Abstract
Metastasis is the cause of over 90% of all human cancer deaths. Early steps in the metastatic process include: the formation of new blood vessels, the initiation of epithelial-mesenchymal transition (EMT), and the mobilization of tumor cells into the circulation. There are ongoing efforts to replicate the physiological landscape of human tumor tissue using three-dimensional in vitro culture models; however, few systems are able to capture the full range of authentic, complex in vivo events such as neovascularization and intravasation. Here we introduce the Prevascularized Tumor (PVT) model to investigate early events of solid tumor progression. PVT spheroids are composed of endothelial and tumor cells, and are embedded in a fibrin matrix containing fibroblasts. The PVT model facilitates two mechanisms of vessel formation: robust sprouting angiogenesis into the matrix, and contiguous vascularization within the spheroid. Furthermore, the PVT model enables the intravasation of tumor cells that is enhanced under low oxygen conditions and is also dependent on the key EMT transcription factor Slug. The PVT model provides a significant advance in the mimicry of human tumors in vitro and may improve investigation and targeting of events in the metastatic process.
Introduction
Survival rates from cancer drop precipitously once a tumor achieves the ability to metastasize. Thus, it is critical to understand the mechanisms that control the early cellular and molecular events within the complex tumor microenvironment that lead to metastasis. The tumor microenvironment involves the symbiotic integration of mechanical, chemical, and biological cues to direct complex processes such as neovascularization, differentiation, and cell migration that are hallmark features of metastatic human cancers.1 In addition to tumor cells, these processes engage a heterogeneous population of normal host cells, including endothelial cells (EC) and fibroblasts.2
It is well established that tumors require neovascularization for continued tumor growth.3 Increasing metabolic demands initiate a cascade of pro-angiogenic signals to drive the formation of new blood vessels (angiogenesis), or the co-option of existing blood vessels,4 which can subsequently become the conduits of transport for metastatic cancer cells.5 Hypoxia is a primary regulator of carcinoma metastasis through the induction of angiogenesis and epithelial-mesenchymal transition (EMT).6, 7 Stabilization of the hypoxia-inducible factor 1 (HIF-1) transcription factor under hypoxic conditions upregulates tumor and stromal cell secretion of pro-angiogenic growth factors such as vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF).8 HIF-1 has also been demonstrated to activate transcription factors such as Snail, Slug, Twist, and SIP1, which regulate gene expression of proteins central to EMT.9–13
The intersection between tissue engineering and tumor biology, recently coined “tumor engineering,”14 has brought about the creation of advanced 3D cell culture models that perform better than current 2D models at capturing complex aspects of in vivo processes within the tumor microenvironment, thereby providing a more relevant platform for both basic research and anti-cancer drug development. Indeed, it is generally accepted that 3D cell cultures better reflect the physiologic environment than traditional monolayer cultures, or “flat biology,”15 and multicellular tumor spheroids are increasingly recognized as a superior model of the structural, chemical, and functional characteristics within the tumor microenvironment.16–18
Co-culture of tumor spheroids with endothelial cells, either as monolayers19–23 or within 3D matrices,24–26 has provided insight into the mechanisms of tumor angiogenesis by probing tumor-directed EC behaviour. For example, human microvascular EC (HMEC-1) have been shown to upregulate T-cadherin, which promotes invasiveness and network formation, when co-cultured as a monolayer with NA8 melanoma spheroids.27 Here, we introduce a convenient and reproducible multicellular model of solid human tumor and microvessels, referred to as the Prevascularized Tumor (PVT) model, and use this system to investigate neovascularization, intravasation, and EMT in a 3D in vitro environment.
Results
PVT model features robust sprouting
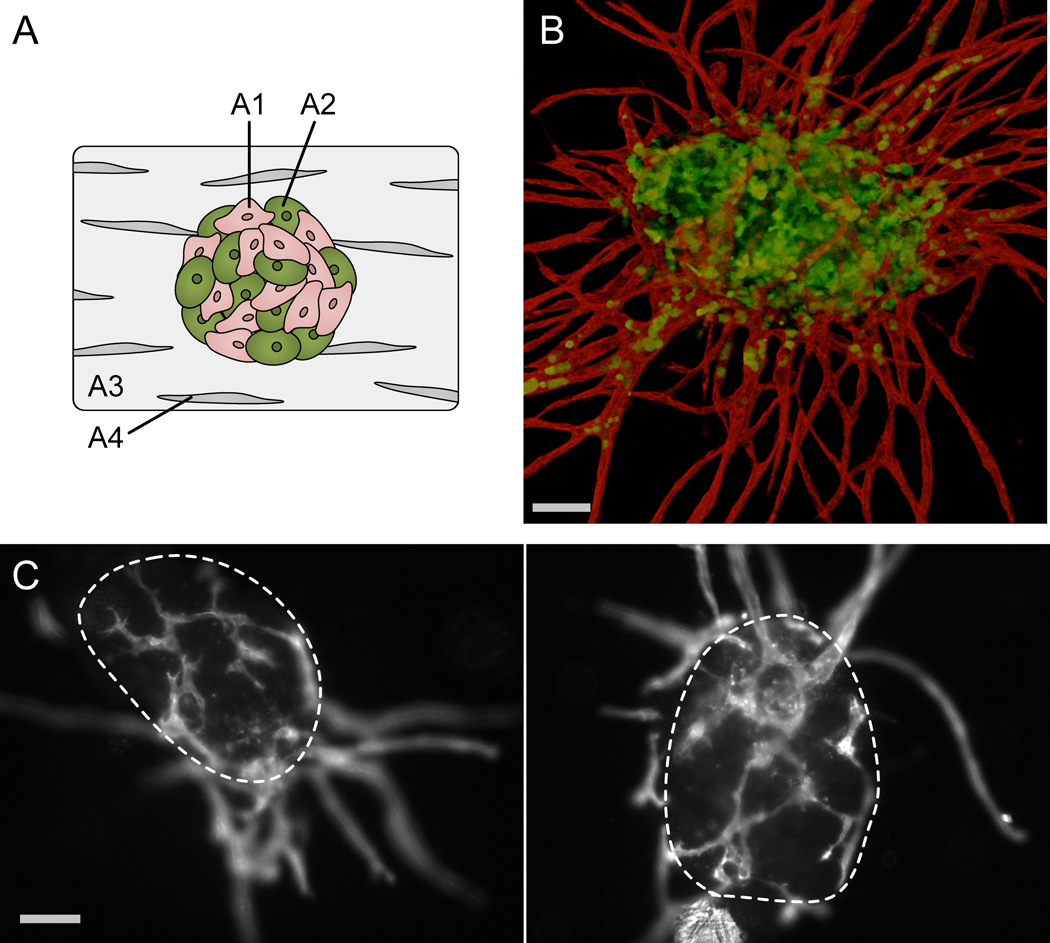
PVT spheroids are formed through the direct co-culture of primary human EC and human tumor cells. These multicellular spheroids are embedded in a fibrin gel distributed with normal human fibroblasts (Fig. 1A). After 7 days in culture, the PVT spheroids exhibit robust sprouting angiogenesis, creating a lumenized vessel network that extends into the surrounding matrix (Fig. 1B). Additionally, the PVT model features a defined and contiguous vessel network that vascularizes the spheroid itself (Fig. 1C). The vessels localized within the spheroid are distinct in morphology, exhibiting a shorter, more branched, and more irregular phenotype compared to the sprouting vessels that extend into the matrix.
