Abstract
Background
Strategies employed by parasites to establish infections are poorly understood. The host-parasite interface is maintained through a molecular dialog that, among other roles, protects parasites from host immune responses. Parasite excretory/secretory products (ESP) play major roles in this process. Understanding the biology of protein secretion by parasites and their associated functional processes will enhance our understanding of the roles of ESP in host-parasite interactions.
Methodology/Principal Findings
ESP was collected after culturing 10 adult female Ascaris suum. Perienteric fluid (PE) and uterine fluid (UF) were collected directly from adult females by dissection. Using SDS-PAGE coupled with LC-MS/MS, we identified 175, 308 and 274 proteins in ESP, PE and UF, respectively. Although many proteins were shared among the samples, the protein composition of ESP was distinct from PE and UF, whereas PE and UF were highly similar. The distribution of gene ontology (GO) terms for proteins in ESP, PE and UF supports this claim. Comparison of ESP composition in A. suum, Brugia malayi and Heligmosoides polygyrus showed that proteins found in UF were also secreted by males and by larval stages of other species, suggesting that multiple routes of secretion may be used for homologous proteins. ESP composition of nematodes is both phylogeny- and niche-dependent.
Conclusions/Significance
Analysis of the protein composition of A. suum ESP and UF leads to the conclusion that the excretory-secretory apparatus and uterus are separate routes for protein release. Proteins detected in ESP have distinct patterns of biological functions compared to those in UF. PE is likely to serve as the source of the majority of proteins in UF. This analysis expands our knowledge of the biology of protein secretion from nematodes and will inform new studies on the function of secreted proteins in the orchestration of host-parasite interactions.
Author Summary
Ascaris lumbricoides, the most prevalent metazoan parasite of humans, is a public health concern in resource-limited countries. Survival of this parasite in its host is mediated at least in part by parasite materials secreted into the host. Little is known about the composition of these secretions; defining their contents and functions will illuminate host-parasite interactions that lead to parasite establishment. Ascaris suum, a parasite of pigs, was used as a model organism because its genome has been sequenced and it is very closely related to A. lumbricoides. Excretory/secretory products (ESP), uterine fluid (UF) and perienteric fluid (PE) were collected from adult A. suum. Proteins were subjected to LC-MS/MS. ESP proteins (the ‘secretome’) included many also present in UF. Proteins in ESP but not in UF had considerably different characteristics than those in PE or UF, which were similar to each other. We conclude that proteins released from the secretory apparatus have distinct patterns of biological function and that UF proteins are likely derived from PE. Comparing the protein composition of A. suum ESP to ESP from B. malayi and H. polygyrus suggests that the secretome is conserved at the level of both phylogeny and host predilection site.
Introduction
Ascaris lumbricoides is an extremely prevalent gastrointestinal nematode parasite of humans. Considered a neglected tropical disease (NTD) and widespread in populations of low-to middle-income countries, especially in tropical and subtropical regions, this parasite is estimated to infect as many as 1.2 billion people (many of whom harbor multiple species) in sub-Saharan Africa, China, South and Central America and East Asia [1]–[5]. Adult parasites reside primarily in the duodenum. Although infections are typically asymptomatic, detectable morbidity occurs in up to 200 million infections, most frequently in chronically infected individuals. Pathology may result when parasites migrate to the bile ducts, causing cholangitis. A high worm burden can obstruct the bowel and lead to volvulus and may cause pain, discomfort and megacolon. Ascaris lumbricoides has also been reported to cause lactose intolerance and to reduce absorption of vitamin A. Severe pathology can occur when larvae migrate through the lungs, causing inflammatory reactions. This may lead to pneumonitis, depending on the number of larvae penetrating the alveolar walls, and to pulmonary eosinophilia, with symptoms of fever and difficulty in breathing.
Chemotherapy remains the primary method of control for ascariasis, most commonly relying on albendazole or mebendazole [6]–[9]. Mass drug administration programs employing these drugs typically target school-age children once or twice a year. A single oral dose of albendazole, mebendazole or pyrantel pamoate is highly efficacious, but the distribution and incidence of this parasite remain very high.
Achieving a better understanding of how nematodes influence host immune responses to establish a chronic infection may lead to the development of novel control methods. The success of a parasite in establishing in a host depends on evading or modulating host immune responses, and understanding the molecular basis underlying this strategy is a compelling area of research [10]–[16]. These strategies are thought to be orchestrated through molecules released by parasites that shape the host-parasite interaction. Recently, the protein composition of nematode excretory/secretory products (ESP) has been characterized in several species using proteomics approaches based on mass spectrometry. Species studied include Brugia malayi [14], [17], [18], Heligmosoides polygyrus [19], [20], Ancylostoma caninum [21], Meloidogyne incognita [22], [23], Strongyloides ratti [24] and Dirofilaria immitis [25].
Despite its importance as a highly prevalent NTD, limited knowledge is available about the biology of ESP in A. lumbricoides. Because A. suum is very closely related to A. lumbricoides and its genome has been published [26], we characterized the protein composition of perienteric fluid (PE), uterine fluid (UF) and total ESP (the secretome) from this parasite. The large size of this species makes it possible to gain insights into the origin of proteins in ESP and the relative contribution of proteins released from the uterus during egg shedding.
Materials and Methods
Parasite culture and fluid collection
Adult A. suum were obtained from JBS Swift and Co. pork processing plant, Marshalltown, Iowa, USA. They were maintained in Locke's solution (NaCl 155 mM, KCl 5 mM, CaCl2 2 mM NaHCO3 1.5 mM, glucose 5 mM) at 32°C. ESP collections were carried out within 24 hr of procurement by maintaining 10 large active females in 500 ml fresh Locke's solution. PE fluid was collected by snipping the head off adult nematodes with a fine scissors so that the turgor pressure discharged the fluid directly into a 1.5 ml screw-capped tube. Approximately, 300 µl of UF was collected by positioning the ovijector over a similar tube and then gently compressing each end of the parasite toward the opening to expel fluid and eggs. The thick nature of UF required 1∶1 dilution with sterile phosphate-buffered saline, pH 7.6, for processing after collection. All solutions were stored in sterile 1.5 ml microtubes and shipped overnight on dry ice to the Institute of Parasitology, McGill University, for further analysis.
Sample preparation
ESP, PE and UF fluids were collected from 10 nematodes and pooled. A mixture of protease inhibitors [bestatin hydrochloride; 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride; N- (trans-epoxysuccinyl)-L- leucine 4-guanidinobutylamide; phosphoramidon disodium salt; pepstatin A; Sigma, St. Louis MO) was added to PE and ESP samples. The ESP sample was sterilized by passage through a 0.22 µm filter and concentrated with an Amicon (MWCO 3000) ultrafiltration as described elsewhere [17]. Only proteins in the ESP sample were precipitated with cold trichloroacetic acid (TCA; final concentration 20%). TCA pellets were washed with cold acetone (−30C) and air-dried. Pellets were suspended in Tris-HCl (20mM, pH 8.0) and concentration measured (Quant-iTTM Protein Assay, Invitrogen). UF and PE were centrifuged at 8, 000×g for 10 min, which pelleted eggs from UF, and the supernatants collected for analysis. All samples were stored at −80C until further analysis.
Gel electrophoresis
Protein pellets were dissolved in Laemmli buffer (50 mM Tris-HCl, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02 % bromophenol blue, pH 6.8). Aliquots of 40 µl were loaded on a precast gradient gel (4–12%, 10×10 cm; Invitrogen). The gel was stained with AgNO3 using standard protocols. ESP, PE and UF lanes were cut into 1 mm×1 cm slices with a razor blade under a laminar flow hood, which were incubated in 30 mM potassium ferricyanide, 100 mM sodium thiosulfate (1∶1) for 5 min. After 3 washes with water, the bands were incubated in ACN for 10 min and prepared for trypsin digestion and mass spectrometry.
