Abstract
MuRF1 is a previously reported ubiquitin-ligase found in striated muscle that targets troponin I and myosin heavy chain for degradation. While MuRF1 has been reported to interact with mitochondrial substrates in yeast two-hybrid studies, no studies have identified MuRF1’s role in regulating mitochondrial function to date. In the present study, we measured cardiac mitochondrial function from isolated permeabilized muscle fibers in previously phenotyped MuRF1 transgenic and MuRF1−/− mouse models to determine the role of MuRF1 in intermediate energy metabolism and ROS production. We identified a significant decrease in reactive oxygen species production in cardiac muscle fibers from MuRF1 transgenic mice with increased alpha-MHC driven MuRF1 expression. Increased MuRF1 expression in ex vivo and in vitro experiments revealed no alterations in the respiratory chain complex I and II function. Working perfusion experiments on MuRF1 transgenic hearts demonstrated significant changes in glucose or oleate oxidation; however, total oxygen consumption was decreased. This data provides evidence for MuRF1 as a novel regulator of cardiac ROS, offering another mechanism by which increased MuRF1 expression may be cardioprotective in ischemia reperfusion injury, in addition to its inhibition of apoptosis via proteasome-mediate degradation of c-Jun. The lack of mitochondrial function phenotype identified in MuRF1−/− hearts may be due to the overlapping interactions of MuRF1 and MuRF2 with energy regulating proteins found by yeast two-hybrid studies reported here, implying a duplicity in MuRF1 and MuRF2’s regulation of mitochondrial function.
Keywords: MuRF1, ubiquitin ligase, mitochondria, metabolism, heart
Introduction
Muscle Ring Finger-1 (MuRF1) is an ubiquitin ligase initially identified for its role in the regulation of skeletal muscle [1] and cardiac atrophy [2] in vivo. In the absence of MuRF1, mice are protected from muscle loss following skeletal muscle denervation, while dexamethasone-induced cardiac atrophy is almost completely inhibited [1,2]. The expression of MuRF1 is striated muscle specific [1] and limited to the M-line and cytoplasm within cardiomyocytes [3,4]. Consistent with its role in degrading sarcomere proteins during atrophy, MuRF1 has been found to interact with numerous sarcomeric proteins, including troponin I, troponin T, titin, telethonin, myosin light chain 2, and nebulin [5] and has been reported to be a bona fide ubiquitin ligase that targets the degradation of troponin I [6] and myosin heavy chain [7]. Taken together, these studies mechanistically link MuRF1 activity to cardiac and skeletal muscle atrophy [1] in regulating the deconstruction of the sarcomere proteins through its poly-ubiquitination and subsequent degradation by the proteasome.
Since ubiquitin ligases exert their activity by placing ubiquitin on specific substrates, early MuRF1 studies focused on determining proteins that MuRF1 interacted with [5]. Previous yeast two hybrid studies using MuRF1 as bait identified >500 clones (of which 110 were sequenced). While it was not surprising that 23 prey clones were derived (and confirmed) from nine different genes coding for myofibrillar proteins, it was, however, intriguing that 27 prey clones from 11 different genes coded for enzymes involved in energy metabolism, including creatine kinase [5]. We have found creatine kinase to be a MuRF1 substrate in the heart, which appears to contribute to the susceptibility to heart failure after pressure-overload induced cardiac hypertrophy in vivo [8]. Of these 11 genes identified in energy metabolism, 4 are involved in mitochondrial oxidative phosphorylation (NADH dehydrogenase, aka ubiquinone, NADH ubiquinone oxidoreductase, and 3-hydroxyisobutyrate dehydrogenase), including the mitochondrial ATP synthase beta-subunit involved in the regeneration of ATP [5]. While these findings demonstrate that MuRF1 interacts specifically with proteins involved in oxidative phosphorylation and mitochondrial biology, the functional significance of these findings has not been reported.