Fig. 1.
Prevascularized Tumor (PVT) spheroid model. (A) Schematic of model shows co-culture spheroids composed of endothelial (A1) and tumor cells (A2) embedded in a fibrin gel (A3) distributed with fibroblasts (A4). (B) Representative fluorescent image of PVT spheroid demonstrates robust radial sprouting of lumenized capillaries. EC are labelled with CD31 antibodies (red), and tumor cells (here, SW620) are transduced with EGFP (green). Additionally, the PVT model features a contiguous vessel network that vascularizes the spheroid itself. Scale bar represents 100 µm. (C) Fluorescent images of vascular network reveal inner capillaries are characteristically shorter, jagged, and more branched compared to radial sprouting capillaries. Boundaries of spheroids are outlined with dashed lines. Scale bar represents 100 µm.
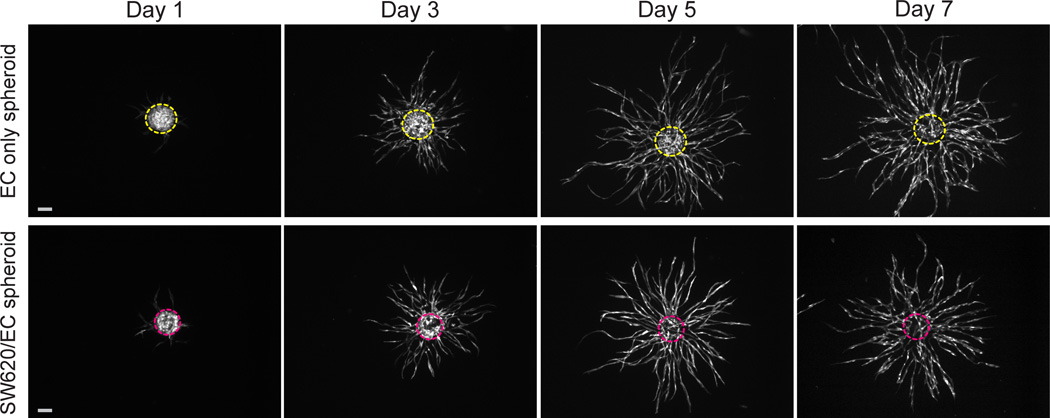
EGFP-transduced EC are used to track vessel development in PVT spheroids composed of SW620 epithelial colon cancer cells and EC (SW620/EC spheroids). EC only spheroids serve as a control. Within 24 hours of tissue construction, EC show signs of early sprout-like structures (Fig. 2), which become robust, highly branched sprouting networks over the course of 7 days. Interestingly, the EC demonstrate significant reorganization to the periphery of the SW620/EC spheroids by Day 3, followed by infiltration of the EC back into the centralized spheroid to form an inner network that is contiguous with the outer sprouting vessels.
Fig. 2.
Development of EC only spheroids and SW620/EC spheroids over 7 days in a fibrin gel. EC only spheroids exhibit robust sprouting angiogenesis (top panel). When combined with SW620, EC initially reorganize to periphery of the spheroids, then infiltrate to form inner network within the boundaries of the spheroid that is contiguous with outer sprouting vessels (bottom panel). EC are transduced with EGFP (white). Dashed lines outline original spheroid boundaries from Day 1. Scale bars represent 500 µm.
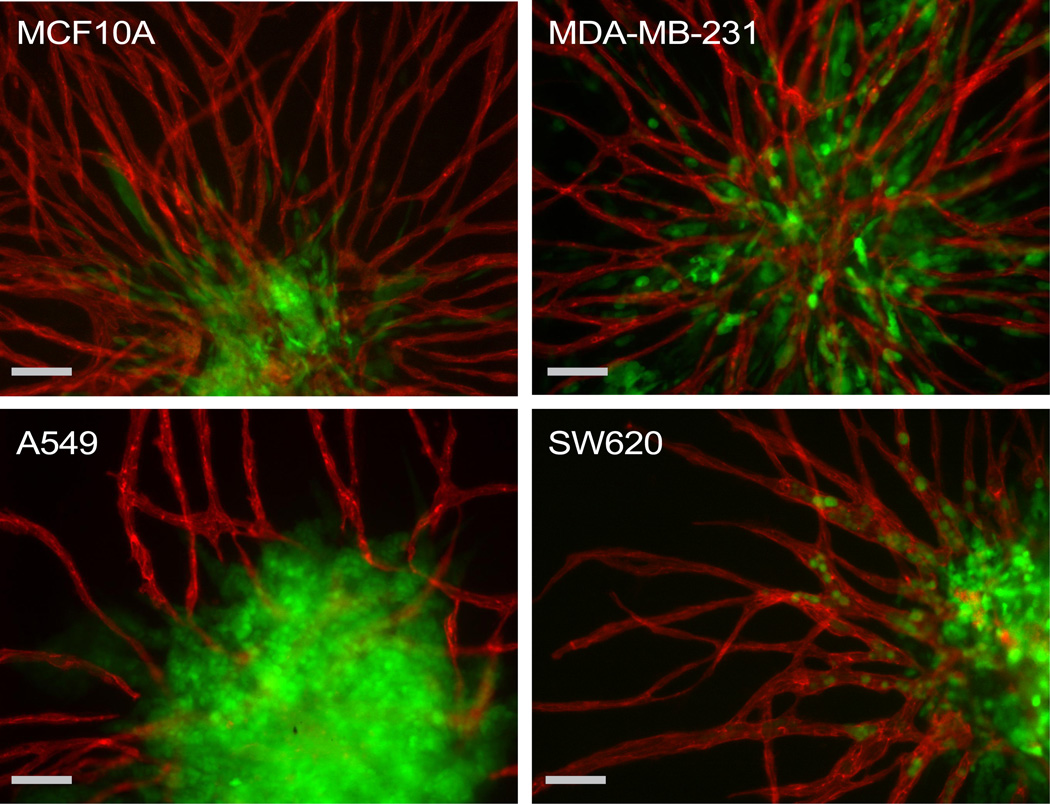
To demonstrate the adaptability of the PVT model to other malignant and non-malignant cell types, spheroids were constructed with EGFP transduced MDA-MB-231, A549, and MCF10A. The model was amenable to all three additional cell lines (Fig. 3).