Mass spectrometry
The gel slices were destained with 50% methanol, reduced in 10 mM DTT for 1 hr at 56 C and alkylated in 55 mM chloroacetamide for 1 hr at room temperature. After washing in 50 mM ammonium bicarbonate, gel pieces were shrunk in 100% ACN. Trypsin (Promega) digestion (100 ng in 50 mM ammonium bicarbonate) was conducted for 8 hr at 37 C. Peptides were extracted in 90% ACN/0.5 M urea and dried in a speed vac. Samples were resolubilized in 5% ACN/ 0.2% formic acid and separated on a laboratory-made C18 column (150 µm×10 cm) using an Eksigent nanoLC-2D system. A 56-min gradient from 5–60% ACN (0.2% FA) was used to elute peptides from a homemade reversed-phase column (150 µm×100 mm) with flow rate = 600 nl/min. The column was directly connected to a nanoprobe interfaced with an LTQ-Orbitrap Elite mass spectrometer (Thermo-Fisher). Each full MS spectrum was followed by 12 MS/MS spectra (13 scan events) from which the 12 most abundant multiply charged ions were selected for MS/MS sequencing. Tandem MS experiments were performed using collision-induced dissociation in the linear ion trap. The data were processed using the 2.3 Mascot (Matrix Science) search engine with tolerance parameters set to 15 ppm and 0.5 Da for the precursor and the fragment ions respectively. The selected variable modifications were carbamidomethyl (C), deamidation (NQ), oxidation (M) and phosphorylation (STY). The database was the Ascaris suum genome draft located at [ftp://ftp.wormbase.org/pub/wormbase/species/a_suum/assemblies/v1/Ascaris_suum_geneset_annotated.pep].
Data analysis
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.3.01). Mascot was set up to search the gs_asc201204 database (selected for All Entries, 201204, 18542 entries) assuming the digestion enzyme, trypsin. Mascot was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 7.0 ppm. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Oxidation of methionine was specified in Mascot as a variable modification. A criterion for protein identification - Scaffold (Version Scaffold_4.2.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet and contained at least 2 identified peptides algorithm [27]. Protein probabilities were assigned by the Protein Prophet Algorithm [28], which within the Scaffold proteome platform predicted a false discovery rate (FDR) of 0 %. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Protein characterization: Bioinformatics
Blast2GO software was used to acquire gene annotations [29]. Using default parameters, a BLASTP search was performed versus the entire non-redundant NCBI amino acid database using 1×10−3 as the minimum expectation value and a 33 cut-off for high-scoring segment pairs. Default parameters were used for annotations while setting the value of pre-eValue-Hit-Filter to 1×10−6, a cut-off value of 55 for annotation and 5 for gene ontology (GO) weight. Annotation Expander (ANNEX), integrated within Blast2GO, was essential to augment annotations [30]. InterPro database scans were performed within Blast2GO to retrieve functional domains and GO terms, both essential to complement annotation. Retrieved GO terms and InterPro results were merged to add domains and motifs to annotations [31]. GO terms were filtered (in percentage) in Blast2GO by the number of sequences collected for each. ESP and UF biological processes (level 2) GO terms were each filtered at 2%. ESP, ESP-UF, PE and UF biological processes (level 4) were filtered at 8%, 5%, 35% and 40%, respectively. UF molecular functions (level 2) were filtered at 2%. ESP, ESP-UF, PE and UF molecular functions (level 4) were filtered at 5%, 5%, 12% and 8%, respectively. Proteins in ESP, PE and UF were scanned for N-terminal signal peptides using SignalP 4.1 [32].
Results
Protein identification
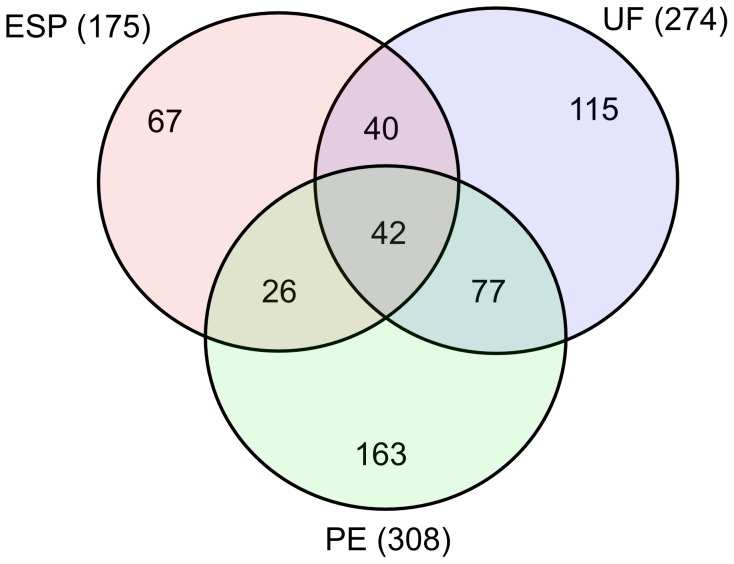
We obtained ∼45 µg protein in the ESP sample, ∼60 µg in UF and ∼55 µg in PE, all of which was used for protein content analysis. We identified 530 distinct proteins in these samples, 175 in ESP, 308 in PE and 274 in UF. The 20 most abundant proteins in these samples are shown in Tables 1–4, with the complete lists provided in Tables S1–S3. In the ESP sample, 67 proteins were specific to ESP only, 26 were shared only between ESP and PE, and 40 were common only to ESP and UF (Table S4). Forty-two proteins (8%) were common among the three compartments; 163 (31%) were appointed solely to PE, 115 (22%) were specific to UF and 77 (15%) were shared between PE and UF (Figure 1).
Table 1. The twenty most abundant proteins in ESP from Ascaris suum.
| # | Accession | Protein Name | SP | NCBI Accession | BlastP result | Organism |
| 1 | GS_20234 | Polyprotein ABA-1 | Y | AAD13651.1 | ABA-1 allergen | A. lumbricoides |
| 2 | GS_08371 | Glb-1 | Y | P28316.2 | Extracellular globin | A. suum |
| 3 | GS_08981 | Protein Y75B7AR.1 | Y | ERG84500.1 | Hypothetical protein ASU_05287 | A. suum |
| 4 | GS_11978 | OV17 | Y | ERG85519.1 | Hypothetical protein ASU_03636 | A. suum |
| 5 | GS_19115 | Serpin B | N | ERG78895.1 | Putative serpin-like protein ma 3388 | A. suum |
| 6 | GS_02689 | Serpin B | N | ERG78895.1 | Putative serpin-like protein ma 3388 | A. suum |
| 7 | GS_06453 | Brugia malayi antigen | N | BAC66614.1 | As16 | A. suum |
| 8 | GS_05746 | Peptidase - Family M1 unassigned peptidases | Y | BAC83078.1 | Aminopeptidase N | A. suum |
| 9 | GS_05201 | Major sperm protein 2 | N | P27439.3 | Major sperm protein isoform alpha | A. suum |
| 10 | GS_13011 | Hypothetical protein | N | - | - | - |
| 11 | GS_07489 | BM20 | Y | ERG87764.1 | Fatty-acid and retinol-binding protein 1 | A. suum |
| 12 | GS_19373 | Vitllogenin-4 (Vit-4) | Y | ERG79133.1 | Vitellogenin-6 | A. suum |
| 13 | GS_11183 | MFP2 | N | AAP94886.1 | MFP2 | A. suum |
| 14 | GS_08343 | Collagen alpha-5 | Y | ERG78661.1 | C-type lectin protein | A. suum |
| 15 | GS_12601 | Brugia malayi antigen | Y | ACJ03764.1 | Ag1 | A. lumbricoides |
| 16 | GS_07456 | Transthyretin-8 (Ttr-8) | Y | ERG85900.1 | Transthyretin-like protein 5 | A. suum |
| 17 | GS_19777 | Probable alpha/beta-glucosidase agdC | N | ERG80708.1 | Sucrase-intestinal | A. suum |
| 18 | GS_15314 | IgGFc-binding protein | N | ERG867007.1 | Apolipophorin | A. suum |
| 19 | GS_11674 | Apolipophorins | N | ERG86007.1 | Apolipophorin | A. suum |
| 20 | GS_16477 | Hypothetical protein | N | - | - | - |
“-“ indicates that a BLASTP search returned no significant match. “SP” refers to signal peptide; “Y” and “N” indicate the presence or absence of a signal peptide, respectively.