Recent studies have used a proteomics approach to identify ubiquitinated proteins in the heart [9]. When the identified proteins were categorized by cellular sub-compartment, the greatest number of ubiquitinated proteins were found in the mitochondria (38.0%), followed by the cytosol (27.3%) [9]. Despite the fact that the mitochondria contain most of the ubiquitinated proteins in the heart, few ubiquitin ligases that drive these processes have been identified in the mitochondria in any cell type. Examples include cell division control protein 53 (Cdc53), mitochondrial distribution and morphology protein 30 (MDM30), MITOL/MARCHV membrane-associated ring finger 5 [10,11], mitochondrial ubiquitin ligase activator of NF-κB (MULAN) [12], ring finger protein 185 (RNF185) [13], and the deubiquitnating enzyme 16 (Ubp16)/ubiquitin specific peptidase 30 (USP30) which broadly play a role in mitochondrial dynamics [14,15]. While two of these mitochondrial ubiquitin ligases are incidentally found in the heart (MARCH5 and MULAN/MAPL), the role of cardiac ubiquitin ligases have not previously been described in the heart [15].
In the present study, we identify for the first time that the striated muscle restricted MuRF1 is prominently present in cardiomyocyte mitochondria to regulate the oxygen consumption and flux through the Krebs cycle without affecting the permeability transition (calcium handling). Most significantly, increasing cardiomyocyte MuRF1 results in significant reductions in the mitochondrial production of reactive oxygen species disproportionately to the decreased oxygen consumption without affecting complex I and complex II activity, the site at which reactive oxygen species are most predominantly formed. These studies indicate mechanism(s) by which increasing MuRF1 may prove to be cardioprotective in clinical scenarios such as cardiac ischemia/reperfusion injury, in addition to MuRF1’s regulation of c-Jun N-terminal protein kinases (JNK) signaling through c-Jun, recently described by our laboratory [16].
Materials and Methods
Muscle Ring Finger-1 (MuRF1) transgenic and MuRF1−/− mouse models
MuRF1 cardiac-specific transgenic (Tg+) and MuRF1−/− mice ~ 12 weeks of age were used in the present studies, as recently described by our laboratory previously [8,17]. The α-myosin heavy chain (MHC) promoter driven murine MuRF1 gene (GenBank NM_001039048) has cardiac MuRF1 transgene expression levels ~45 fold WT levels [8]. The creation of MuRF1−/− mice has previously been described [1] to have no apparent cardiac phenotype at a baseline state [17]. No obvious developmental defects have been detected compared to wildtype (WT) littermates in size, activity, or longevity. All experiments used male WT littermates as controls. The mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) review boards at the University of North Carolina and East Carolina University, and were performed in accordance with federal guidelines.
Preparation of permeabilized cardiac myofibers
The use of permeabilized cardiac myofibers has been previously described [18,19]. Approximately 30 mg of left ventricular tissue was dissected and placed in ice-cold buffer X [50 mM MES, 7.23 mM K2EGTA, 2.77 mM CaK2EGTA, 20 mM imidazole, 0.5 mM DTT (dithiothreitol), 20 mM taurine, 5.7 mM ATP, 14.3 mM PCr (phosphocreatine) and 6.56 mM MgCl2·6H2O (pH 7.1, 290 mOsm)], then trimmed of connective and vascular tissue. Approximately 5 muscle bundles were prepared from each animal and gently separated along their longitudinal axis with needle-tipped forceps, treated with 50 μg/ml saponin in ice-cold buffer X, incubated for 30 min at 4°C [20]. The fibers were then used for mO2 (mitochondrial O2) consumption experiments and placed in buffer Z [105 mM K-Mes, 30 mM KCl, 1 mM EGTA, 10 mM K2HPO4, 5 mM MgCl2·6H2O, 0.005 mM glutamate and 0.002 mM malate with 5.0 mg/ml BSA (pH 7.4, 290 mOsm)]. The fibers used for mH2O2 (mitochondrial H2O2) were placed in ice-cold buffer Y (250 mM sucrose, 10 mM Tris/HCl, 20 mM Tris base, 10 mM KH2PO4, 2 mM MgCl2·6H2O and 0.5 mg/ml BSA). Permeabilized fibers in buffer Z were rotated at 4°C until analysis (<100 min maximum).
