate, pH 5.6, 20% polyethylene glycol 4000, and 20% isopropyl alcohol was equilibrated against 0.5 ml of the reservoir. Crystals suitable for data collection (see Table 1) grew within 1 week at 25°C to ≈50 μm maximum dimension. Several crystals were washed in reservoir solution, dissolved, and subjected to SDS/PAGE. The silver-stained gel revealed that the GAT domain protein but not ARF1 was present in the crystals. Native crystals were cryoprotected by adding 2 μl of 0.1 M sodium citrate, pH 5.6/10% polyethylene glycol 4000/30% polyethylene glycol 400/1 M NaBr directly to the droplet. After soaking for 10 s, the crystals were immediately frozen under N2 vapor at 95 K. SeMet crystals were cryoprotected by adding 2 μl of 0.1 M Na-citrate, pH 5.6/10% polyethylene glycol 4000/20% polyethylene glycol 400/20% isopropyl alcohol directly to the droplet and were rapidly frozen in liquid propane. A three-wavelength multiwavelength anomalous dispersion data set was collected from frozen crystals of SeMet GGA1 GAT at beamline 8.2.1, Advanced Light Source (Berkeley, CA). Data were collected for 1.5° and 10 s per exposure in 15° wedges, with a total of 90° direct and 90° inverse beam collected for each of the wavelengths. Data were collected on an ADSC four-panel charge-coupled device and processed with HKL2000, keeping the processed intensities unmerged. Se positions were located with SOLVE (28) and the map was improved with RESOLVE (29). The resulting 3.0-Å map was interpreted with o (30), and the assignment of sequence was initiated by using the identified SeMet positions. Native data were collected at beamline 19-ID, Advanced Photon Source, using a Mar charge-coupled device, and processed to 2.4 Å with HKL2000. Native and SeMet crystals were nonisomorphous. After preliminary refinement of the SeMet structure, the model was placed in the unit cell of the native protein with the fast direct rotation search and the translation search implemented in cns (31), and the structure was refined by using REFMAC5 (32).
Table 1.
Crystallographic data, phasing, and refinement statistics
| SeMet | Native | |
|---|---|---|
| Data collection and phasing | ||
| Space group | R3 | R3 |
| Unit cell, Å | a = b = 84.34, | a = b = 76.60 |
| c = 59.02 | c = 60.60 | |
| α = β = 90° | α = β = 90° | |
| γ = 120° | γ = 120° | |
| Wavelength, Å* | 0.9791 | 0.9199 |
| Resolution, Å | 50–2.8 (2.98–2.8)† | 50–2.4 (2.49–2.4) |
| Unique reflections | 3776 (385) | 5195 (474) |
| Completeness, % | 98.1 (98.7) | 98.7 (90.1) |
| Rmerge‡, % | 6.0 (22.5) | 5.5 (29.6) |
| Anomalous differences, % | 4.3–6.1 | |
| Dispersion differences, % | 3.4–5.3 | |
| Figure of merit-SOLVE | 0.53 (0.29) | |
| Refinement statistics | ||
| Resolution range, Å | 29.1–2.40 | |
| No. of reflections | 4,936 | |
| R§, % | 0.253 | |
| Rfree¶, % | 0.286 | |
| Crossvalidated Luzatti error | 0.298 | |
| rms deviations | ||
| Bond length, Å | 0.015 | |
| Bond angle, ° | 1.33 | |
| Average B factor, Å2 | 47.6 | |
| Protein atoms, no. | 854 | |
| Solvent atoms, no. | 39 | |
| Residues in core Φ–Ψ region | 100% | |
Statistics are shown for the peak wavelength of selenomethionine MAD data sets. Statistics for the inflection wavelength (0.9793 Å) and remote wavelength (0.95000 Å) were similar, except that the Rmerge values were lower and the anomalous differences varied in the range shown.
Statistics shown in parentheses are for the highest resolution shell (Å).
Rmerge = ∑|I(k) − 〈I(k)〉|/∑I(k).
R = ∑|Fobs − kFcalc|/∑|Fobs|.
Rfree is the R value calculated for a test set of reflections, composed of a randomly selected 5% of the data, not used during refinement.
Results
Structure of the GGA1 GAT Domain.