Table 4. The twenty most abundant proteins in ESP-UF from Ascaris suum.
| # | Accession | Protein Name | SP | NCBI Accession | BlastP result | Organism |
| 1 | GS_06453 | Brugia malayi antigen | N | BAC66614.1 | As16 | A. suum |
| 2 | GS_05746 | Peptidase - Family M1 unassigned peptidases | Y | BAC83078.1 | Aminopeptidase N | A. suum |
| 3 | GS_12601 | Brugia malayi antigen | Y | ACJ03764.1 | Ag1 | A. lumbricoides |
| 4 | GS_07456 | Transthyretin-8 (Ttr-8) | Y | ERG85900.1 | Transthyretin-like protein 5 | A. suum |
| 5 | GS_17977 | Probable alpha/beta-glucosidase agdC | N | ERG85169.1 | T family of potassium channels protein 12 | A. suum |
| 6 | GS_16078 | Protein Y75B7AR.1 | N | - | - | - |
| 7 | GS_07759 | Protein C28C12.4 | Y | ERG83483.1 | Hypothetical protein ASU_06904 | A. suum |
| 8 | GS_23855 | Zinc Finger protein 420 | Y | ERG80621.1 | Hypothetical protein ASU_11569 | A. suum |
| 9 | GS_08951 | Hypothetical protein | N | ERG86760.1 | Hypothetical protein ASU_01744 | A. suum |
| 10 | GS_21887 | Protein F55H12.4 | Y | XP_003112257.1 | Hypothetical protein CRE_29767 | C. remanei |
| 11 | GS_17143 | Protein T18B10.2 | Y | ERG79357.1 | Hypothetical protein ASU_13792 | A. suum |
| 12 | GS_14400 | Hypothetical protein | Y | - | - | - |
| 13 | GS_02066 | Fab1, identical | Y | P55776.1 | Fatty acid-binding protein homolog | A. suum |
| 14 | GS_01881 | Transthyretin-8 (Ttr-8) | Y | ERG85900.1 | Transthyretin-like protein | A. suum |
| 15 | L3E_02632 | Transthyretin-8 (Ttr-8) | Y | ERG85638.1 | Transthyretin-like protein | A. suum |
| 16 | GS_18848 | Glutamate dehydrogenase | N | ERG83952.1 | Glutamate dehydrogenase | A. suum |
| 17 | GS_20676 | Annexin | N | ERG80393.1 | Annexin A6 | A. suum |
| 18 | GS_05584 | Aminopeptidase N | N | ERG83077.1 | Aminopeptidase N | A. suum |
| 19 | GS_02555 | Aminopeptidase N | N | ERG83079.1 | Aminopeptidase N | A. suum |
| 20 | GS_04373 | L-isoaspartate O-methyltransferase | N | XP_001902004.1 | L-isoaspartate O-methyltransferase | B. malayi |
“-“ indicates that a BLASTP search returned no significant match. “SP” refers to signal peptide; “Y” and “N” indicate the presence or absence of a signal peptide, respectively.
Figure 1. Distribution of proteins among ESP, PE and UF.
The Venn diagram shows the numbers of proteins shared between and among the three compartments based on common Accession Number.
Table 2. The twenty most abundant proteins in UF from Ascaris suum.
| # | Accession | Protein Name | SP | NCBI Accession | BlastP result | Organism |
| 1 | GS_20234 | Polyprotein ABA-1 | Y | AAD13651.1 | ABA-1 allergen | A. lumbricoides |
| 2 | GS_15314 | IgGFc-binding protein | N | ERG867007.1 | Apolipophorin | A. suum |
| 3 | GS_19373 | Vitllogenin-4 (Vit-4) | Y | ERG79133.1 | Vitellogenin-6 | A. suum |
| 4 | GS_01900 | Methylmalonyl-CoA decarboxylase | N | ERG82671.1 | Propionyl-carboxylase alpha | A. suum |
| 5 | GS_15475 | Phosphoenolpyruvate carboxykinase | N | ERG79462.1 | Phosphoenolpyruvate carboxykinase | A. suum |
| 6 | GS_14231 | Glutamate dehydrogenase | N | ERG83952.1 | Glutamate dehydrogenase | A. suum |
| 7 | GS_06246 | Phosphoglycerate kinase | N | ERG83027.1 | Putative phosphoglycerate kinase | A. suum |
| 8 | GS_21097 | Fumarate hydratase | N | ERG84110.1 | Putative fumarate hydratase | A. suum |
| 9 | GS_21295 | Enolase | N | ERG79934.1 | Enolase | A. suum |
| 10 | GS_11183 | MFP2 | N | AAP94886.1 | MFP2 | A. suum |
| 11 | GS_08789 | Probable citrate synthase | N | ERG83688.1 | Putative citrate synthase | A. suum |
| 12 | GS_14262 | P40 | N | AAP94889.1 | MFP2b | A. suum |
| 13 | GS_10682 | P40 | N | AWW29197.1 | MFP2c | A. suum |
| 14 | GS_11674 | Apolipophorin | N | ERG86007.1 | Apolipophorin | A. suum |
| 15 | GS_09975 | Fructose-bisphosphate aldolase 1 | N | ERG82018.1 | Fructose-bisphosphate aldolase 1 | A. suum |
| 16 | GS_14525 | Methylmalonyl-CoA mutase | N | ERG80125.1 | Putative Methylmalonyl-CoA mutase | A. suum |
| 17 | GS_03297 | Protein disulfide-isomerase a3 | Y | ERG84937.1 | Protein disulfide-isomerase a3 | A. suum |
| 18 | GS_09492 | L-threonine 3-dehydrogenase | N | ERG80509.1 | Sorbitol dehydrogenase | A. suum |
| 19 | GS_24173 | Peptidase - subfamily S1A unassigned peptidases | N | ERG83341.1 | Trypsin family protein | A. suum |
| 20 | GS_23848 | Glutamate dehydrogenase | N | ERG82853.1 | Glutamate dehydrogenase | A. suum |
“SP” refers to signal peptide; “Y” and “N” indicate the presence or absence of a signal peptide, respectively.
Protein abundance
Estimates of protein abundance are based on the number of peptides and assigned spectra and peptides from Scaffold [33]. For comparison, we subtracted proteins found in UF from those detected in ESP (ESP-UF) to segregate proteins presumably released through the ES system from those that originate from egg shedding. In support of this assumption, UF proteins detected in ESP were generally among the most abundant in UF (Table S3). Screening the ESP tryptic peptide data against all GenBank proteins identified hits against mammalian keratin and a few bacterial proteins in low abundance in ESP, PE and UF (data not shown), which were automatically excluded from the results.
Polyprotein ABA-1 (UniProt ID: F1KUF9) was among the most abundant proteins in all samples (Table 1–3). A protein related to the Onchocerca volvulus OV-17 antigen was abundant in both ESP and UF, suggesting that it may be derived from UF, whereas several homologs of B. malayi antigens were only present in ESP-UF. The high relative abundance of these antigens suggests that proteins in adult female ESP are derived from both the release of UF during egg shedding and the ES apparatus, a finding supported by the appearance of OV-17 and major sperm protein 2 in the ESP fraction. Significant overlap in the list of most abundant proteins between UF and PE suggests that the former is derived from the latter compartment and that both can be distinguished from ESP per se (ESP-UF). The ESP-UF subset was characterized by high abundance of hypothetical proteins compared to PE and UF. In contrast, glycolytic enzymes were much more abundant in PE and UF than in ESP-UF.