Measurement of mO2, mH2O2 and mCa2+ in permeabilized cardiac myofibers
Mitochondrial measurements were made using the O2K Oxygraph system (Oroboros Instruments) at 30°C. Measurements of mH2O2 and mCa2+ measurements were obtained using a spectrofluorometer (Photon Technology Instruments), equipped with a thermo-jacketed cuvette chamber. Mitochondrial oxygen consumption experiments were performed in buffer Z. In mH2O2 measurements, buffer Y contained 10 μM Amplex Red (Invitrogen), 1 unit/ml horseradish peroxidase, 5 mM pyruvate, 2 mM malate, and SOD (25 U/mL); mH2O2 emission was calculated as outlined previously [19].
Ex Vivo Assessment of Myocardial Contractile Function
Myocardial contractile function was determined ex vivo through isolated working mouse heart perfusions, as described previously [21]. All hearts were perfused in the working mode in a non-recirculating manner with a preload of 12.5 mmHg and an afterload of either 50 mm Hg (baseline) or 80 mm Hg (high). Standard Krebs-Henseleit buffer was supplemented with 8 mM glucose, 0.4 mM oleate conjugated to 3% BSA (fraction V, fatty acid-free; dialyzed), 0.05 mM L-carnitine, 0.13 mM glycerol, and 10 μUnit/ml insulin. Following perfusion under baseline conditions (30 minutes), a workload challenge was performed, wherein responsiveness to 1 μM epinephrine plus an elevated afterload of 80 mmHg was assessed for 30 minutes. Measures of cardiac function (i.e., cardiac power, cardiac efficiency, heart rate, developed pressure, and rate pressure product) were determined at 5-minute intervals, as previously described [21]. At the end of the perfusion period, hearts were snap-frozen in liquid nitrogen and stored at −80°C prior to analysis. Data are presented as steady state values (i.e., the mean of the last two time points during a distinct perfusion condition for each individual heart).
Mitochondrial isolation and calcium studies
Cardiac mitochondria were isolated from the left ventricle of hearts utilizing a modified Chappell-Perry protocol adapted from Sloan et al. [22]. Briefly, hearts were excised and immersed in 10 mL ice cold mitochondrial isolation medium [23] containing (in mM): 300 sucrose, 10 sodium-hepes and 0.2 EDTA. The left ventricle was isolated, weighed, and rinsed in fresh MIM buffer. Hearts were minced finely and subjected to 2 min of digestion using 0.25 mg trypsin, diluted in 2 mL of MIM (pH=7.2). Following digestion, 1.3 mg of trypsin inhibitor was diluted into 2mL of MIM buffer+BSA (1 mg/mL) at pH=7.4. The minced tissue was resuspended in 3 mL MIM buffer+BSA and homogenized with a Teflon® Potter homogenizer. The heart homogenate was centrifuged at 600 g for 10 min, and the supernatant was then centrifuged at 8000 g for 15 min. The supernatant was discarded, and the pellet re-suspended in 2 mL MIM buffer+BSA. This step was repeated one more time, and the final pellet was stored on ice in ~75 μL MIM buffer. All steps were performed at 4°C and all instruments were chilled for 4°C prior to use. Mitochondrial protein content was determined using a BCA protein assay. For mCa2+ measurements, buffer Y contained 300ug of isolated mitochondria as determined by BCA, 5 μM Calcium Green 5-N (Invitrogen) with either 5 mM pyruvate and 2 mM malate or 5mM succinate. At the start of mCa2+ experiments, with 10 μM EGTA added to chelate residual Ca2+ and to establish Fmin. Pulses of 15 nmol of Ca2+ (CaCl2) were sequentially added and Ca2+ uptake was followed until mPTP (mitochondrial permeability transition pore) opening, as described previously [24]. At the end of the experiment, 1 mM CaCl2 was added to saturate the probe and establish Fmax. Changes in free Ca2+ in the cuvette during mCa2+ uptake were then calculated using the known Kd for Calcium Green 5-N and the equations established by Tsien [25] for calculating free ion concentrations using ion-sensitive fluorophores. At the conclusion of all experiments, fibers were rinsed in double-distilled water, freeze-dried for >2 h and weighed on a micro-scale. The data are expressed as pmol·min−1·(mg of dry weight)−1 (for mH2O2) or nmol·(mg of μg protein)−1 (for mCa2+).
