Abstract
Background
A significant fraction of cases of diarrhea, a leading cause of childhood mortality worldwide, remain unexplained.
Objectives
To identify viruses in unexplained cases of diarrhea using an unbiased metagenomics approach.
Study design
Viral nucleic acids were enriched from the feces from 48 cases of unexplained diarrhea from Burkina Faso, sequenced, and compared against all known viral genomes.
Results
The full genome of a highly divergent astrovirus was sequenced in a sample co-infected with parechovirus 1. RT-PCR identified a single astrovirus infection in these 48 patients indicating a low prevalence. Human astrovirus-BF34 was most closely related to mamastrovirus species 8 and 9 also found in human with which it shared 62%, 74%, and 57% amino acid identities over its protease, RNA dependent RNA polymerase and capsid proteins, respectively.
Conclusions
Burkina Faso astrovirus is proposed as prototype for a novel species in the genus Mamastrovirus, here tentatively called Mamastrovirus 20, representing the fifth human astrovirus species.
Keywords: astrovirus, species, diarrhea, metagenomics, children
Background
Astroviruses are small non-enveloped viruses with a characteristic star-like structure whose RNA genomes are 6.4–7.7 Kb in size and contains three ORFs designated ORF1a, 1b, and 2, coding for protease, protease-RdRp fusion, and capsid proteins, respectively. The first astrovirus was identified by electron microscopy in human fecal samples in 1975 1. The family Astroviridae is currently classified into two distinct genera, Mamastrovirus and Avastrovirus infecting mammals and birds respectively.
According to the ICTV different strains of the same astrovirus species should share >75% identity in their capsid proteins. The genus Mamastrovirus currently consists of at least 19 species. The prototypic astrovirus species from human is highly diverse consisting of 8 genotypes which together form the species HAstV or mamastrovirus 1. The second species of human astrovirus, prototype strain MLB1, was characterized by Finkbeiner et al in 2008 2 is now called mamastrovirus 6. The third human astrovirus species (mamastrovirus 8) was reported in 2009 with strains VA2 3 and HMO-A 4. The fourth human astrovirus species (mamastrovirus 9), also described in 2009, includes strains VA1 5, VA3 6, HMO-B and HMO-C 4. HAstV-VA1 was reported as the causative agent for a diarrhea outbreak in a child care center in Virginia 5. Serological studies showed HAstV-HMO-C to be a highly prevalent human infection 7 with approximately 65% of adult in the US showing antibody reactivity. Recently HAstV-VA4 was discovered in diarrhea from Nepalese children with a prevalence of 1% (2/196) 6. The HAstV-VA4’s capsid shared the highest identity of 77.5% to HAstV-VA2 6, suggesting that it also belonged to Mamastrovirus 8.
Objectives
To analyze the fecal virome in unexplained cases of diarrhea using an unbiased metagenomics approach and genetically characterized novel viruses.
Study design
Feces from Burkina Faso children with unexplained acute gastroenteritis were analyzed by deep sequencing. These specimens had been previously screened for rotavirus, norovirus, pathogenic bacteria, parasites, and yeasts and non-reactive samples were selected here for further viral metagenomics analysis 8, 9.. Fecal viral particles from 48 patients were enriched by filtration of stool supernatants and nuclease treatment of the filtrate to reduce the concentration of naked host and bacteria nucleic acids. Briefly, the samples were clarified by 15,000 × g centrifugation for 10 min. A total of 200 μl of supernatants was filtered through a 0.45-μm filter (Millipore) to remove bacterium-sized particles. The filtrate was then treated with a cocktail of DNases (Turbo DNase from Ambion, Baseline-ZERO from Epicentre, and Benzonase from Novagen) and with RNase (Fermentas) to digest unprotected nucleic acids. Nuclease resistant nucleic acids were then extracted 10. A DNA library was constructed using ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre) according to the manufacturer’s instructions, and sequenced using the Illumina MiSeq platform. The Illumina kit generated sequences were compared to the GenBank nonredundant protein databases using BLASTx.