Fig. 3.
The adaptability of the PVT spheroid model extends to cell lines of breast (MCF10A, MDA-MB-231), lung (A549), and colon (SW620) origin. Intravasation events were only observed for SW620 cells. EC are labelled with CD31 antibodies (red). MCF10A, MDA-MB-231, A549, and SW620 are transduced with EGFP (green). Scale bars represent 100 µm.
Decreased oxygen increases tumor cell intravasation
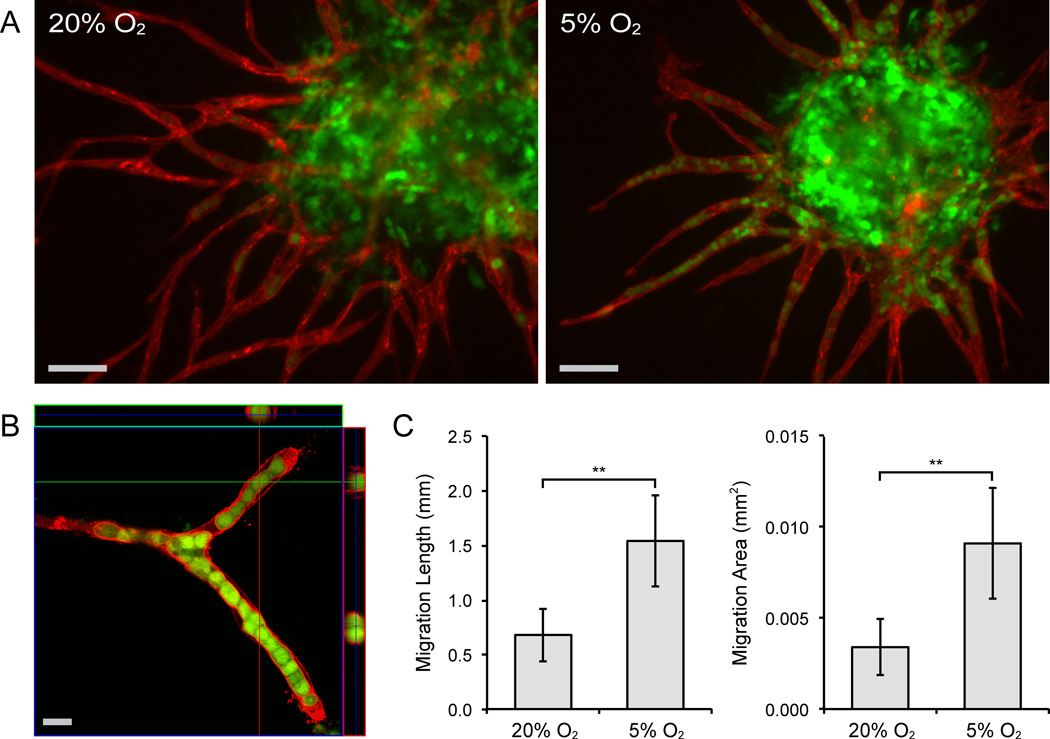
In all PVT spheroids, individual tumor cells broke away from the primary mass and migrated or invaded the surrounding matrix (Fig. 3). In a feature unique to the SW620/EC spheroids, however, the tumor cells exhibited localized migration within the lumens of the sprouting vascular network (Fig. 4A). Confocal imaging of individual capillaries demonstrated SW620 cells within the vessel lumens (Fig. 4B).
Fig. 4.
Decreased oxygen tension increases intravasation of SW620 cells. (A) Fluorescent images of PVT spheroids cultured under 20% O2 and 5% O2 demonstrates that intravasation of SW620 is oxygen dependent. EC are labelled with CD31 antibodies (red). SW620 cells are transduced with EGFP (green). Scale bar represents 100 µm. (B) Confocal immunofluorescence analysis of single vessel branch reveals localization of tumor cells within lumens. Scale bar represents 20 µm. (C) Quantification of Migration Length and Migration Area shows the significantly enhanced intravasation of SW620 under conditions of low oxygen. Error bars represent standard deviation for each condition. **p < 0.001
To examine the effects of oxygen tension on the intravasation of the SW620 cells, spheroids were cultured in either 20% or 5% O2. Interestingly, intravascular tumor cell migration was significantly enhanced under lowered oxygen (5%) conditions. Intravasation was characterized by Migration Length and Migration Area parameters, defined as the total distance and area, respectively, travelled by all tumor cells beyond the surface boundary of the spheroid (Supplementary Fig. 1). Specifically, we saw an approximate doubling in both parameters at 5% O2 compared to 20% O2 conditions (Fig. 4C). This effect was found to be independent of SW620 proliferation, for which there was no significant difference between 20% and 5% conditions (data not shown).