Mitochondrial isolation for complex I/complex II functional studies
Mitochondria were isolated from mouse hearts by differential centrifugation [26,27]. Ventricles were minced in ice cold homogenization buffer (0.25M sucrose, 10mM Hepes, pH 7.4, 1mM EDTA, 1mM DTT plus protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). Minced tissue was then homogenized in Potter-Elvehjem homogenizer for 6 passes then centrifuged at 600xg for 5 min to remove unbroken cells and nuclei. The crude mitochondrial fraction was isolated from the supernatant by centrifugation for 15 min at 12,000xg. The pellet was then resuspended in homogenization buffer, layered over a 30% Percoll gradient, and centrifuged at 95,000xg for 30 min to separate mitochondria from other membranes. The lower band was collected, washed 2 times to remove Percoll, leaving a final mitochondrial fraction resuspended in homogenization buffer and stored at −80°C.
Complex I and Complex II functional assays
Mitochondrial complex I and II activities were measured in HL1 cell lysates and both whole heart homogenates and isolated heart mitochondria from MuRF1 Tg+ mice and WT control mice using microplate assay kits purchased from Abcam (Mitosciences) according to the manufacturer’s protocols. In each assay kit, the enzyme complex was immunocaptured within the wells of the microplate. For all measurements, total protein was determined prior to immunocapture and activity data was normalized per ug of total protein input. The Complex I Microplate Assay Kit (ab109721) measures the activity of immunocaptured mitochondrial Complex I by following the oxidation of nicotinamide adenine dinucleotide (NADH) to NAD+ and the simultaneous reduction of a dye leading to increased absorbance at 450 nm. In the Complex II Microplate Assay Kit (ab109908), the activity of immunocaptured mitochondrial complex II is measured by following the reduction of ubiquinone to ubiquinol and recording the associated decrease in absorbance at 600 nm. Prior to measurement of complex I and complex II activity in HL1 cells, cells were transduced with adenovirus expressing CMV GFP-tagged myc-MURF1 or CMV GFP control (n=6/condition) at MOI=30 for 24hrs. Cells were then collected by scraping and low-speed centrifugation prior to processing according to the kit protocols. Whole heart homogenates from WT and MuRF1 Tg+ mice (n=5/genotype) were homogenized in 0.5 mL ice-cold PBS containing HALT protease/phosphatase inhibitors (Pierce). Prior to complex I and II activity determination mitochondria were isolated from WT and MuRF1 Tg+ mouse hearts (n=7 WT, n=5 Tg+) by differential centrifugation in buffer containing 0.25M sucrose, 10mM Hepes, pH 7.4, 1mM EDTA, 1mM DTT with protease inhibitors (Roche 11 697 498 001) and phosphatase inhibitor cocktail (Calbiochem Cat. # 524625).