Results
All 48 samples were analyzed in 5 pools using one Illumina MiSeq run of 250 bases paired-end reads. One pool showed a single read encoding an astrovirus-like protein segment with a best BLASTx E-score of 10−11 to human astrovirus HMO-A (GenBank NC_013443). The specific fecal sample within the pool containing this sequence was identified using RT-PCR with primers complementary to this astrovirus sequence and then re-analyzed individually using the same metagenomics approach. A total of 127 unique sequences covering 57% of the astrovirus genome were identified. Also detected were 233 unique sequences covering 82% of a parechovirus 1 genome indicating a viral co-infection. The complete genome of the astrovirus was then determined by filling gaps between sequence fragments by RT-PCR, and the RNA genome extremities were amplified by 5′ and 3′ RACE. PCR amplicons were directly sequenced by primer walking.
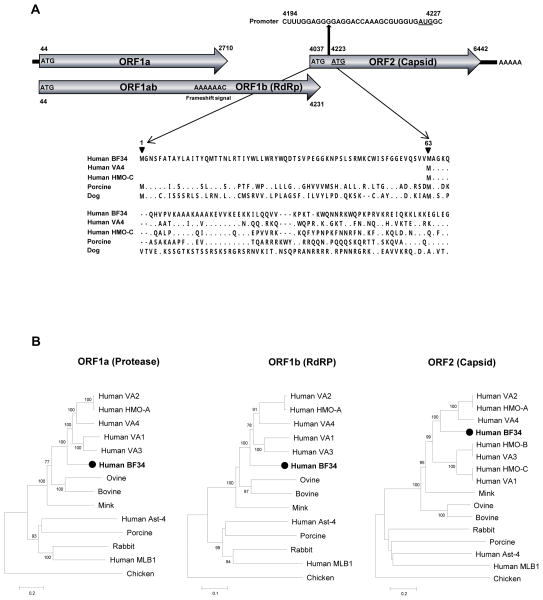
The complete genome of the human astrovirus named Burkina Faso 34 (HAstV-BF34, GenBank: KF859964) is 6561-bp in length excluding the polyadenylated tract, with a GC content of 43%. HAstV-BF34 has a 5′ UTR of 43 bases, three overlapping open reading frames (ORFs), and a 3′ UTR of 119 bases. ORF1a encoded the nonstructural polyprotein (888-aa) with a conserved trypsin-like serine protease domain with best BLASTx match to that of mamastrovirus 8, sharing 60% aa-identities. ORF1b encoded RdRp (523-aa) expressed via a ribosomal frameshift caused by a conserved “slippery heptamer” sequence [AAAAAAC]. Pair-wise analysis showed that the RdRp region had the highest identity of 73–74% to those of mamastrovirus 8 strains. NCBI ORF finder and protein alignments revealed that HAstV-BF34 had an alternative ORF2 start codon M1GNS upstream of the putative start codon of M63AGKQ seen in other genetically-related human astroviruses (Figure 1A). An unusual long N-terminus for ORF2 was also observed in porcine and dog astroviruses sharing the same M1GNS with HAstV-BF34 (Figure 1A). The consensus promoter initiating ORF2 subgenomic RNA synthesis in HAstV-BF34 was identified as CUUUGGAGGGGAGGACCAAAGCGUGGUGAUGGC (M63 start codon underlined) 11. We selected the second methionine codon (M63AGKQ) as the putative start codon for this analysis. The shorter ORF2 encoded a viral capsid precursor protein 739-aa in size. It was most closely related to those of HAstV-HMO-A, HAstV-VA2, and HAstV-VA4 (mamastrovirus 8) with 57% aa-identity and to HAstV-C (mamastrovirus 9) with 52% aa-identity. Because the identity over the capsid protein is less than 75% to those of previously reported astroviruses, HAstV-BF34 is proposed as prototype for a novel species in the genus Mamastrovirus, here tentatively called Mamastrovirus 20 pending ICTV review, representing the fifth human astrovirus species.
Figure 1. Astrovirus genome and phylogeny.
(A) Organization of the astrovirus genome. Locations for the conserved “slippery heptamer” and promoter sequences were indicated. The alignment of the ORF2 N-terminus regions of human [Genbank NC_019027 and GQ415662], porcine [Genbank NC_019494], and dog [Genbank GU376736] astroviruses were shown. Dots indicated identity to the amino acid sequence of HAstV-BF34 while dash denoted deletion. (B) Phylogenetic trees generated with protease, RdRp, and capsid proteins of HAstV-BF34 and genetically-related astroviruses in the genus Mamastrovirus. The scale indicated amino acid substitutions per position. Genbank numbers for astrovirus references used in the phylogenetic trees are as follows: Human astrovirus VA1 [FJ973620], VA2 [GQ502193], VA3 [NC_019026], VA4 [NC_019027], HMO-A [NC_013443], HMO-B [GQ415661], HMO-C [GQ415662], MLB1 [JQ086552], Ast-4 [DQ070852]; Mink [AY179509]; Ovine [NC_002469]; Bovine [KF233994]; Rabbit [JF729316]; Porcine [JF713713]; Chicken [JF414802];
Sequence alignment was performed using CLUSTAL X with the default settings 12. Phylogenetic tree with 100 bootstrap resamples of the alignment data sets was generated using MEGA version 5 13. Bootstrap values (based on 100 replicates) for each node are shown if >70 (Figure 1B). Phylogenetic analysis showed that HAstV-BF34 was most closely related to Mamastrovirus 8 and 9 infecting humans (Figure 1B).