After screening several different combinations of the N- and C-terminal boundaries of the GAT domains of human GGAs 1–3 for their ability to crystallize either alone or in complex with ARF1-GTPγS, we obtained suitable crystals of the GGA1 GAT domain comprising residues 166–303. The crystals were obtained from a mixture of the ARF1 Q71L mutant in complex with GTPγS, which had been copurified as a complex on gel filtration chromatography. This complex was much more soluble than the GAT domain alone. The complex had a solubility limit of >20 mg/ml, as compared with 2 mg/ml for GGA1-GAT alone. The GGA1-GAT crystal form obtained grew only from the ARF1 mixture and never with the GAT domain alone. Nevertheless, the crystals obtained did not contain bound ARF1-GTPγS. We believe that the complex dissociated either during dialysis against crystallization buffer or subsequently during the incubation of crystallization trials. The ARF1-GTPγS appears to solubilize transiently the GAT domain. We believe that, as the complex dissociated, the less soluble GAT domain was slowly released from the complex and crystallized.
The structure was determined by multiwavelength anomalous dispersion phasing using SeMet GAT protein (Fig. 1). The SeMet GGA-GAT1 domain crystallized in the same space group but with two cell constants different by 8 Å from the native protein. The GGA1-GAT domain contains seven Met residues, but only three were located in the automated Patterson search. The phasing power from these three SeMet residues was sufficient to obtain an interpretable solvent-flattened map at 3.0-Å resolution. The other four Met residues were subsequently located, but all of these were solvent-exposed and had high B factors, explaining the failure of the Patterson search to detect them and their lack of contribution to phasing. The native protein was refined against a 2.4-Å-resolution data set for native crystals soaked into a NaBr-containing solution that had been obtained in the course of an attempt to determine the structure by multiwavelength anomalous dispersion phasing with solvent Br ions, because the 2.4-Å-resolution data set is the best native data set available. Residues 190–302 could be visualized in electron density, with the exception of a break from 238 to 243. The N-terminal 24 residues, together with five residues derived from the TEV cleavage site and the expression vector, appear to be completely disordered.
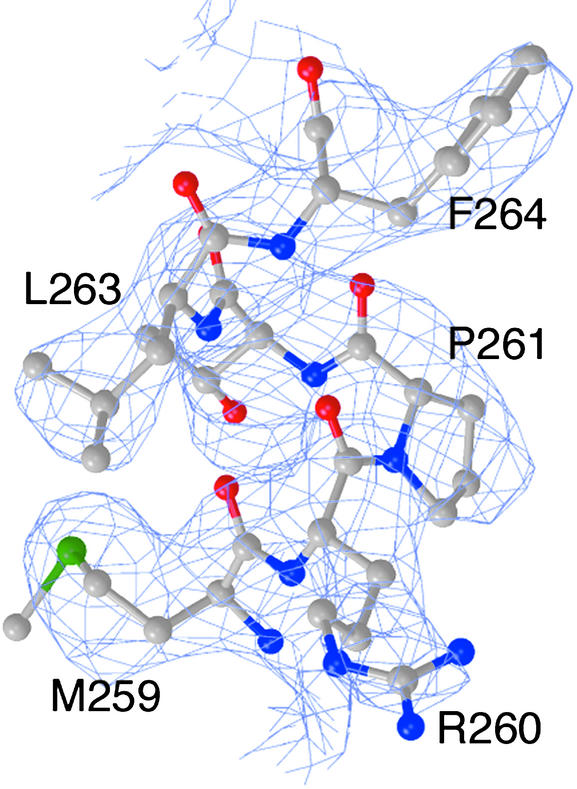
Fig 1.
Experimental electron density. Electron density from the density-modified 3.0-Å multiwavelength anomalous dispersion map contoured at 1.4 σ in the region of the putative Rabaptin-5 binding site near Phe-264.
The GGA1-GAT domain is a three-helical bundle in the shape of a long rod (Fig. 2). The first helix α1 extends 70 Å from the most N-terminal ordered residue, Leu-190, to Ser-236. The second helix α2 runs from Glu-246 to Ser-268. This helix is bent by ≈20° near the middle, at Pro-261. Of the linker between α1 and α2, only residues 244–245 are visible. In contrast, the loop connecting α2 and α3 is well ordered throughout. Helix α3 runs from Asp-274 to the C terminus. The first helix is longer than the other two, so that the first five turns of the helix do not participate in the bundle. As a result, this region is highly mobile and has higher B factors than the rest of the domain. The N terminus of α1 makes crystal lattice contacts, which stabilize its conformation enough to visualize in electron density.