Preparation of HL-1 mitochondria and cytosol and Western blotting
Cardiac cell lines HL-1 [28] and H9C2 (CRL1446, ATCC®, Manassas, VA) [29] were plated in 15 cm dishes, transduced with Ad.MuRF1 or Ad.GFP, and whole cell lysate prepared in lysis buffer (Cell Signaling, Cat#9803) including protease inhibitor (Roche, Cat.#04 906 837 001) and beta-glycerol phosphate. C2C12 (ATCC, Manassas, VA, Cat# CRL-1772) myoblasts were differentiated to myotubes as described by the manufacturer and similarly transduced with Ad.MuRF1 and Ad.GFP and harvested. Mitochondria and cytosol fractions were obtained using the Thermo Scientific Mitochondria Isolation Kit for Cultured Cells (Cat#89874). Briefly, cells were homogenized on ice using a syringe with a 27g needle (26 strokes), 800uL of Mitochondria Isolation Reagent C and 200uL Mitochondria Isolation Reagent A were added, and the homogenate was centrifuged at 700g for 10 min at 4°C. The mitochondria pellet was lysed following 10 min incubation in RIPA buffer (Sigma, Cat#R0278) including protease inhibitor (Roche, Cat#04906837001) and beta-glycerol phosphate. Twenty μg of lysate was resolved on a 4–12% Bis-Tris gel, transferred to PVDF membrane, blocked in 5% nonfat milk/TBS-Tween, and incubated with primary antibodies overnight at 4°C (using anti-c-Myc-Peroxidase antibody (1:5,000, Sigma, Cat#A5598), anti-cytochrome c (1:1,000, Cell signaling, Cat#4272), anti-protein disulfide isomerase (1:1,000, Cell Signaling, Cat#3501), and anti-GAPDH (1:1,000, Millipore, Cat#MAB374)). Anti-Rabbit IgG-Peroxidase (1:10,000, Sigma, Cat#A9169) or anti-Mouse IgG-Peroxidase (1:60,000, Sigma, Cat#A9917) were incubated for 1 hr at RT and developed using Amersham ECLSelect (Cat#RPN 2235) for chemiluminescence detection.
Statistical analysis
A One Way ANOVA was performed to determine significance in work heart assays of baseline and high workload conditions, followed by multiple comparison procedures (Holm-Sidak) to determine significance between groups. A Student’s t-test was performed to determine differences between experimental and control groups in each strain of mouse line in all other experiments. Both One Way ANOVA and Student’s t-test analyses were performed using Sigma Plot 11 (Systat Software, Inc., San Jose, CA). Statistical significance was defined as p<0.05 unless otherwise noted.
Results
Recent studies have reported that MuRF1−/− hearts do not have a cardiac phenotype (wall thickness and systolic function) different from their wildtype siblings by conscious echocardiography at <6 months of age, unless challenged with cardiac stresses such as pressure overload-induced cardiac hypertrophy [17]. MuRF1 Tg+ mice with constitutively increased cardiomyocyte MuRF1 have a non-progressive decrease in wall thickness (15–20%) and mildly decreased fractional shortening % (25%), a measure of systolic, with conscious echocardiography [2,8]; the MuRF1 Tg+ cardiac function is unimpaired function by ex vivo Langendorff assays [16]. Neither phenotype is consistent with progressive muscle wasting. To build upon these previous studies, we sequentially challenged these mouse models to dynamic changes in workload using the physiologocially relevant working heart perfusion model to determine for the first time the role of cardiac MuRF1 in both cardiac performance and mitochondrial function.
MuRF1 activity is present in mitochondria fractions
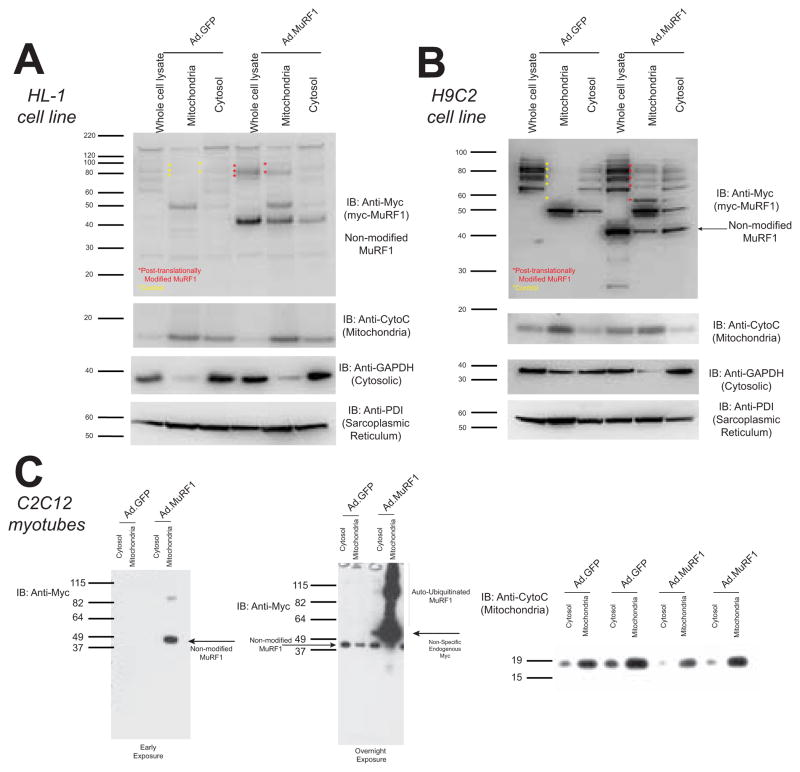