Table 3. The twenty most abundant proteins in PE from Ascaris suum.
| # | Accession | Identified Proteins (308) | SP | NCBI Accession | BlastP result | Organism |
| 1 | GS_20234 | Polyprotein ABA-1 | Y | AAD13651.1 | ABA-1 allergen | A. suum |
| 2 | GS_11674 | Apolipophorin | N | ERG86007.1 | Apolipophorin | A. suum |
| 3 | GS_10956 | Vitellogenin-5 | N | ERG83573.1 | Vitellogenin-6 | A. suum |
| 4 | GS_15314 | IgGFc-binding protein | Y | ERG867007.1 | Apolipophorin | A. suum |
| 5 | GS_19373 | Vit-4 protein | N | ERG79133.1 | Vitellogenin-6 | A. suum |
| 6 | GS_04737 | Vitellogenin | N | ERG86007.1 | Apolipophorin | A. suum |
| 7 | GS_03797 | Heat shock 70 kDa protein 1 | Y | ADI54942.1 | Heat shock protein 70 | D. medinensis |
| 8 | GS_17449 | Molecular chaperone HtpG | N | ACO55134.1 | Heat shock protein-90 | A. suum |
| 9 | GS_21295 | Enolase | N | ERG79934.1 | Enolase | A. suum |
| 10 | GS_08371 | GLB-1 protein | N | P28316.2 | Hemoglobin | A. suum |
| 11 | GS_06246 | Phosphoglycerate kinase | Y | ERG83027.1 | Putative phosphoglycerate kinase | A. suum |
| 12 | GS_05301 | Phosphoenolpyruvate carboxykinase [GTP] | N | ERG87334.1 | Phosphoenolpyruvate carboxykinase | A. suum |
| 13 | GS_19115 | Serpin | Y | ERG78895.1 | Putative serpin-like protein ma 3388 | A. suum |
| 14 | GS_19276 | Fructose-bisphosphate aldolase | Y | ERG79663.1 | Fructose-bisphosphate aldolase 1 | A. suum |
| 15 | GS_11347 | Starch phosphorylase | Y | ERG79023.1 | Glycogen muscle form | A. suum |
| 16 | GS_18098 | Aldo/keto reductase family protein | N | XP_003143212.1 | Oxidoreductase | Loa loa |
| 17 | GS_23993 | Tubulin beta | N | AGM37949.1 | Beta-tubulin | P.s equorum |
| 18 | GS_17648 | Phosphomannomutase | N | EJW88303.1 | Phosphoglucomutase/phosphomannomutase domain-containing protein | W. bancrofti |
| 19 | GS_07489 | BM20 | N | ERG87764.1 | Fatty-acid and retinol-binding protein 1 | A. suum |
| 20 | GS_05590 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | N | AHJ11135.1 | 14-3-3 zeta | A. suum |
“SP” refers to signal peptide; “Y” and “N” indicate the presence or absence of a signal peptide, respectively.
Gene ontology
Blast2GO was used to extract GO terms for 134 proteins in ESP, 291 in PE, 264 in UF and 65 in ESP-UF.
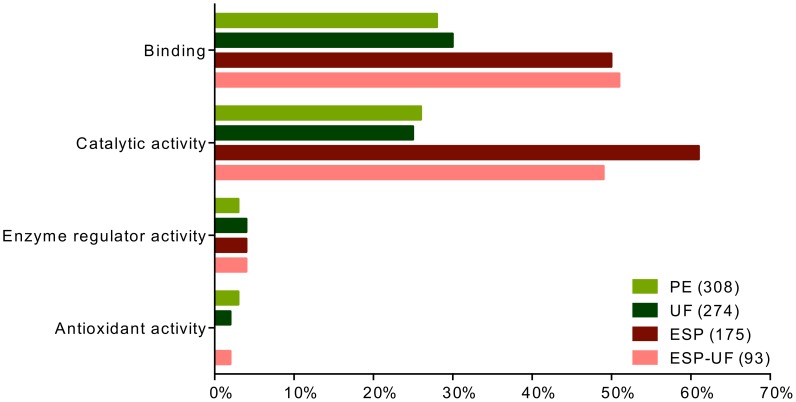
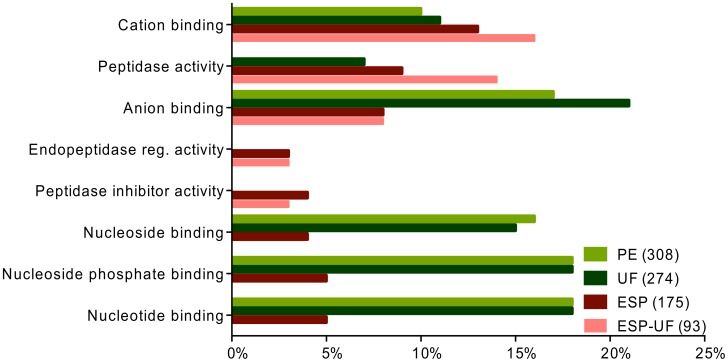
The molecular functions “binding” and “catalytic activity” were commonly predicted for proteins in UF, PE, ESP and ESP-UF (Figure 2). Higher-level molecular functions GO terms revealed subcategories of binding activity, such as “anion binding”, “nucleotide binding”, “nucleoside binding” and “nucleoside phosphate binding”, which dominated in PE and UF compared to ESP and ESP-UF (Figure 3). Although “peptidase activity” and “cation binding” were shared between the three sets of proteins, they were much more common in ESP-UF. Interestingly, “peptidase inhibitor activity” and “endopeptidase regulatory activity” were only found in ESP and ESP-UF.
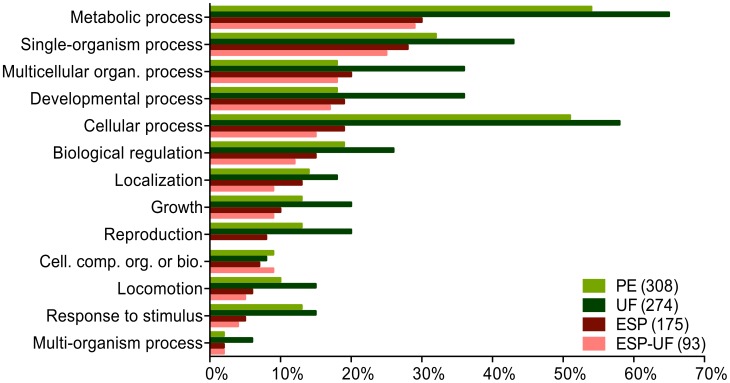
Figure 2. Distribution of level 2 molecular functions GO terms for proteins from UF, PE, ESP and ESP-UF.
The three fractions are represented by different colors. ESP-UF refers to the proteins found in ESP but not in UF.
Figure 3. Distribution of level 4 molecular functions GO terms for proteins from UF, PE, ESP and ESP-UF.
The three fractions are represented by different colors. ESP-UF refers to the proteins found in ESP but not in UF.
Major biological function categories were “metabolic process”, “cellular process” and “single-organism process” in PE and UF (Figure 4). ESP and ESP-UF were similar in almost all GO categories. On a higher level of biological function analysis, most GO terms fell under cellular and metabolic processes (Figure 5), especially those from PE and UF. Cellular processes were allocated to “small molecule metabolic process”, “organic substance biosynthetic process” and similar GO terms with cellular processes activity (Figure 5).
Figure 4. Distribution of level 2 biological processes GO terms for proteins from UF, PE, ESP and ESP-UF.
The three fractions are represented by different colors. ESP-UF refers to the proteins found in ESP but not in UF.
Figure 5. Distribution of level 4 biological processes GO terms for proteins from UF, PE, ESP and ESP-UF.
The three fractions are represented by different colors. ESP-UF refers to the proteins found in ESP but not in UF.
Signal peptide
All proteins in ESP, PE and UF were scanned for N-terminal signal peptide sequence using Signal P 4.1 [32]. Seventy protein sequences (40%), 57 (19%) and 39 (14%) had a signal peptide in ESP, PE and UF, respectively (Tables S1–S3).
Comparative analysis
We previously analyzed ESP obtained in similar protocols from cultures of the Clade III nematode B. malayi [18] and the Clade V species H. polygyrus, which resides in the same general intestinal niche as A. suum [20]. We considered two samples to contain the same protein based on the name assigned to it in the Scaffold annotation, as confirmed by BlastP. The presence of >1 sequence with the same annotation in a sample was not a factor in the analysis. Because A. suum larval ESP was not investigated, we restricted the analysis to ESP from adult B. malayi.