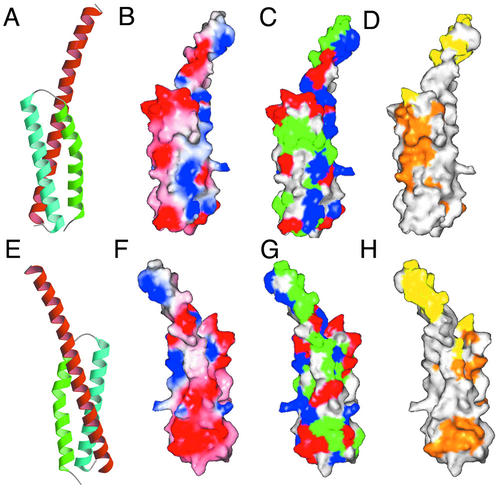
Fig 2.
Overall structure and molecular surface of the GAT domain. (A and E) Overall structure of the GAT domain in two views related by a rotation of 180° about the y axis. Helices α1, α2, and α3 are orange, green, and cyan, respectively. (B and F) Molecular surface colored by electrostatic potential with saturating blue and red corresponding to potentials of +10 kT/e and −10 kT/e, respectively, at T = 297 K. (C and G) Molecular surface colored according to residue property, with hydrophobic residues being green, basic blue, acidic red, and uncharged polar white. (D and H) Molecular surface colored according to sequence conservation. Residues identical or similar in all GAT domains are orange, residues identical or similar in GGAs, but not other GAT domains, are yellow, and nonconserved residues are white.
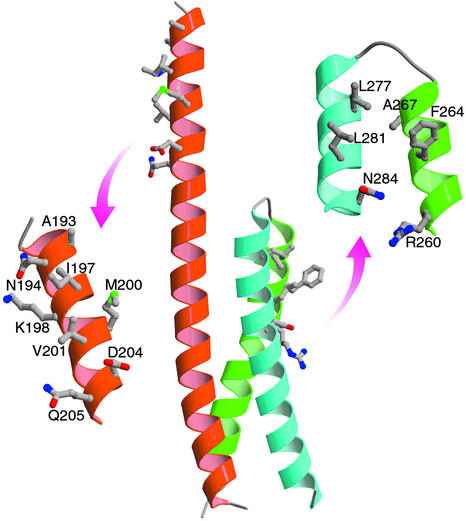
The overall bundle is a right-handed supercoil held together by packing of small and medium hydrophobic residues. The packing core is virtually devoid of aromatic or polar residues, except for two Tyr residues at the end. Helices α1 and α3 are parallel to each other, whereas α2 crosses over the other two and spans the width of both of them. The bend appears to facilitate the crossing of α2 over the other two helices with minimal linkers. The surface contains numerous charged residues, but the overall charge is relatively balanced, and no large electropositive or negative patches exist. The helical core contains exposed hydrophobic residues that suggest potential protein-binding site. The side chains of Phe-264, Leu-277, and Leu-281, in particular, are adjacent to each other and highly exposed to solvent. Several other conserved residues adjoin this hydrophobic patch, including Arg-260, Ala-267, and Asn-284. The conservation and unusual degree of exposed hydrophobic surface suggest that this region is the most likely protein–protein interaction site on the surface of the three-helix bundle (Figs. 2 and 3).
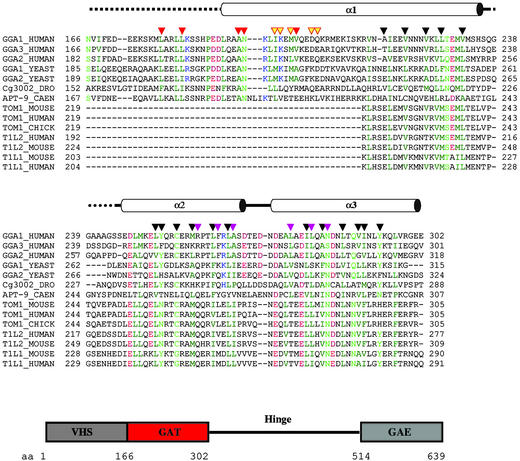
Fig 3.