In three different cardiac and skeletal muscle cell lines, we investigated the location of MuRF1’s ubiquitin ligase activity using a MuRF1 adenovirus (and control GFP) construct as previously described [30]. We identified MuRF1 activity in mitochondria, determined by MuRF1 auto-ubiquitination in the HL-1 atrial cardiomyocyte-derived cell line (Figure 1A), the H9C2 ventricular cardiomyocyte cell line (Figure 1B), and C2C12 skeletal muscle cell line myotubes (Figure 1C). As a ubiquitin ligase, MuRF1’s importance to date has relied on its ability to ubiquitinate substrates, so its interesting to note that MuRF1’s critical activity, represented by auto-ubiquitination, was found primarily in the mitochondrial fraction in each of these myocyte cells lines (Figure 1).
Figure 1. MuRF1 is found present in the mitochondria.
Western analysis of MuRF1 expression in mitochondrial and cytosolic fractions in HL-1 and H9C2 cells. Cells were transduced with Ad.mycMuRF1 or Ad.GFP and lysed to obtain whole cell lysate or differential centrifugation to isolate mitochondria and cytosol fractions. Immunoblot for A. MuRF1 (anti-myc), cytochrome c, GAPDH, and PDI illustrate MuRF1’s presence in the mitochondria and auto-ubiquitination. These unique auto-ubiquitinated forms of (myc)MuRF1 can be seen in the mitochondria and cytosol fractions in both the HL-1 atrial cardiomyocyte derived, B. H9C2 ventricular cardiomyocyte derived cell line, as well as the C. C2C12 undifferentiated myotubes. MuRF1 (anti-myc) and cytochrome c (mitochondria), illustrated at both early (left, 30 second exposure) and extended (right-overnight) exposures.
MuRF1−/− working hearts exhibit enhanced function in the working heart
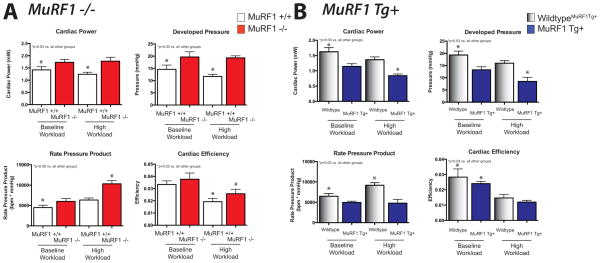
Isolated working MuRF1−/− and MuRF1 Tg+ hearts were first assayed for their functional characteristics ex vivo. MuRF1−/− hearts exhibited significantly increased cardiac power and developed pressure at both baseline and high workload conditions (Figure 2A). At the same time, sibling-matched wildtype control hearts exhibit a decreased cardiac power and developed pressure with high workload. This enhanced contractility is amplified when the amount of work per time (rate pressure product) in the MuRF1−/− hearts, being largest with the high workload (Figure 2A). MuRF1−/− hearts maintain more cardiac efficiency, a measure of output as a function of oxygen consumption, compared to wildtype in high workload conditions, consistent with their enhanced function (Figure 2A). Recent studies have identified that MuRF1−/− mice exhibit an increased cardiac capacity for aerobic activity (voluntary running). In these studies, untrained MuRF1−/− mice ran farther and faster than wild type controls mice, an advantage which was enhanced with subsequent training [31]. MuRF1 Tg+ hearts, in contrast, exhibited significantly less cardiac function than their wildtype counterparts. Specifically, MuRF1 Tg+ hearts had decreased cardiac power and developed pressure at both baseline and high workload conditions (Figure 2B). When heart rate is taken in to consideration, MuRF1 Tg+ rate pressure product was significantly less than wildtype hearts at both baseline and high workload conditions. At baseline workload, MuRF1 Tg+ hearts exhibited less cardiac efficiency, a change that was not seen in the high workload (Figure 2B). Taken together, MuRF1 expression correlated inversely with the contractile function of the heart, an effect that was exaggerated when challenged with a high workload.