Tumor cell intravasation is Slug-dependent
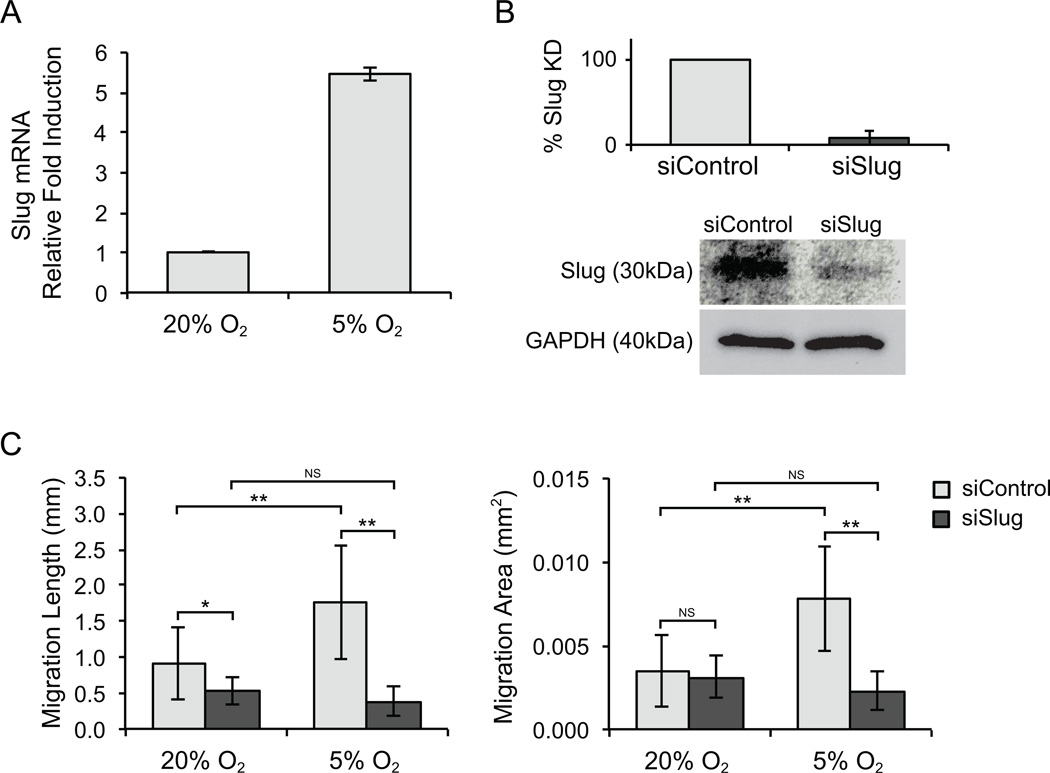
To examine whether the oxygen-dependent phenomenon of intravascular SW620 migration was related to EMT, we measured expression of the transcription factor Slug, a major driver of EMT. SW620 cells incubated at 5% O2 exhibited over a 5-fold increase in Slug expression compared to those incubated at 20% O2 (Fig. 5A). We then used siRNA to reduce Slug expression (confirmed with qPCR and Western Blot, Fig. 5B). SW620 treated with control or Slug siRNA were then used to construct SW620/EC spheroids, and intravasation was assessed at both 20% and 5% O2. Strikingly, intravasation of SW620 cells was strongly reduced in Slug knockdown spheroids under 5% O2 conditions (Fig. 5C), confirming a critical role for Slug in this oxygen-regulated EMT-like response. There was no observable difference in vessel growth or branching between PVT spheroids that contained either siSlug or siControl transfected SW620 at a given oxygen level (Supplementary Fig. 2).
Fig. 5.
SW620 intravasation is Slug-dependent. (A) SW620 cells incubated at 5% O2 showed more than a 5-fold increase in Slug expression compared to cells incubated at 20% O2. Values were normalized to GAPDH expression levels. (B) Slug knockdown efficiency in SW620 cells was validated by qPCR (top) and western blot (bottom) up to 72 hours post-transfection. (C) Quantification of Migration Length and Migration Area at day 7 confirms the attenuation of intravascular cell migration by siSlug compared to siControl. Error bars represent standard deviation for each condition. *p < 0.05 and **p < 0.001. NS: no statistically significant difference between conditions.
Discussion
Cancer biologists and engineers are increasingly collaborating to examine the fundamental physical characteristics of the tumor microenvironment that contribute to disease progression, including mechanical forces, transport of fluid and soluble mediators, growth kinetics, and reaction kinetics.28 This has yielded the development of advanced in vitro technologies to study, quantify, and manipulate these phenomena. While there have been significant advances in “tumor engineering,” few models have been able to recapitulate the complex in vivo processes of tumor angiogenesis and intravasation– critical and early steps in tumor growth and metastasis. We have developed a novel in vitro model of the tumor microenvironment that captures features of both tumor angiogenesis and intravasation simultaneously. The PVT model can be used to create a network of vessels that surround the tumor and that is contiguous with a network of vessels within the tumor. Furthermore, we have used the model to demonstrate that tumor cell intravasation can be easily visualized, depends on the tumor cell line, is enhanced at low oxygen levels, and depends on expression of the transcription factor Slug. Importantly, the PVT model begins to reflect the cellular heterogeneity of native tumors, in which normal cells such as EC and fibroblasts communicate directly with cancer cells to influence the progression of the cancer.
Co-culture of tumor and EC into multicellular spheroids is not a new concept. Such models have been used to investigate the effects of tumor-EC interactions on gene expression, morphogenesis, cell survival, and response to anti-cancer therapies26, 29, 30. Some co-culture models have even been demonstrated to enhance tumor growth and angiogenesis when implanted in vivo.30 Nonetheless, the tumor/EC spheroid has been underutilized as a model to investigate mechanisms of tumor angiogenesis and intravasation, due in part to the necessity of a supporting stromal cell population to promote robust vessel sprouting, and in particular, lumen formation.31 The importance of fibroblasts in capillary network formation has been extensively studied,32, 33 and its inclusion in in vitro assays is important as it encourages the production of capillaries more representative of those found in vivo.31, 34
Additionally, an appreciation for the role of fibroblasts in cancer development is increasing. As the main cellular component of the tumor stroma, fibroblasts play a key role in defining the rate and scale of cancer progression, and are considered to be an intriguing target for anti-cancer therapies.35 Here, we advance the tumor/EC spheroid model significantly by embedding them in a fibrin matrix distributed with fibroblasts. Fibrin was used in these studies because it is present in the tumor microenvironment, and because it represents a physiological ECM that supports both normal and tumor angiogenesis in vivo.36 The combination of EC and NHLF in a fibrin matrix has also been previously shown to support robust capillary formation in vitro.37, 38 With the addition of fibroblasts, the PVT model is able to support radially sprouting, lumenized capillaries.