Structure-based sequence alignment of GAT domains and domain organization of GGA1. (A) GAT domain alignment with helices shown above. Disordered regions in the GAT domain are marked by dashed lines. Conserved hydrophobic residues are dark green, conserved charged residues are red (acidic) and blue (basic), and the remaining conserved residues are light green. Residues known to be involved in interaction with ARF1 are marked by solid orange triangles, and those predicted to interact with ARF1 on the basis of the structure are marked by yellow triangles with orange borders. Residues involved in protein packing are marked by black triangles. Other possible protein–protein interaction sites are marked by magenta triangles. (B) Domain organization of human GGA1.
ARF1-Binding Site.
The primary binding site for ARF small GTPases maps to the first half of α1 (Figs. 3 and 4). The GAT domain was previously shown to contain critical ARF1-binding residues in a sequence near its N terminus that is conserved in the GGAs but absent in TOM1/TOM1-like (11). The sequence contains conserved residues Leu-178, Leu-182, Ala-193, Asn-194, and Val-201 (GGA1 numbering) that are involved in ARF1 binding (11, 33, 34). The importance for Leu-182 was shown in a study of the corresponding residue in yeast Gga2p (33). However, Leu-178 and Leu-182 are outside of the ordered region in this structure. The remaining residues are on the same exposed face of the N-terminal portion of α1 that does not participate in the three-helix bundle. The structure shows that Ile-197, Lys-198, Met-200, Asp-204, and Gln-205 adjoin the established ARF-binding residues and are therefore likely to participate in the ARF interface.
Fig 4.
Location of the ARF-binding site and possible Rabaptin-5-binding sites on the GGA1-GAT domain. (Left) A close-up view of the ARF binding site. (Right) The putative protein–protein interaction site.
It was previously shown that deletion of the C-terminal half of the GAT domain does not impair ARF1 binding, suggesting that the N-terminal region could function independently of the rest of the structure (11). The GAT domain structure suggests that the ARF1-binding functionality of the GGA GAT domains results from the appendage of a N-terminal ARF-binding segment that acts largely independently of the three-helix core.
Structural Similarity to Syntaxin.
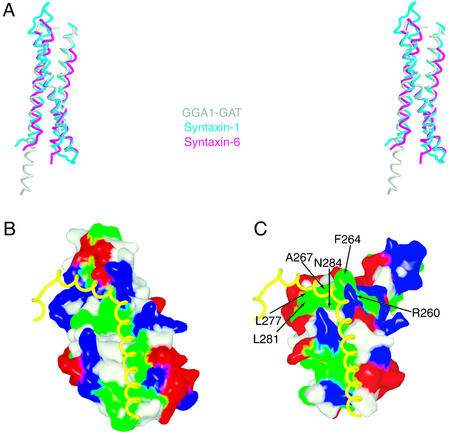
We searched the protein databank by using VAST (35) for structural homologs of the GAT domain. Despite the lack of significant sequence similarity, we found that the structures of the N-terminal domains of syntaxin 1a (36–38) and syntaxin-6 (39) could be overlaid with the majority of the GAT domain. These two domains are three-helix bundles of the same topology as the GAT domain. All three of the helices can be superimposed (Fig. 5A). The syntaxin-6 N-terminal domain superimposes with an rms deviation of 1.7 Å over 68 Cα positions of the GAT domain. For comparison, the crystal structures of the homologous syntaxin 1a and syntaxin 6 N-terminal domains can be superimposed with an rms deviation of 1.8 Å over 92 Cα positions. Thus the region of similarity comprises essentially the entire three-helical core of the GAT domain, and the differences between GAT and syntaxin N-terminal domains are only slightly greater than the differences among different syntaxin N-terminal domain family members. Only the N-terminal extension of α1 and the flexible linkers are not superimposable.
Fig 5.
Structural similarity between the GAT domain and the syntaxin N-terminal domain. (A) Stereoview of the overlaid structures of the GAT domain (gray), the N-terminal domain of syntaxin 1a (x-ray structure; cyan), and the structure of the N-terminal domain of syntaxin-6 (magenta). (B) The N-terminal domain of syntaxin 1a (residues 27–144) from the crystal structure of full-length syntaxin 1a complexed with nSec1 (38) is shown in a molecular surface representation colored as in Fig. 3 C and G, and the C-terminal helical region (residues 186–232) is shown in a worm representation. (C) The GGA1 GAT domain is shown in the same orientation as syntaxin 1a with the same color scheme. The C-terminal domain of syntaxin 1a is docked based on the superposition of the N-terminal domain of syntaxin 1a with GAT to show how a helical protein region could potentially interact with the GAT molecular surface. The side-chain dihedral angles of three basic side chains of GAT were adjusted to avoid collisions with the syntaxin 1a C-terminal domain.