Comparison of A. suum UF and ESP to B. malayi ESP from female and male (separately and together) based on protein composition revealed that female B. malayi ESP was more similar to UF (UF-ESP: 14%) than to ESP (ESP-UF: 7%) from A. suum (Figure 6). Interestingly, protein composition of ESP from male B. malayi equally related to that of UF (UF-ESP: 18%) and ESP (ESP-UF: 18%) from A. suum. Overall, B. malayi adult ESP was more similar to A. suum UF (UF-ESP: 16%) than ESP (ESP-UF: 6%).
Figure 6. Percentage of protein similarity among UF, UF-ESP, ESP and ESP-UF from female A. suum and ESP from adult B. malayi.
Percent similarity among UF, UF-ESP, ESP and ESP-UF from female A. suum and female, male and total adult B. malayi ESP. The comparison is based on annotation names for proteins in the various compartments, confirmed by BlastP analysis.
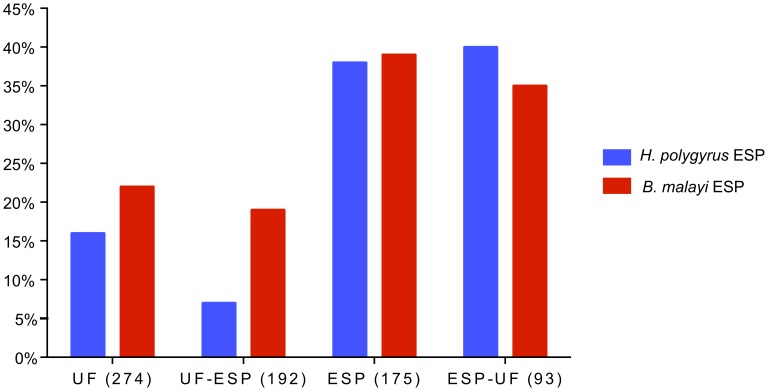
The same approach was used to compare ESP from A. suum, B. malayi and H. polygyrus (Figure 7). UF (UF-ESP) from A. suum was more similar to ESP from B. malayi (19%) than to ESP from H. polygyrus (7%) (Figure 7). In contrast, ESP-UF from A. suum was more similar to ESP from H. polygyrus (25%) than to ESP from B. malayi (13%) (Figure 7).
Figure 7. Cross-species comparisons of secreted proteins.
Depicted is percent similarity of proteins in UF, UF-ESP, ESP and ESP-UF from A. suum with ESP from B. malayi and H. polygyrus. Comparisons are based on annotation names for proteins in the various samples, confirmed by BlastP analysis.
Discussion
Establishment or resolution of a nematode infection is determined by molecular negotiations at the host-parasite interface, and it is a long-standing goal of parasitologists to identify the parasite and host molecules which determine the outcome of infection. It is generally accepted that parasite proteins make up at least part of the tactical strategy used to overcome host immune responses. Parasite secreted proteins also play essential roles in migration through the host, establishment in a preferred location for reproduction, and acquisition of nutrients. Nonetheless, we understand very little about the roles of specific parasite-derived proteins in the infection process. The very large number of proteins detected in nematode secretomes precludes a systematic evaluation of their biological roles, necessitating additional analyses to prioritize them for intensive experimental investigation. In this regard, A. suum is a useful experimental model, as its large size permits the selective determination of proteins in ESP based on their anatomical origin. The very close phylogenetic relationship of this species to the human-selective species A. lumbricoides, the most widespread helminth parasite of humans, further justifies this analysis.
Parasite proteins released into the host, excluding those derived from damaged or dying specimens, may be derived from classical excretory/secretory structures, from cuticle turnover, from the products of digestion in the parasite intestinal tract, and from the release of fluids during egg shedding and insemination. As immunomodulatory effects of helminth infections may be detected at larval stages which are not reproductively competent, identifying the source of proteins in the A. suum secretome could help prioritize proteins for more intensive study: proteins released in uterine fluid, for example, may be hypothesized to be present for maintenance of egg and 1st stage larval viability more than for their effects on the host.
Protein identification in A. suum ESP, PE and UF was made more straightforward by the publication of the A. suum genome [26]. Comparisons among the compartments illuminate some fundamental aspects of the secretome; in addition, comparison of the current results with data obtained from our work on secretomes of other nematode species that parasitize mammals using the same methods [18] permits a deeper understanding of the biology of proteins secreted by parasitic nematodes.
Anatomical sources of secreted proteins
Very few tryptic peptides derived from bacteria or of host origin were identified, suggesting that extensive washing eliminated non-nematode proteins from the sample, and that the culture conditions were effectively sterile. The parasites remained fully motile and appeared healthy during the incubation period, suggesting that proteins released from degenerating nematodes were not major contributors to the dataset. Experience in our laboratories suggests that A. suum can be maintained without decreases in ATP content or physiological decay for several days under these conditions. Although it is possible that protein release changes upon removal from the host, similar periods of incubation have been used in all other reports of secretome composition; whether the secretome detected in culture is a faithful reproduction of the in situ secretome is an area of high priority for research.
Whether any of the nematode proteins detected in ESP were excreted from the intestinal tract cannot be determined based on this analysis. Several types of proteases are abundant in both UF and ESP, but a homolog of a metallo-endoprotease (MEP) protease localized in the intestinal tract of hookworms [34] was found in ESP but not UF, suggesting the possibility that proteins released from the parasite intestine may contribute to the secretome. No cuticle proteins were found in ESP. While release of proteins from the nematode surface has been reported [35], the origin of these proteins is not clear. It seems unlikely that pathways exist for the export of proteins from the hypodermis through the cuticle. Instead, surface-associated proteins may be released from the ES system and adhere to the cuticle after secretion [18]. The absence of classical cuticular proteins in ESP mirrors what has been reported in other nematode secretomes and we conclude that turnover of parasite cuticle is at best a minor source of protein release from healthy nematodes under these conditions.
Inter-compartmental comparison
Since the secretome contains proteins derived from UF and anatomical ES pathways, we compared the protein composition of PE, UF and ESP. The composition of these three samples was highly overlapping (Figure 1). Significantly, the ESP fraction included a high proportion of proteins also detected in UF. As the majority of UF proteins detected in ESP were among the most abundant in the UF proteome, we assume that all UF proteins would appear in the ESP fraction if a large enough sample was analyzed. It is important to note that non-UF proteins were prominent in the ‘most abundant’ subset of ESP (Table 4), confirming the hypothesis that anatomical secretory pathways provide an important contribution to the secretome. Although 28% of proteins in the ESP-UF subset were also identified in the PE fraction, there was no overlap among the 20 most abundant proteins in these two sets (Table 3–4).
Almost 40% of the proteins detected in UF were present in PE. Based on GO term analysis, the protein functional compositions of PE and UF were much more similar to each other than either was to ESP-UF (Figures 2–5). These results suggest that UF is primarily derived from PE, but that the ESP-UF fraction is derived differently. Proteins unique to ESP also have distinct GO term profiles compared to PE and UF. Unique ESP components include homologs of filarial antigens, some proteases, antioxidant enzymes, hypothetical proteins and some proteins involved in glycogenesis. In contrast, PE unique proteins include other enzymes involved in glycogenesis and antioxidant activities, very few proteases, and proteins involved in binding activity (not shown).
Cross-species comparisons
To avoid complications arising from the use of different methods, we only included datasets generated in our laboratory. Comparisons were based on the primary annotation of proteins. Based on conservation of proteins, UF from A. suum was more closely related to ESP from B. malayi than to ESP from H. polygyrus. In contrast, ESP from A. suum was more closely related to ESP from H. polygyrus than to ESP from B. malayi. Although the number of species is obviously limited, we suggest that secretome composition may be determined by both phylogeny and predilection site in the host. The figure for B. malayi includes ESP from adult females (11%) and males (18%). Interestingly, the protein composition of female B. malayi ESP had little similarity to A. suum ESP-UF. Male B. malayi ESP was as closely related to UF as to ESP from A. suum in protein composition. It thus appears that a given protein may be secreted through more than one pathway. The entirety of the secretome, including proteins derived from UF, must be considered when functional roles in the modulation of host responses are investigated.