Oxygen and nutritional limitations restrict the size of avascular tumors. Hence, the establishment of a vascular network within the tumor mass is an important feature to replicate when modelling tumor progression towards a metastatic state. Previous efforts have demonstrated rudimentary vascularization of tumor spheroids either via contact with endothelial cell monolayers21 or co-incubation with differentiated embryoid bodies.39 However, the PVT model advances these efforts significantly by supporting a branched, lumenized, and extensive EC network that not only penetrates the spheroid mass, but also remains contiguous with the radially sprouting vessel network. It is interesting to note the altered morphology of the vessels that develop within the tumor mass. These vessels are of heterogeneous diameter, show multiple filopodia, and are generally reminiscent of tumor vasculature seen in vivo.4
A unique and potentially valuable feature of the PVT model is its ability to support tumor cell intravasation through lumenized vessels. PVT spheroids cultured in lowered oxygen (5% O2) showed significantly enhanced intravascular migration of SW620 compared to those cultured under ambient oxygen (20% O2).The stabilization of HIF-1 has previously been shown to induce migration and invasion of SW620 cells during hypoxia.40 However, to our knowledge, hypoxia-induced intravasation of tumor cells through engineered microvasculature has never been demonstrated. This represents a novel model of in vitro cancer cell migration in 3D. We devised two quantitative parameters of intravasation (Migration Length and Migration Area) to describe our observations, and in both cases, intravasation of SW620 was markedly greater at 5% O2 compared to 20% O2. When factoring in the analysis that the 5% O2 conditioned vessel networks are significantly less developed than the 20% O2 networks (hence having less available space through which tumor cells may intravasate), this differential becomes even more impressive.
The observed phenomenon of intravascular cell migration is consistent with EMT, a key mechanistic feature of metastasis in which primary cancer cells of epithelial origin are transformed into migratory mesenchymal cells capable of invading the extracellular matrix.41 Because cancer cell migration is promoted by EMT, which in turn is highly controlled by the transcription factor Slug in human carcinomas,42 we hypothesized that the observed behaviour was controlled in part by oxygen-regulated Slug expression in SW620. Slug, a member of the Snail family of transcription factors, is known to repress E-cadherin expression and increase the motility of cancer cells.42, 43 Cells from the highly metastatic cell line SW620 repeatedly demonstrate a high level of Slug expression.44, 45 In a study of 8 different colorectal carcinoma cell lines cultured under normoxia (20% O2), SW620 maintained the highest level of Slug expression, the lowest level of E-cadherin expression, and showed the greatest capacity for cell migration and invasion.46
We quantified Slug expression in SW620 using qRT-PCR, and detected an approximate 5-fold increase in cells maintained at 5% O2 compared to those maintained at 20% O2. These results are consistent with studies that showed increased Slug expression of some carcinomas cultured under hypoxia.47 Suppression of Slug by siRNA significantly reduced the intravasation of the SW620, particularly under 5% O2 conditions. We conclude that either supra-induction of Slug is required for intravasation, or that low oxygen conditions induce a cooperating factor(s) for Slug-dependent migration.
We additionally demonstrated the flexibility of the PVT model for assaying multiple cancer cell types of different origin. Thus far, we have incorporated tumor cells of colon, lung, and breast origin into the system. Furthermore, the significant initial remodelling of EC within the PVT spheroids may provide insight into the early stages of solid tumor progression that are cancer-specific.
Our results demonstrate the utility of a new 3D tissue engineered model that may be used to investigate cell-cell communication patterns in the tumor microenvironment. In particular, we have shown the unique ability to investigate mechanisms of tumor cell intravasation, which was enhanced at low oxygen and dependent on the expression of the transcription factor Slug. There is ample room for future investigation in this area, such as dependence on other EMT regulators like Snail or Twist. This flexible, simple, and reproducible experimental platform has the potential to improve the clinical relevance of 3D in vitro culture systems by providing a more accurate simulation of in vivo tumor angiogenesis and metastatic processes.
Materials and methods
Cell culture
Endothelial colony forming cell-derived endothelial cells (EC) were isolated from human umbilical cord blood as previously described.48 Previous work in our lab has demonstrated the ability of these cells to i) form lumenized vessel structures when co-cultured with normal human lung fibroblasts (NHLF) in a fibrin matrix,49, 50 ii) anastomose with a host (e.g. mouse) circulation upon implantation,48, 51 and iii) be perfused with cell culture media using a microfluidic platform.52 These studies have demonstrated in vivo function including pericyte-like recruitment, retention of 70 kDa dextran, and physiological flow and shear. EC were cultured on 1% gelatin coated tissue culture flasks and fed with endothelial growth medium-2 (EGM-2; Lonza, Wakersfield, MD). NHLF (Lonza) were cultured in fibroblast growth medium-2 (FGM-2; Lonza). SW620 colon cancer cells were cultured in DMEM high glucose (Invitrogen, Carlsbad, CA) supplemented with 2mM glutamine, 100U/mL antibiotic-antimycotic (Invitrogen), and 10% fetal bovine serum (FBS; Invitrogen). A549 lung cancer cells were fed with DMEM high glucose containing 10% FBS. MDA-MB-231 and MCF10A breast cancer cells were fed with mammary epithelial growth medium (MEGM; Lonza). All cells were maintained at 37°C in humidified air containing 5% CO2 and the medium was changed every 48 hours. Cells were passaged at 80% confluence with 0.025% trypsin. EC were used at passages 4-5, and NHFL were used at passages 6-7.