Discussion
General interest in the GAT domain has centered on its role in the Arf-dependent targeting of GGAs to the trans-Golgi network. The structure of the GGA1 GAT domain yields two surprising observations that require a rethinking of the function of this domain. The first major observation is that the primary ARF-binding site is located on an N-terminal extension that is removed from the three-helical core of the GAT domain. This does not rule out an auxiliary role for the GAT domain core in ARF binding. Indeed, the VHS domain of the GGAs also plays a role in modulating ARF affinity, because the ARF affinity is ≈10-fold higher for the VHS-GAT combination than for GAT alone. The overall picture is that of a structurally isolated and independent ARF-binding site located in between the VHS domain and the core of the GAT domain, whose function may be modulated and enhanced by the surrounding domains, which also suggests that ARF binding is not the primary function of the core GAT domain fold.
The second major observation in this study, which is the unexpected structural homology between the GAT domain and the N-terminal domains of syntaxin-1a and its relatives, suggests a hypothesis for the primary function of the GAT domain that is distinct from its function in ARF binding. Syntaxin 1a is the founding member of one of four major classes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). SNAREs are found on vesicles and on the membranes with which they fuse. SNAREs direct the association of the surfaces of two membranes as they prepare to fuse, and they participate in many membrane-trafficking pathways (40–43). Four major SNARE families exist, those defined by VAMP2, the N- and C- terminal domains of SNAP-25 (S25N and S25C), and syntaxin 1a. SNAREs of different families associate with one another to form the complexes that mediate membrane fusion, as illustrated by the structure of the neuronal core SNARE complex (44).
Syntaxin 1a is a key mediator of exocytosis in neurons (45). Its N-terminal domain is the structural archetype for the N-terminal domains of a larger family of SNAREs with diverse biological roles. Syntaxin 6, for example, is found in endosomal transport vesicles. Four structures of syntaxin 1a N-terminal domain family members have been solved: syntaxin 1a (36–38), Sso1p (46), Vam3p (47), and syntaxin 6 (39). All of them contain the same type of three-helix bundle seen in the GAT structure. These domains bind C-terminal SNARE motifs at a site located in a groove in between the second and third helices. This site coincides with the exposed hydrophobic and conserved site that we have noted on the surface of the GAT core. The finding that a putative functional site on the GAT domain overlays the known C-terminal SNARE motif-binding site of the syntaxin 1a family strongly supports a common ancestry and related function for these domains.
The structural similarity between the GAT domain core and a family of SNARE suggests a role for the GAT domain in membrane fusion and protein–protein interactions with an elongated helical protein partner. Rabaptin-5 was recently identified as a new ligand for the GGA GAT domain (16). Rabaptin-5 is involved in endosome fusion (48). Rabaptin-5 interacts with both the Rab4 and Rab5 small GTPases and forms a complex with the Rab5 guanine nucleotide exchange factor Rabex-5 (48–50). Rabaptin-5 interacts with the GGA GAT domain through its C-terminal coiled-coil domain. By the same token, the GAT domain of Lsb5p interacts with a 244-aa central region of Sla1p (25) that includes a strongly predicted coiled-coil region from residues 584 to 618 (51, 52). We speculate that these coiled coils could form complexes with the associated GAT domains analogous to the intramolecular complex between the syntaxin 1a N-terminal domain and its C-terminal SNARE motif (Fig. 5 B and C). The SNARE-motif interaction site within The N-terminal domain of syntaxin 1a contains a binding site between helices 2 and 3 for the C-terminal SNARE motif of the same protein (38); this binding site is structurally equivalent to the putative helical protein–protein binding site of the GAT domain.
The structure of the GAT domain shows how it could mediate bivalent interactions between the GGAs and both ARF and other proteins, such as Rapabtin-5. The interaction sites for these two proteins are 35 Å apart and there is no overlap between them. The structure suggests tantalizing hints about the formation of a complex molecular assembly as trans-Golgi network-derived and endosomal vesicles dock with each other.
Acknowledgments
We thank J. S. Bonifacino and C. Jackson for DNA constructs and discussions; P. Randazzo for discussions; B. Canagarajah for assistance with computing; and T. Earnest and the staffs of beamline 8.2.1, Advanced Light Source, Lawrence Berkeley Laboratory, and the SER-CAT beamline 22-ID, Advanced Photon Source, for assistance with data collection.