Although the three species could be differentiated based on protein composition of ESP, they are difficult to distinguish by GO term classification. GO term distribution is dependent on the number of proteins in the secretome, the proportion of hypothetical proteins and the level of annotation. Although the ESP protein composition had somewhat limited similarity among these 3 species based on annotation, they were quite similar in GO terms, suggesting that the general functions associated with proteins in ESP are conserved.
Exosomes
Exosomes, membrane-bound vesicles produced by eukaryotic cells, are commonly found in extracellular compartments. Considerable evidence points to the involvement of exosomes in cell-cell communication, immune system modulation and tumor progression. Exosomes contain mRNA, microRNA and proteins [36]–[38]. Exosomes have been detected in the secretory system of C. elegans [39], but work has not been reported in parasitic species. Exosome-like vesicles produced by the trematodes Echinostoma caproni and Fasciola hepatica are proposed to modulate host-parasite interactions through their uptake by host cells [40]. Interestingly, the protein composition of F. hepatica ESP shares some similarity with A. suum ESP and UF, particularly with regard to proteins typically associated with exosomes [41].
Many exosome-associated proteins are present in high abundance in A. suum ESP and other parasitic nematodes [see [25]], including proteases, cellular communication proteins, structural proteins, glycolytic enzymes and detoxifying enzymes. In this regard, the protein compositions of A. suum PE and UF were very similar, but were distinct from ESP. The number of proteins in PE and UF with signal peptides was similar and low (14% and 19%, respectively), in contrast to those in ESP (40%; Tables S1–S3). Interestingly, PE and UF were relatively enriched in proteins associated with exosomes compared to ESP (Table S5). This could imply that an exosomal secretion is process associated with egg expulsion, or that the protein content of UF is derived, at least in part, by exosome pathways. The lower abundance of exosome-associated proteins in the ESP-UF fraction may indicate that, although exosomes are involved in protein release from anatomical secretory pathways, classical signal peptide-mediate processes are also important. We propose that these data support the previous assertion that protein secretion from parasitic nematodes occurs primarily through exosomal pathways [25]; further work is needed to verify or refute this hypothesis.
Functional considerations
Lipid binding proteins
Parasites must acquire lipids from their host, probably by using lipid-binding proteins [42]–[43]. These proteins are thought to play key roles in small hydrophobic molecule transportation, membrane trafficking, signaling pathways, and regulatory processes [44]. Lipid-binding proteins have been classified as either nematode polyprotein allergens (NPAs) or fatty acid and retinol-binding (FAR) proteins [45]–[48]. The polyprotein ABA-1 allergen, a member of the NPA family [49], was the most abundant protein identified in ESP, PE and UF; its abundance in UF suggests that this may be the primary source of ABA-1 in ESP, although we also detected a close homolog in the secretome of male B. malayi (but not microfilariae) [17]. Close homologues are secreted by many other parasitic nematodes (see [25]). In contrast to NPAs, FAR proteins bind small lipids such as vitamin A (among other ligands) and may sequester them [50]. Locally depressed levels of vitamin A may impede renewal of the intestinal mucosa and interfere with functions of neutrophils, natural killer cells and macrophages. Vitamin A deficiency also affects adaptive immunity by hindering antibody production and differentiation of T helper cells and B cells [50].
Vitellogenins (vtgs) are large lipoproteins that function in nematode oogenesis and larval development [51]. The presence of vtgs in the A. suum secretome accounts in part for the high percentage of the GO terms “embryo development” and “regulation of growth” in the biological processes category. Although vtgs were more abundant in A. suum PE fluid and UF, they were also detected in ESP and in other nematodes, including H. polygyrus [19], [20], M. incognita [23] and D. immitis [25], but perhaps surprisingly not in ESP from female B. malayi [17] or S. ratti [24]. The source of secreted vtgs is presumed to be UF; whether these proteins act in the host-parasite interface is unknown.
Heat shock proteins
Heat shock proteins (HSPs) were among the most abundant proteins in PE and UF and to a lesser extent in ESP and ESP-UF. HSP 70 and other HSPs are commonly found in nematode secretomes [25]. Genes encoding HSPs are highly conserved in parasites, are inducible, and appear to be crucial for parasite survival [52]. Secreted parasite HSPs are immunogenic [53], but the biology in the host of the HSPs detected in the A. suum secretome is unknown. It is possible that keeping parasitic nematodes in culture could cause the release of HSPs (and other proteins) due to stress. Alternatively, release of parasite HSPs into the host may modulate host responses to enable and sustain an infection.
Other abundant proteins
Although the medium used to culture the parasites was changed twice, major sperm proteins (MSPs) [54] were abundant in ESP and UF. This observation suggests that the worms may have been reproductively competent during the 24 hr incubation. MSPs have commonly been reported from secretomes of other parasitic nematodes; whether this reflects unusual stability of these proteins or their release from storage by adult females fertilized prior to being placed in culture is unknown. It is also possible that sperm were present in UF and not removed by centrifugation; however, we did not detect mitochondrial proteins in the UF fraction, suggesting that disrupted sperm were not a common contaminant to this proteome.
Nematode transthyretin-like proteins show modest homology to transthyretin proteins from vertebrates [55], [56]. Although vertebrate transthyretins are involved in transport of thyroid hormones and vitamin A, transthyretin-like proteins of nematodes are likely to have diverse and different functions [55], [56]. Transthyretin-like proteins were abundant in A. suum ESP, PE and UF. Related proteins were reported in ESP of all parasitic nematodes [25]. Multiple genes encoding these proteins are present in C. elegans [55], [56], but phenotypes derived from loss-of-function mutations are uninformative. Their roles in initiating and/or maintaining an infection remain obscure.
Hypothetical proteins were present in each compartment at low frequency. Eight hypothetical proteins were shared between ESP and PE and 8 were found in ESP and UF. Two were shared between ESP, PE and UF. The majority of hypothetical proteins had no homologs in other species. Very few matches were found between hypothetical proteins of A. suum and B. malayi and H. polygyrus through BlastP analyses. Much remains to be learned about the basic biology of parasitic nematodes, but we lack convenient platforms to investigate roles that secreted hypothetical proteins play at the host-parasite interface. It will be interesting to discover if the proportion of proteins with no close homologs decreases as genomes are reported from other species of parasitic nematodes.
Parasite proteases are involved in degradation of host proteins during invasion and migration and also take part in establishment and maintenance of long term infections, at least in part through immunomodulation [57], [58]. Aspartyl-, serine-, cysteine-, and metalloproteinases are found in secretions of GI nematodes. The papain superfamily of cysteine proteinases and zinc metalloproteinases is prevalent in nematode secretions [58]. Cysteine and aspartic proteases were previously identified in A. suum PE fluid in low abundance, whereas an aminopeptidase was highly secreted [59]. We found metallopeptidases and serine proteases to be abundant in ESP-UF. Aminopeptidase N was the most abundant protease in the secretome, although the neprilysin (M13) family of peptidases and serine carboxypeptidase A were also found in ESP-UF extracts. Because A. suum resides primarily in the duodenum, aminopeptidase N might facilitate digestion of proteins which have been partially digested by gastric enzymes [59], enhancing uptake of small peptides by the parasite. A leucine aminopeptidase (LAP; M17 family of metalloendopeptidases; MEP) in ESP was previously identified as secreted by A. suum; it was also reported in PE fluid and extracts of various tissues [59]. A homolog of LAP in filarial nematodes, ES-62, is a secreted phosphorylcholine-containing glycoprotein of 62 kDa [60]. ES-62 alters T and B cell proliferation and suppresses cytokines and interferon involved in TH1 immune responses through the phosphorylcholine moiety.