EGFP transduction of cells
EC, SW620, MDA-MB-231, A549, and MCF10A cells were transduced using a lentivirus in order to express enhanced green fluorescent protein (EGFP). To prepare the lentiviral vector, HEK293T cells were plated at 5×105 cells per well of a 6-well plate in DMEM without sodium pyruvate and with 10% FBS. Cells were maintained for 24 hours at 37°C in humidified air containing 5% CO2. 3 µg of plasmid DNA that consisted of 1.5 µg pRRLSIN.cPPT.PGK-GFP.WPRE, 0.75 µg pMDLg/pPRE, 0.3 µg pRSV-Rev, and 0.45 µg pMD2.G (Addgene, Cambridge, MA) was diluted with 250µL of Opti-MEM (Invitrogen), and incubated at room temperature for 25 minutes. 7.5µL of Lipofectamine 2000 (Invitrogen) was diluted with an additional 250µL of Opti-MEM, and incubated at room temperature for 5 minutes. The two solutions were combined and added drop wise to each well of HEK293T cells. After 24 hours, each well was replaced with fresh medium. After 48 hours, the viral supernatant was collected, centrifuged to remove debris, and frozen at −80°C for further use.
To transduce cells, 2mL of viral titer and 6µL of 10 mg/mL polybrene (Millipore, Billerica, MA) was added to 8mL culture medium for each of the respective cell lines (as described in Cell culture section), and was used to feed approximately 40% confluent cells in 75 cm2 flasks. The cells were cultured overnight, after which the medium was replaced with fresh culture medium. Transduction efficiency was greater than 90% for all cells. EGFP transduction of EC was performed to visualize EC re-organization over the course of the assay to qualitatively inform the model; however, non-transduced EC were used for all quantified experiments.
PVT spheroid preparation
A stock solution of methocel was prepared by dissolving 6 g of methylcellulose in 500 mL of DMEM. EC and tumor cells were trypsinized, and suspended in EGM-2 containing 15% methocel (v/v). 150µL of the cell suspension was seeded to each well of a non-adherent round-bottom 96-well plate (Greiner, Frickenhausen, Germany). Co-culture (PVT) spheroids were composed of a 3:1 EC to tumor cell ratio. EC only spheroids were also generated to serve as a control. All PVT and control spheroids contained a total of 1000 cells. After incubation overnight in 37°C, the cell suspension solution contributed to the formation of a single spheroid per well. The spheroids were harvested for experiments within 24 hours.
Fibrin tissue assembly and oxygen conditioning
For each tissue construct, 8-10 spheroids were suspended in 50 µL of bovine fibrinogen (Sigma-Aldrich, St. Louis, MO) solution that contained NHLF at a concentration of 1×106 cells/mL. To make the fibrinogen solution, fibrinogen was dissolved in Dulbecco’s phosphate buffered saline (DPBS, Invitrogen) to a final concentration of 2.5 mg/mL. The fibrinogen-NHLF-spheroid solution was mixed with 2.5 µL FBS and 1 µL thrombin (50 units/mL; Sigma-Aldrich), and pipetted onto a 12 mm circular glass cover slip with an affixed polydimethylsiloxane (PDMS) retaining ring. The PDMS rings have a diameter of 8 mm and a height of 0.8 mm, for a final tissue thickness of ~1 mm. The tissues were left undisturbed for 5 minutes at room temperature, and then moved to a 37°C incubator for 30 minutes to completely polymerize. The tissues were then transferred to 24-well plates (one tissue per well), and fed with EGM-2. Assembled fibrin tissues were maintained at 20% O2 for 24 hours following polymerization. Tissues were then assigned to one of two groups (20% or 5% O2) and kept in culture for an additional 6 days. All conditions were maintained with 5% CO2 and balance N2. Medium was changed every two days.
Immunofluorescent staining and visualization of EC
To visualize the vessel networks formed by the EC, the tissues were immunofluorescently stained with mouse anti-human CD31 antibody (Dako, Carpinteria). First, the tissues were fixed in 10% formalin, and then blocked with a 2% bovine serum albumin (BSA, Sigma-Aldrich) and 0.1% Tween 20 (Sigma-Aldrich) solution. Overnight incubation with CD31 antibody diluted at 1:200 was followed by overnight incubation with Alexa Fluor 555-conjugated goat anti-mouse IgG (Invitrogen) diluted at 1:500. Washes with a 0.1% Tween 20 solution followed both incubation periods. Images of the stained tissues used for vessel quantification purposes were acquired using an inverted fluorescence microscope (Nikon Eclipse TE300). Images to confirm lumen formation and intravascular cell migration were acquired using laser scanning confocal microscopy (Zeiss LSM 510 Meta).
Quantification of sprouting angiogenesis and intravasation
Vessel (+CD31) networks that emanate beyond the surface boundary of the spheroid were manually selected using ImageJ (NIH, Bethesda, MD). These images were then analysed using AngioTool,53 software that provides automated measurements of the Total vessel network length and the Total number of junctions per spheroid.
Two parameters were used to quantify tumor cell intravasation, and were calculated manually using ImageJ. The Migration Length was defined as the total distance travelled by all tumor cells beyond the surface of the spheroid (dotted lines in Supplementary Fig. 1A). The Migration Area was defined as the total area of tumor cells past the surface of the spheroid (black area in Supplementary Fig. 1B). Both parameters were calculated for each individual spheroid, and then averaged over the entire experimental group. For statistical analysis, five spheroids from each of 9 fibrin tissues (45 spheroids total over 3 experiments) were analysed for each condition.
Small interfering RNA transfection
SW620 at 80% confluency were transfected with 50 nM (Invitrogen) siRNA using Lipofectamine 2000 in Opti-MEM (Invitrogen), incubated overnight with transfection mixture, and recovered in EGM-2 media the following day. The non-targeting stealth RNAi negative control high GC duplex #2 (Invitrogen) was used as a control for sequence independent effects of siRNA delivery. Transfection efficiencies were determined by qPCR and Western blot analysis. Stealth siRNAs targeting Slug were purchased from Invitrogen and sequences were as follows: 5`-GGCUCAUCUGCAGACCCAUUCUGAU-3` (sense) and 5`-AUCAGAAUGGUCUGCAGAUGAGCC-3` (antisense). Cells transfected with siRNAs targeting Slug or a non-specific sequence serving as a control, were used to make PVT spheroids, which were then divided into two experimental groups: 20% or 5% O2 (for a total of four conditions). For characterization of intravasation, five spheroids from each of 9 fibrin tissues (45 spheroids total over 3 experiments) were analysed for each condition.