Abbreviations
ARF, ADP-ribosylation factor
GGA, Golgi-associated, γ-adaptin homologous, ARF-interacting protein
GAT domain, GGA and TOM1 proteins
VHS domain, Vps27, HRS, and STAM proteins, SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor
GTPγS, guanosine 5′-[γ-thio]triphosphate
SeMet, selenomethionyl
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Boman A. L., Zhang, C., Zhu, X. & Kahn, R. A. (2000) Mol. Biol. Cell 11, 1241-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirst J., Lui, W. W., Bright, N. A., Totty, N., Seaman, M. N. & Robinson, M. S. (2000) J. Cell Biol. 149, 67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dell'Angelica E. C., Puertollano, R., Mullins, C., Aguilar, R. C., Vargas, J. D., Hartnell, L. M. & Bonifacino, J. S. (2000) J. Cell Biol. 149, 81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poussou A., Lohi, O. & Lehto, V. P. (2000) J. Biol. Chem. 275, 7176-7183. [DOI] [PubMed] [Google Scholar]
- 5.Takatsu H., Yoshino, K. & Nakayama, K. (2000) Biochem. Biophys. Res. Commun. 271, 719-725. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S. & Traub, L. M. (2003) Annu. Rev. Biochem. 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed]
- 7.Puertollano R., Aguilar, R. C., Gorshkova, I., Crouch, R. J. & Bonifacino, J. S. (2001) Science 292, 1712-1716. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y., Doray, B., Poussu, A., Lehto, V. P. & Kornfeld, S. (2001) Science 292, 1716-1718. [DOI] [PubMed] [Google Scholar]
- 9.Takatsu H., Katoh, Y., Shiba, Y. & Nakayama, K. (2001) J. Biol. Chem. 276, 28541-28545. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen M. S., Madsen, P., Christensen, E. I., Nykjaer, A., Gliemann, J., Kasper, D., Pohlmann, R. & Petersen, C. M. (2001) EMBO J. 20, 2180-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puertollano R., Randazzo, P. A., Presley, J. F., Hartnell, L. M. & Bonifacino, J. S. (2001) Cell 105, 93-102. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson J. G. & Jackson, C. L. (2000) Curr. Opin. Cell. Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- 13.Costaguta G., Stefan, C. J., Bensen, E. S., Emr, S. D. & Payne, G. S. (2001) Mol. Biol. Cell 12, 1885-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullins C. & Bonifacino, J. S. (2001) Mol. Cell. Biol. 23, 7981-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doray B., Bruns, K., Ghosh, P. & Kornfeld, S. A. (2002) Proc. Natl. Acad. Sci. USA 99, 8072-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattera R., Arighi, C. N., Lodge, R., Zerial, M. & Bonifacino, J. S. (2003) EMBO J. 22, 78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiba T., Takatsu, H., Nogi, T., Matsugaki, N., Kawasaki, M., Igarashi, N., Suzuki, M., Kato, R., Earnest, T., Nakayama, K., et al. (2002) Nature 415, 937-941. [DOI] [PubMed] [Google Scholar]
- 18.Misra S., Puertollano, R., Kato, Y., Bonifacino, J. S. & Hurley, J. H. (2002) Nature 415, 933-937. [DOI] [PubMed] [Google Scholar]
- 19.Evans P. R. & Owen, D. J. (2002) Curr. Opin. Struct. Biol. 12, 814-821. [DOI] [PubMed] [Google Scholar]
- 20.Nogi T., Shiba, Y., Kawasaki, M., Shiba, T., Matsugaki, N., Igarashi, N., Suzuki, M., Kato, R., Takatsu, H., Nakayama, K., et al. (2002) Nat. Struct. Biol. 9, 527-531. [DOI] [PubMed] [Google Scholar]
- 21.Kent H. M., McMahon, H. T., Evans, P. R., Benmerah, A. & Owen, D. J. (2002) Structure (London) 10, 1139-1148. [DOI] [PubMed] [Google Scholar]
- 22.Bateman A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacques K. M., Nie, Z., Stauffer, S., Hirsch, D. S., Chen, L. X., Stanley, K. T. & Randazzo, P. A. (2002) J. Biol. Chem. 277, 47235-47241. [DOI] [PubMed] [Google Scholar]