A nested PCR assay was used to determine the prevalence of this virus in the 48 diarrhea feces from Burkina Faso children. Primers BF34-F1 (5′-GTC CTG AAG ATT ACA GCA AGT CCT-3′) and BF34-R1 (5′-GAC CCA TCC GAG TGA GTG TG-3′) were used for the first round of PCR, and primers BF34-F2 (5′-GAA CCA TTG ACT AAC ATA AAA GCC A-3′) and BF34-R2 (5′-TCC TCA AAA ACA CAG CCT ATT CT-3′) for the second round of PCR, resulting in an expected amplicon of ~350 bp. The PCR conditions was as follows: denaturation at 95°C for 5 min, 35 cycles of 95°C for 30 s, 51°C or 49°C (for the first or second round, respectively) for 30 s and 72°C for 1 min, a final extension at 72°C for 10 min, and then held at 4°C. No other samples except the one initially detected by deep sequencing were PCR positive yielding a low prevalence of detection of <2% (1/48) in this population.
Discussions
The classic human astrovirus (mamastrovirus 1) has been associated with acute gastroenteritis in humans worldwide 11, 14–17. Chronic infections of astrovirus were reported in immunodeficient patients 18, 19. Mamastrovirus 9 (strain HAstV-PS) was also identified as the causative agent in a fatal case of encephalitis in a child with agammaglobulinemia 20 and HAstV-MLB2 (whose capsid is 74% identical to mamastrovirus 6 strain MLB1 and may therefore also qualify as a distinct species) has also been reported in the plasma of a febrile child 21. Astroviruses were also found in the brain of minks with a shaking symptoms 22 and in the brain and spinal cord of cows with neurologic symptoms 23 indicating that astroviruses are not exclusively enteric infections but can also affect other organ systems. Varied astroviruses have also been detected in feces from a wide variety of farm, wild, and laboratory aminals 11 indicating that the known diversity of this viral family is likely to rapidly expand.
In this study, fecal shedding of a previously uncharacterized astrovirus was detected in an unexplained case of diarrhea. The 14-month old female patient has significant malnutrition indicators such as severe underweight, severe wasting, and moderate stunting. This patient showed typical symptoms of acute gastroenteritis including fever (38°C), vomiting, abdominal pain, liquid feces (4 times/day) and moderate dehydration. The simultaneous detection of parechovirus 1 in this fecal sample provides an alternative explanation for the patient’s symptoms although a primary or aggravating role for the astrovirus is also conceivable. Because the parechovirus and astrovirus genomes both consist of ssRNA of similar length their relative viral loads are likely reflected by the respective number of sequence reads (233/127) or nearly twice as many parchovirus as astrovirus RNA genomes. The absence of detected recombination between mamastrovirus 8, 9, and proposed mamastrovirus 20 genomes, using SimPlot (data not shown), would support the presence of different astrovirus species. The phylogenetic clustering of now three species of human astrovirus (mamastrovirus 8, 9, and proposed 20) to the exclusion of animal astroviruses may also reflect their monophyletic origin and these species could potentially be subsumed into a single, more diverse group. It may also be concluded that astrovirus evolution within human has occurred over long enough period of time to yield multiple divergent clades and that yet more intermediate forms between these clades may be anticipated. In a similar fashion the three bat mamastroviruses 17, 18, 19 species also appear monophyletic 24. Wider geographic sampling of human fecal samples will likely continue to increase the known diversity of human astroviruses. Case-control studies of unexplained diarrhea will help determine whether any of these novel astroviruses has pathogenic potential 25 although their occasional pathogenicity in highly susceptible individuals 20 will remain a possibility irrespective of their associations or lack thereof with diarrhea or other enteric condition.