Western Blot
Protein was harvested at 72 hours post transfection from SW620 transfected with control or Slug siRNA. SW620 were lysed directly from the culture dish using lysis buffer (20 mM Tris pH 7.9, 137 mM NaCl, 5mM EDTA, and 1% NP-40) supplemented with 1X protease inhibitor cocktail, 1 µM PMSF, and 1x phosphatase inhibitor. SW620 were lysed on ice with supplemented lysis buffer in the culture dishes for 10 minutes. Dishes were scrapped and the cellular contents were added to a microfuge tube and allowed to lyse for an additional 10 minutes on ice. Lysates were then sonicated at 15 W for 15 seconds and then spun at 14,000 rpm for 10 minutes at 4°C. Protein concentrations were determined using bicinchoninic acid assay (Sigma-Aldrich) according to manufacturer’s instructions. Samples were mixed 3:1 with Laemmli 4X sample buffer (BioRad), boiled for 5 minutes at 95°C, and equal amounts of protein (100 µg) were loaded and electrophoresed in 4-20% Mini-PROTEAN TGX polyacrylamide gels (BioRad) under denaturing and reducing conditions. Proteins were transferred to a polyvinylidene fluoride membrane (Millipore). Membranes were blocked for 1 hour in TBS/0.1% Tween 20 (0.1% TBST) containing 5% non-fat dry milk. Membranes were then incubated overnight at 4°C in primary rabbit monoclonal anti-Slug (1:750; Cell Signaling, 9585) diluted in 0.1% TBST containing 5% bovine serum albumin (BSA). The following day, membranes were washed with TBS/0.2% Tween 20 (0.2% TBST) before secondary antibody was added. HRP-conjugated goat anti-rabbit secondary antibody (1:5000; Abcam) was diluted in 0.1% TBST containing 5% BSA and added to the blot for 1 hours at RT. Protein expression was detected using Amersham ECL Western Blotting Detection Reagent (GE Healthcare) and membranes were imaged using a Nikon AF 50 mm f/1.4D camera (Nikon). To ensure equal loading, membranes were stripped using restore stripping buffer (Thermo Scientific), blocked for 1 hour in 0.1% TBST containing 5% BSA and re-probed overnight with HRP-conjugated GAPDH (1:5000; Abcam, ab9482) antibody.
Quantification of Slug induction
Monolayers of SW620 were prepared in 100 mm tissue culture treated petri dishes. Cells were fed with EGM-2, and incubated at 37°C in either 20% or 5% O2 conditions for 24-48 hours. Total RNA was isolated from SW620 using TriZOL reagent (Invitrogen) according to the manufacturer’s protocol. Isolated RNA was then treated with RQ1 DNase (Promega) for 1 hour. 1 µg of total RNA was used for cDNA synthesis using iScript cDNA Synthesis Kit (BioRad). A BioRad iCycler was used to perform qRT-PCR using SYBR Green (Molecular Probes) and HotStarTaq DNA Polymerase (Qiagen). Average CT values were normalized to GAPDH expression levels and all samples were measured in triplicate. Primers were synthesized by Integrated DNA Technologies and sequences are as follows: Slug, 5`-AGATGCATATTCGGACCCAC-3` (forward) and 5`-CCTCATGTTTGTGCAGGAGA-3` (reverse), GAPDH, 5`-TCGACAGTCAGCCGCATCTTC-3` (forward) and 5`GCGCCCAATACGACCAAATCC-3` (reverse). Three replicates were performed for each condition.
Statistical analysis
Statistical analyses were performed using one-way analysis of variance (ANOVA) with SigmaStat (Systat Software, San Jose, CA). Comparisons between groups were made with the Tukey test for multiple comparisons. All data are presented as the mean ± standard deviation. Results are considered statistically significant for p < 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 CA170879 and UH2 TR000481 to SCG, and RO1 HL60067 to CCWH). CCWH receives support from the Chao Family Comprehensive Cancer Center (CFCCC) through an NCI Center Grant award P30A062203.KMW-R is supported by a pre-doctoral award from the American Heart Association. The authors would like to thank Linda McCarthy for her assistance with the EGFP transduction.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/b000000x/
Notes and references
- 1.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Joyce JA, Pollard JW. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkman J, Bach M, Rowe JW, Davidoff F, Lambert P, Hirsch C, Goldberg A, Hiatt HH, Glass J, Henshaw E. N. Engl. J. Med. 1971;285:1182–1186. [Google Scholar]
- 4.Carmeliet P, Jain RK. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 5.Fidler IJ. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 6.Bergers G, Benjamin LE. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 7.Cannito S, Novo E, Compagnone A, Valfre di Bonzo L, Busletta C, Zamara E, Paternostro C, Povero D, Bandino A, Bozzo F, Cravanzola C, Bravoco V, Colombatto S, Parola M. Carcinogenesis. 2008;29:2267–2278. doi: 10.1093/carcin/bgn216. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 9.Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, Konishi I. The American journal of pathology. 2003;163:1437–1447. doi: 10.1016/S0002-9440(10)63501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurrey NK, Amit K, Bapat SA. Gynecol. Oncol. 2005;97:155–165. doi: 10.1016/j.ygyno.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 11.Peinado H, Cano A. Nat. Cell Biol. 2008;10:253–254. doi: 10.1038/ncb0308-253. [DOI] [PubMed] [Google Scholar]
- 12.Evans AJ, Russell RC, Roche O, Burry TN, Fish JE, Chow VWK, Kim WY, Saravanan A, Maynard MA, Gervais ML, Sufan RI, Roberts AM, Wilson LA, Betten M, Vandewalle C, Berx G, Marsden PA, Irwin MS, Teh BT, Jewett MAS, Ohh M. Mol. Cell. Biol. 2007;27:157–169. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Nat. Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 14.Ghajar CM, Bissell MJ. Tissue Eng. Part A. 2010;16:2153–2156. doi: 10.1089/ten.tea.2010.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada KM, Cukierman E. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland RM. Science. 1988;240:177–184. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 17.Kunz-Schughart LA, Kreutz M, Knuechel R. Int. J. Exp. Pathol. 1998;79:1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pampaloni F, Reynaud EG, Stelzer EHK. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 19.Walter-Yohrling J, Pratt BM, Ledbetter S, Teicher BA. Cancer Chemother. Pharmacol. 2003;52:263–269. doi: 10.1007/s00280-003-0664-2. [DOI] [PubMed] [Google Scholar]