Protease inhibitors are also commonly found in nematode secretomes (see summary table in [25]) and play important roles in immunomodulation among other functions [61]–[63]. Protease inhibitors are well-known from A. suum [64], including inhibitors of elastase, chymotrypsin, trypsin, carboxypeptidase A and B, pepsin and gastricsin. Many protease inhibitors were detected in the current study; their roles in internal compartments are unknown, but the biology of protease inhibitors in parasitic nematodes appears to extend beyond possible effects in the host.
A. suum uses fermentative metabolism for energy generation. Energy metabolism mainly occurs through pairing phosphorylation to electron transport to drive anaerobic energy generation [65]. This process yields carbohydrate end products [65] and accounts at least in part for the high number related GO terms (Figure 5). Several glycolytic enzymes are associated with exosome-mediated secretion pathways [36], [38] and are typical components of parasitic nematode secretomes [25]. Perhaps surprisingly, glycolytic enzymes were also abundant in UF and to a lesser extent in PE. As noted, the parasites were fully viable during the incubation, so we do not believe that the glycolytic enzymes came from unhealthy worms. Glycolytic enzymes were among the 20 most abundant proteins in UF. The function of secreted parasite glycolytic enzymes demands further experimental investigation, although some are thought to be involved in modulation of host immune responses [66] and many are commonly associated with exosome pathways of protein release.
A recent report describing the protein composition of ESP from larval stages of A. suum [66] shows considerable stage-dependent variation. Interestingly, ESP protein composition from intestinal L4 larvae is much more similar to the composition of adult ESP reported here than is the case for other larval stages, suggesting again that there is a strong niche dependence of ESP content, probably reflecting functional roles of ES proteins in modulating local host responses.
Conclusions
We exploited the unique sensitivity of mass spectrometry combined with the genome of A. suum to identify secreted proteins and proteins in the primary internal fluid compartments of the parasite. Using bioinformatic tools to mine GO terms, molecular and biological functions were retrieved and compared between the sources of these proteins. The protein composition profile of ESP differed from those of PE and UF, which were similar to each other. We suggest that proteins in UF are primarily derived from PE, and that a considerable proportion of secreted proteins originate from sources other than the classical secretory system, at least in female parasites. Some proteins in nematode ESP appear to be secreted through multiple pathways.
Supporting Information
Distribution of level 2 biological processes GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 4 biological processes GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 2 molecular functions GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 4 molecular functions GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Proteins identified in A. suum excretory/secretory products (ESP). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Proteins identified in A. suum perienteric fluid (PE). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Proteins identified in A. suum uterine fluid (UF). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Common and unique proteins in A. suum ESP.
(XLSX)
The presence of common exosome-associated proteins in A. suum ESP. Exosome proteins were obtained from [http://www.exocarta.org/exosome_markers]. Comparisons were made using the protein name. Matches were confirmed by sequence alignment.
(XLSX)
Acknowledgments
The authors thank Dr. Eric Bonneil, Manager de la plateforme de protéomique, Iric-Université de Montréal, QC, Canada, for mass spectrometry analysis, and the Clinical Proteomics Platform, McGill University [ http://www.clinprot.org/ ] for data analysis support.
Funding Statement
This research was supported by grants from the Canada Research Chairs program and the Natural Sciences and Engineering Research Council of Canada to TGG. This work was also supported by the Fonds Québécois de la Recherché sur la Nature et les Technologies (FQRNT) through the Centre for Host-Parasite Interactions. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI047194 to RJM and R21AI092185-01A1 to APR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, et al. (2006) Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367: 1521–1532. [DOI] [PubMed] [Google Scholar]
- 2. Crompton D (2001) Ascaris and ascariasis. Adv Parasit 48: 285–375. [DOI] [PubMed] [Google Scholar]
- 3. Dold C, Holland CV (2011) Ascaris and ascariasis. Microbes Infect 13: 632–637. [DOI] [PubMed] [Google Scholar]
- 4. Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, et al. (2008) Helminth infections: the great neglected tropical diseases. J Clin Invest 118: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott ME (2008) Ascaris lumbricoides: a review of its epidemiology and relationship to other infections. Ann Nestlé 66: 7–22. [Google Scholar]
- 6. Abbas A, Newsholme W (2011) Diagnosis and recommended treatment of helminth infections. Prescriber 22: 56–64. [Google Scholar]
- 7. Albonico M, Crompton D, Savioli L (1999) Control strategies for human intestinal nematode infections. Adv Parasit 42: 277–341. [DOI] [PubMed] [Google Scholar]
- 8. Georgiev VS (2001) Pharmacotherapy of ascariasis. Exp Opin Pharmacol 2: 223–239. [DOI] [PubMed] [Google Scholar]
- 9. Keiser J, Utzinger J (2008) Efficacy of current drugs against soil-transmitted helminth infections. J Amer Med Assoc 299: 1937–1948. [DOI] [PubMed] [Google Scholar]
- 10. Allen JE, Maizels RM (2011) Diversity and dialogue in immunity to helminths. Nat Rev Immunol 11: 375–388. [DOI] [PubMed] [Google Scholar]
- 11. Anthony RM, Rutitzky LI, Urban JF Jr, Stadecker MJ, Gause WC (2007) Protective immune mechanisms in helminth infection. Nat Rev Immunol 7: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Artis D (2006) New weapons in the war on worms: identification of putative mechanisms of immune-mediated expulsion of gastrointestinal nematodes. Int J Parasitol 36: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooke A (2012) Parasitic worms and inflammatory disease. Curr Opin Rheumatol 24: 394–400. [DOI] [PubMed] [Google Scholar]
- 14. Hewitson JP, Grainger JR, Maizels RM (2009) Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol 167: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McSorley HJ, Hewitson JP, Maizels RM (2013) Immunomodulation by helminth parasites: Defining mechanisms and mediators. Int J Parasitol 43: 301–310. [DOI] [PubMed] [Google Scholar]
- 16. Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, et al. (2008) The secretome of the filarial parasite, Brugia malayi: Proteomic profile of adult excretory–secretory products. Mol Biochem Parasitol 160: 8–21. [DOI] [PubMed] [Google Scholar]
- 17. Moreno Y, Geary TG (2008) Stage-and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl Trop Dis 2: e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, et al. (2009) Brugia malayi excreted/secreted proteins at the host/parasite interface: stage-and gender-specific proteomic profiling. Plos Neglect Trop Dis 3: e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hewitson JP, Harcus Y, Murray J, van Agtmaal M, Filbey KJ, et al. (2011) Proteomic analysis of secretory products from the model gastrointestinal nematode Heligmosomoides polygyrus reveals dominance of Venom Allergen-Like (VAL) proteins. J Proteomics 74: 1573–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moreno Y, Gros PP, Tam M, Segura M, Valanparambil R, et al. (2011) Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl Trop Dis 5: e1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, et al. (2009) Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum . Mol Cell Proteomics 8: 109–121. [DOI] [PubMed] [Google Scholar]
- 22. Bellafiore S, Shen Z, Rosso MN, Abad P, Shih P, et al. (2008) Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog 4: e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X-R, Moreno YA, Wu HR, Ma C, Li YF, et al. (2012) Proteomic profiles of soluble proteins from the esophageal gland in female Meloidogyne incognita . Int J Parasitol 42: 1177–1183. [DOI] [PubMed] [Google Scholar]
- 24. Soblik H, Younis AE, Mitreva M, Renard BY, Kirchner M, et al. (2011) Life cycle stage-resolved proteomic analysis of the excretome/secretome from Strongyloides ratti—Identification of stage-specific proteases. Mol Cell Proteomics 10: M111.010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geary J, Satti M, Moreno Y, Madrill N, Whitten D, et al. (2012) First analysis of the secretome of the canine heartworm, Dirofilaria immitis . Parasit Vect 5: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jex AR, Liu S, Li B, Young ND, Hall RS, et al. (2011) Ascaris suum draft genome. Nature 479: 529–533. [DOI] [PubMed] [Google Scholar]
- 27. Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii, A.I., et al., A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem, 2003. )75(: 17p. 4646–4658. [DOI] [PubMed]
- 29. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 30. Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petersen TN, Brunak S, Heijne Gv, Nielson H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 33. Searle BC (2010) Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10: 1265–1269. [DOI] [PubMed] [Google Scholar]
- 34. Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A (2003) Digestive proteases of blood-feeding nematodes. Trends Parasitol 19: 417–423. [DOI] [PubMed] [Google Scholar]
- 35. Zhan B, Liu Y, Badamchian M, Williamson A, Feng J, et al. (2003) Molecular characterisation of the Ancylostoma secreted protein family from the adult stage of Ancylostoma caninum . Int J Parasitol 33: 897–907. [DOI] [PubMed] [Google Scholar]
- 36. Johnstone RM (2006) Exosomes biological significance: a concise review. Blood Cell Mol Dis 36: 315–321. [DOI] [PubMed] [Google Scholar]
- 37. Keller S, Sanderson MP, Stoeck A, Altevogt P (2006) Exosomes: from biogenesis and secretion to biological function. Immunol Lett 107: 102–108. [DOI] [PubMed] [Google Scholar]
- 38. Raimondo F, Morosi L, Chinello C, Magni F, Pitto M (2011) Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics 11: 709–720. [DOI] [PubMed] [Google Scholar]
- 39. Liégeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M (2006) The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans . J Cell Biol 173: 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marcilla A, Trelis M, Cortés A, Sotillo J, Cantalapiedra F, et al. (2012) Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One 7: e45974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mathivanan S, Simpson RJ (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9: 4997–5000. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy MW (2001) Structurally novel lipid-binding proteins. In Kennedy MW, Harnett, editors. Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology. Wallingford, CABI Publishing. p. 309–330.