Highlights.
New astrovirus found in feces from unexplained diarrhea case in Burkina Faso
Genetic divergence indicates new species related to mamastrovirus 8 and 9
Mamastrovirus 20 found in co-infection with parechovirus 1
Virus detected in only 1 out of 48 unexplained diarrhea cases
Acknowledgments
Funding
This work was supported by NHLBI grant R01 HL105770 to E.L.D and the Blood Systems Research Institute.
Footnotes
The GenBank accession numbers for human astrovirus BF34 is KF859964
Competing interests
None for all authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Madeley CR, Cosgrove BP. Letter: 28 nm particles in faeces in infantile gastroenteritis. Lancet. 1975;2:451–2. doi: 10.1016/s0140-6736(75)90858-2. [DOI] [PubMed] [Google Scholar]
- 2.Finkbeiner SR, Kirkwood CD, Wang D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virology journal. 2008;5:117. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, et al. Human stool contains a previously unrecognized diversity of novel astroviruses. Virology journal. 2009;6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, et al. Multiple novel astrovirus species in human stool. The Journal of general virology. 2009;90:2965–72. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. Journal of virology. 2009;83:10836–9. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, et al. Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology. 2013;436:8–14. doi: 10.1016/j.virol.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo PD, Ching KH, Esper F, Iadarola MJ, Delwart E, Lipkin WI, et al. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PloS one. 2011;6:e22576. doi: 10.1371/journal.pone.0022576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitiema LW, Nordgren J, Ouermi D, Dianou D, Traore AS, Svensson L, et al. Burden of rotavirus and other enteropathogens among children with diarrhea in Burkina Faso. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2011;15:e646–52. doi: 10.1016/j.ijid.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PloS one. 2013;8:e69557. doi: 10.1371/journal.pone.0069557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. Journal of virology. 2009;83:4642–51. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11:1529–44. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guix S, Bosch A, Pinto RM. Human astrovirus diagnosis and typing: current and future prospects. Letters in applied microbiology. 2005;41:103–5. doi: 10.1111/j.1472-765X.2005.01759.x. [DOI] [PubMed] [Google Scholar]
- 15.Afrad MH, Karmakar PC, Das SK, Matthijnssens J, Ahmed F, Nahar S, et al. Epidemiology and genetic diversity of human astrovirus infection among hospitalized patients with acute diarrhea in Bangladesh from 2010 to 2012. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2013;58:612–8. doi: 10.1016/j.jcv.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto T, Wangchuk S, Tshering K, Yahiro T, Zangmo S, Dorji T, et al. Complete Genome Sequences of Two Astrovirus MLB1 Strains from Bhutanese Children with Diarrhea. Genome announcements. 2013:1. doi: 10.1128/genomeA.00485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li Y, Jin Y, Li DD, Li X, Duan ZJ. Recently identified novel human astroviruses in children with diarrhea, China. Emerging infectious diseases. 2013;19:1333–5. doi: 10.3201/eid1908.121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wunderli W, Meerbach A, Gungor T, Berger C, Greiner O, Caduff R, et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PloS one. 2011;6:e27483. doi: 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood DJ, David TJ, Chrystie IL, Totterdell B. Chronic enteric virus infection in two T-cell immunodeficient children. Journal of medical virology. 1988;24:435–44. doi: 10.1002/jmv.1890240410. [DOI] [PubMed] [Google Scholar]
- 20.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerging infectious diseases. 2010;16:918–25. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtz LR, Wylie KM, Sodergren E, Jiang Y, Franz CJ, Weinstock GM, et al. Astrovirus MLB2 viremia in febrile child. Emerging infectious diseases. 2011;17:2050–2. doi: 10.3201/eid1711.110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. Journal of clinical microbiology. 2010;48:4392–6. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, et al. Divergent astrovirus associated with neurologic disease in cattle. Emerging infectious diseases. 2013;19:1385–92. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu HC, Chu DK, Liu W, Dong BQ, Zhang SY, Zhang JX, et al. Detection of diverse astroviruses from bats in China. The Journal of general virology. 2009;90:883–7. doi: 10.1099/vir.0.007732-0. [DOI] [PubMed] [Google Scholar]
- 25.Holtz LR, Bauer IK, Rajendran P, Kang G, Wang D. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PloS one. 2011;6:e28647. doi: 10.1371/journal.pone.0028647. [DOI] [PMC free article] [PubMed] [Google Scholar]