- 20.Offner FA, Wirtz HC, Schiefer J, Bigalke I, Klosterhalfen B, Bittinger F, Mittermayer C, Kirkpatrick CJ. Am. J. Pathol. 1992;141:601–610. [PMC free article] [PubMed] [Google Scholar]
- 21.Timmins NE, Dietmair S, Nielsen LK. Angiogenesis. 2004;7:97–103. doi: 10.1007/s10456-004-8911-7. [DOI] [PubMed] [Google Scholar]
- 22.Offner FA, Bigalke I, Schiefer J, Wirtz HC, Klosterhalfen B, Feichtinger H, Kirkpatrick CJ. Int. J. of Cancer. 1993;54:506–512. doi: 10.1002/ijc.2910540325. [DOI] [PubMed] [Google Scholar]
- 23.Knuchel R, Feichtinger J, Recktenwald A, Hollweg HG, Franke P, Jakse G, Rammal E, Hofstadter F. J. Urol. 1988;139:640–645. doi: 10.1016/s0022-5347(17)42550-x. [DOI] [PubMed] [Google Scholar]
- 24.Szot CS, Buchanan CF, Freeman JW, Rylander MN. Tissue Eng. Part C. 2013;19 doi: 10.1089/ten.tec.2012.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ZJ, Htay A, Dos Santos W, Gillies GT, Fillmore HL, Sholley MM, Broaddus WC. J. Neurooncol. 2009;92:121–128. doi: 10.1007/s11060-008-9742-y. [DOI] [PubMed] [Google Scholar]
- 26.de Sampaio PC, Auslaender D, Krubasik D, Failla AV, Skepper JN, Murphy G, English WR. PLoS ONE. 2012;7:e30753. doi: 10.1371/journal.pone.0030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh S, Josh MB, Ivanov D, Feder-Mengus C, Spagnoli GC, Martin I, Erne P, Resink TJ. FEBS Lett. 2007;581:4523–4528. doi: 10.1016/j.febslet.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 28.Delnero P, Song YH, Fischbach C. Biomedical microdevices. 2013 doi: 10.1007/s10544-013-9763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapp V, Parmakhtiar B, Papazian V, Willmott L, Fruehauf JP. Angiogenesis. 2010;13:305–315. doi: 10.1007/s10456-010-9187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upreti M, Jamshidi-Parsian A, Koonce NA, Webber JS, Sharma SK, Asea AAA, Mader MJ, Griffin RJ. Transl. Oncol. 2011;4 doi: 10.1593/tlo.11187. 365-U138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman AC, Chou W, Welch-Reardon KM, Fong AH, Popson SA, Phan DT, Sandoval DR, Nguyen DP, Gershon PD, Hughes CCW. Arterioscler. Thromb. Vasc. Biol. 2013;33:513–522. doi: 10.1161/ATVBAHA.112.300782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman AC, Nakatsu MN, Chou W, Gershon PD, Hughes CC. Mol. Biol. Cell. 2011;22:3791–3800. doi: 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berthod F, Germain L, Tremblay N, Auger FA. J. Cell. Physiol. 2006;207:491–498. doi: 10.1002/jcp.20584. [DOI] [PubMed] [Google Scholar]
- 34.Donovan D, Brown NJ, Bishop ET, Lewis CE. Angiogenesis. 2001;4:113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 35.Kalluri R, Zeisberg M. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 36.Dvorak HF, Flier J, Frank H. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 37.Chen XF, Aledia AS, Ghajar CM, Griffith CK, Putnam AJ, Hughes CCW, George SC. Tissue Eng. Part A. 2009;15:1363–1371. doi: 10.1089/ten.tea.2008.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatsu MN, Sainson RCA, Aoto JN, Taylor KL, Aitkenhead M, Perez-del-Pulgar S, Carpenter PM, Hughes CCW. Microvasc. Res. 2003;66:102–112. doi: 10.1016/s0026-2862(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 39.Wartenberg M, Donmez F, Ling FC, Acker H, Hescheler J, Sauer H. FASEB J. 2001;15:995–1005. doi: 10.1096/fj.00-0350com. [DOI] [PubMed] [Google Scholar]
- 40.Zhao XY, Chen TT, Xia L, Guo M, Xu Y, Yue F, Jiang Y, Chen GQ, Zhao KW. Carcinogenesis. 2010;31:1367–1375. doi: 10.1093/carcin/bgq116. [DOI] [PubMed] [Google Scholar]
- 41.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. J. Cell. Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 43.Hajra KM, Chen DYS, Fearon ER. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 44.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Br. J. Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinacher-Schick A, Baldus SE, Romdhana B, Landsberg S, Zapatka M, Monig SP, Holscher AH, Dienes HP, Schmiegel W, Schwarte-Waldhoff I. J. Pathol. 2004;202:412–420. doi: 10.1002/path.1516. [DOI] [PubMed] [Google Scholar]
- 46.Camp ER, Findlay VJ, Vaena SG, Walsh J, Lewin DN, Turner DP, Watson DK. J. Surg. Res. 2011;170:56–63. doi: 10.1016/j.jss.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, Wu KJ, Yang MH. Neoplasia. 2009;11 doi: 10.1593/neo.91326. 1371-U1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen XF, Aledia AS, Popson SA, Him L, Hughes CCW, George SC. Tissue Eng. Part A. 2010;16:585–594. doi: 10.1089/ten.tea.2009.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghajar CM, Kachgal S, Kniazeva E, Mori H, Costes SV, George SC, Putnam AJ. Exp. Cell Res. 2010;316:813–825. doi: 10.1016/j.yexcr.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghajar CM, Chen X, Harris JW, Suresh V, Hughes CCW, Jeon NL, Putnam AJ, George SC. Biophys. J. 2008;94:1930–1941. doi: 10.1529/biophysj.107.120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White SM, Hingorani R, Arora RP, Hughes CCW, George SC, Choi B. Tissue Eng. Part C. 2012;18:697–709. doi: 10.1089/ten.tec.2011.0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moya ML, Hsu YH, Lee AP, Hughes CCW, George SC. Tissue Eng. Part C. 2013;19:730–737. doi: 10.1089/ten.tec.2012.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zudaire E, Gambardella L, Kurcz C, Vermeren S. PLoS ONE. 2011;6:e27385. doi: 10.1371/journal.pone.0027385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.