- 43.Behm C (2002) Metabolism. In Lee DL, editor. The Biology of Nematodes. London, Taylor and Francis. p. 261–290.
- 44. Lin HH, Han LY, Zhang HL, Zheng CJ, Xie B, et al. (2006) Prediction of the functional class of lipid binding proteins from sequence-derived properties irrespective of sequence similarity. J Lipid Res 47: 824–831. [DOI] [PubMed] [Google Scholar]
- 45. Garofalo A, Kennedy MW, Bradley JE (2003) The FAR proteins of parasitic nematodes: their possible involvement in the pathogenesis of infection and the use of Caenorhabditis elegans as a model system to evaluate their function. Med Microbiol Immunol 192: 47–52. [DOI] [PubMed] [Google Scholar]
- 46. Meenan NA, Ball G, Bromek K, Uhrín D, Cooper A, et al. (2011) Solution structure of a repeated unit of the ABA-1 nematode polyprotein allergen of Ascaris reveals a novel fold and two discrete lipid-binding sites. PLoS Negl Trop Dis 5: e1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tezuka H, Imai S, Hidano S, Tsukidate S, Fujita K (2003) Various types of Dirofilaria immitis polyproteins selectively induce a Th2-Type immune response. Infect Immun 71: 3802–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xia Y, Spence HJ, Moore J, Heaney N, McDermott L, et al. (2000) The ABA-1 allergen of Ascaris lumbricoides: sequence polymorphism, stage and tissue-specific expression, lipid binding function, and protein biophysical properties. Parasitology 120: 211–224. [DOI] [PubMed] [Google Scholar]
- 49. Kennedy MW (2011) The polyprotein allergens of nematodes (NPAs)–Structure at last, but still mysterious. Exp Parasitol 129: 81–84. [DOI] [PubMed] [Google Scholar]
- 50. Stephensen CB (2001) Vitamin A, infection, and immune function. Ann Rev Nutr 21: 167–192. [DOI] [PubMed] [Google Scholar]
- 51. Cantacessi C, Zou FC, Hall RS, Zhong W, Jex AR, et al. (2009) Bioinformatic analysis of abundant, gender-enriched transcripts of adult Ascaris suum (Nematoda) using a semi-automated workflow platform. Mol Cell Probes 23: 205–217. [DOI] [PubMed] [Google Scholar]
- 52. Him NA, Gillan V, Emes RD, Maitland K, Devaney E (2009) Hsp90 and the biology of nematodes. BMC Evol Biol 9: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maresca B, Kobayashi G (1994) Hsp70 in parasites: as an inducible protective protein and as an antigen. Cell Mol Life Sci 50: 1067–1074. [DOI] [PubMed] [Google Scholar]
- 54. Ma X, Zhao Y, Sun W, Shimabukuro K, Miao L (2012) Transformation: how do nematode sperm become activated and crawl? Protein Cell 3: 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richardson SJ, Hennebry SC, Smith BJ, Wright HM (2005) Evolution of the thyroid hormone distributor protein transthyretin in microbes, C. elegans, and vertebrates. Ann NY Acad Sci 1040: 448–451. [DOI] [PubMed] [Google Scholar]
- 56. Saverwyns H, Visser A, Van Durme J, Power D, Morgado I, et al. (2008) Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi – Comparison with other strongylid nematodes and Caenorhabditis elegans . Int J Parasitol 38: 1545–1556. [DOI] [PubMed] [Google Scholar]
- 57.Donnelly S, Dalton JP, Robinson MW (2011) How pathogen-derived cysteine proteases modulate host immune responses. In Robinson MW, Dalton JP editors. Cysteine Proteases of Pathogenic Organisms. New York, Springer Science.p. 192–207. [DOI] [PMC free article] [PubMed]
- 58. Tort J, Brindley PJ, Knox D, Wolfe KH, Dalton JP (1999) Proteinases and associated genes of parasitic helminths. Adv Parasitol 43: 161–266. [DOI] [PubMed] [Google Scholar]
- 59. Rhoads M, Fetterer R (1998) Purification and characterisation of a secreted aminopeptidase from adult Ascaris suum . Int J Parasitol 28: 1681–1690. [DOI] [PubMed] [Google Scholar]
- 60. Harnett W, Harnett MM (2010) Helminth-derived immunomodulators: can understanding the worm produce the pill? Nat Rev Immunol 10: 278–284. [DOI] [PubMed] [Google Scholar]
- 61. Gregory WF, Maizels RM (2008) Cystatins from filarial parasites: evolution, adaptation and function in the host–parasite relationship. Int J Biochem Cell B 40: 1389–1398. [DOI] [PubMed] [Google Scholar]
- 62. Knox D (2007) Proteinase inhibitors and helminth parasite infection. Parasite Immunol 29: 57–71. [DOI] [PubMed] [Google Scholar]
- 63. Molehin AJ, Gobert GN, McManus DP (2012) Serine protease inhibitors of parasitic helminths. Parasitology 139: 681–695. [DOI] [PubMed] [Google Scholar]
- 64. Hawley J, Martzen M, Peanasky R (1994) Proteinase inhibitors in Ascarida . Parasitol Today 10: 308–313. [DOI] [PubMed] [Google Scholar]
- 65.Komuniecki R, Tielens AGM (2002) Carbohydrate and energy metabolism in parasitic helminths. In Marr JJ, Nilsen TW, Komuniecki RW editors. Molecular Medical Parasitology. San Diego, Elsevir Science.p. 339–358.
- 66. Wang T, Van Steendam K, Dhaenens M, Vlaminck J, Deforce D, et al. (2013) Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLoS-Negl Trop Dis 7: e2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of level 2 biological processes GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 4 biological processes GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 2 molecular functions GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Distribution of level 4 molecular functions GO terms between ESP from A. suum , B. malayi and H. polygyrus .
(TIF)
Proteins identified in A. suum excretory/secretory products (ESP). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Proteins identified in A. suum perienteric fluid (PE). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Proteins identified in A. suum uterine fluid (UF). “SP” = signal peptide. “Y” indicates the presence of signal peptide while “N” indicates a lack of signal peptide. “PIP” and “SC” indicate protein identification probability and sequence coverage, respectively.
(XLSX)
Common and unique proteins in A. suum ESP.
(XLSX)
The presence of common exosome-associated proteins in A. suum ESP. Exosome proteins were obtained from [http://www.exocarta.org/exosome_markers]. Comparisons were made using the protein name. Matches were confirmed by sequence alignment.
(XLSX)