- 24.Seykora J. T., Mei, L., Dotto, G. P. & Stein, P. L. (2002) J. Biol. Chem. 277, 2812-2822. [DOI] [PubMed] [Google Scholar]
- 25.Dewar H., Warren, D. T., Gardiner, F. C., Gourlay, C. G., Satish, N., Richardson, M. R., Andrews, P. D. & Ayscough, K. R. (2002) Mol. Biol. Cell 13, 3646-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffield P., Garrard, S. & Derewenda, Z. (1999) Protein Expression Purif. 15, 34-39. [DOI] [PubMed] [Google Scholar]
- 27.Randazzo P. A., Terui, T., Sturch, S., Fales, H. M., Ferrige, A. G. & Kahn, R. A. (1995) J. Biol. Chem. 270, 14809-14815. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger T. C. & Berendzen, J. (1996) Acta Crystallogr. D 52, 749-757. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjelgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 32.Bailey S. & the Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 33.Boman A. L., Salo, P. D., Hauglund, M. J., Strand, N. L., Rensink, S. J. & Zhdankina, O. (2002) Mol. Biol. Cell 13, 3078-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takatsu H., Yoshino, K., Toda, K. & Nakayama, K. (2000) Biochem. J. 365, 369-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibrat J. F., Madej, T. & Bryant, S. H. (1996) Curr. Opin. Struct. Biol. 6, 377-385. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez I., Ubach, J., Dulubova, I., Zhang, X., Südhof, T. C. & Rizo, J. (1998) Cell 94, 841-849. [DOI] [PubMed] [Google Scholar]
- 37.Lerman J. C., Robblee, J., Fairman, R. & Hughson, F. M. (2000) Biochemistry 39, 8470-8479. [DOI] [PubMed] [Google Scholar]
- 38.Misura K. M. S., Scheller, R. H. & Weis, W. I. (2000) Nature 404, 355-362. [DOI] [PubMed] [Google Scholar]
- 39.Misura K. M. S., Bock, J. B., Gonzalez, L. C., Jr., Scheller, R. H. & Weis, W. I. (2002) Proc. Natl. Acad. Sci. USA 99, 9184-9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothman J. E. & Warren, G. (1994) Curr. Biol. 4, 220-233. [DOI] [PubMed] [Google Scholar]
- 41.Weber T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T. H. & Rothman, J. E. (1998) Cell 92, 759-772. [DOI] [PubMed] [Google Scholar]
- 42.Jahn R. & Sudhof, T. C. (1999) Annu. Rev. Biochem. 68, 863-911. [DOI] [PubMed] [Google Scholar]
- 43.Bock J. B., Matern, H. T., Peden, A. A. & Scheller, R. H. (2001) Nature 409, 839-841. [DOI] [PubMed] [Google Scholar]
- 44.Sutton R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. (1998) Nature 395, 347-353. [DOI] [PubMed] [Google Scholar]
- 45.Li C., Ullrich, B., Zhang, J. Z., Anderson, R. G., Brose, N. & Sudhof, T. C. (1995) Nature 375, 594-599. [DOI] [PubMed] [Google Scholar]
- 46.Munson M., Chen, X., Cocina, A. E., Schultz, S. M. & Hughson, F. M. (2000) Nat. Struct. Biol. 7, 894-902. [DOI] [PubMed] [Google Scholar]
- 47.Dulubova I., Yamaguchi, T., Wang, Y., Sudhof, T. C. & Rizo, J. (2001) Nat. Struct. Biol. 8, 258-264. [DOI] [PubMed] [Google Scholar]
- 48.Lippe R., Miaczynska, M., Rybin, V., Runge, A. & Zerial, M. (2001) Mol. Biol. Cell 12, 2219-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horiuchi H., Lippe, R., McBride, H. M., Rubino, M., Woodman, P., Stenmark, H., Rybin, V., Wilm, M., Ashman, K., Mann, M., et al. (1997) Cell 90, 1149-1159. [DOI] [PubMed] [Google Scholar]
- 50.de Renzis S., Sonnichsen, B. & Zerial, M. (2002) Nat. Cell Biol. 4, 124-133. [DOI] [PubMed] [Google Scholar]
- 51.Koretke K. K., Russell, R. B., Copley, R. R. & Lupas, A. N. (1999) Proteins 3, 141-148. [DOI] [PubMed] [Google Scholar]
- 52.Rost B. (2001) J. Struct. Biol. 134, 204-218. [DOI] [PubMed] [Google Scholar]