Figure 2. MuRF1−/− hearts exhibit increase power and developed pressure at baseline and high workload conditions while MuRF1 Tg+ hearts have impaired.
Functional characteristics of A. MuRF1−/− and B. MuRF1 Tg+ heart function ex vivo. Hearts were isolated and immediate perfused in the working mode at basal (50 mm Hg) and high (80 mm Hg) workloads. Cardiac power (a derivative of cardiac output), cardiac efficiency (ratio of cardiac work and myocardial oxygen consumption), and rate pressure product (heart rate*developed pressure) were calculated as previously described [21]. Values are expressed as mean ± SEM and represent 5–7 mice per group. Groups were analyzed by a One Way ANOVA vs. sibling match wildtype controls at baseline and high workload conditions. n.s.= not significant. p<0.05, unless otherwise noted in the panel.
Glucose and fatty acid oxidation in the working heart
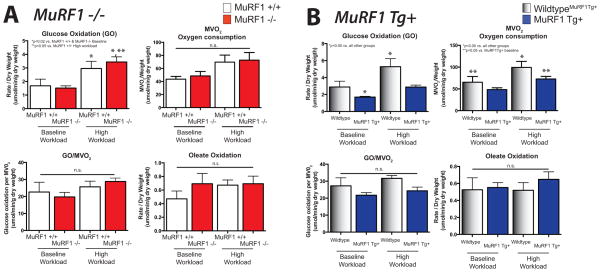
MuRF1 has been reported to bind the pyruvate dehydrogenase complex (PDH) via pyruvate dehydrogenase (lipoamide) beta and pyruvate dehydrogenase kinase, which regulates PDH activity in yeast two hybrid studies [5]. Since PDH catalyzes the conversion of pyruvate to acetyl-CoA and glucose oxidation, we investigated the role of MuRF1 in regulating glucose oxidation in our MuRF1−/− and MuRF1 Tg+ models using a working heart assay. In the present study, we identified that neither MuRF1 Tg+ hearts or MuRF1−/− hearts have any differences in glucose oxidation in the working heart ex vivo both at baseline, or in response to increased workload compared to sibling-matched WT controls (Figure 3A, top). While MuRF1 Tg+ hearts did not significantly differ in their glucose oxidation in the working heart ex vivo at baseline, glucose oxidation trended to be attenuated in the face of increased workload compared to sibling WT controls, although not significantly (Figure 3B, top). The oxidation of the long-chain fatty acid oleate generally did not differ between MuRF1−/− and sibling WT mice (Figure 3A, lower left). Similarly, oleate oxidation by MuRF1 Tg+ hearts did not differ from wildtype controls (Figure 3B, lower left), even when normalized for MVO2 consumption (Figure 3, lower left). Minor differences in oxygen consumption could be seen in the working MuRF1 Tg+ hearts had both at baseline and after challenge with a high workload (Figure 3, lower right).
Figure 3. MuRF1 Tg+ hearts exhibits alterations in glucose oxidation and oxygen consumption ex vivo.
Metabolic (glucose and oleate oxidation) and oxygen consumption utilization of A. MuRF1−/− and B. MuRF1 Tg+ hearts compared to strain matched wildtype controls. Hearts were isolated and immediate perfused in the working mode at basal (50 mm Hg) and high (80 mm Hg) workloads. Values are expressed as mean ± SEM and represent 5–7 mice per group. Groups were analyzed by a One Way ANOVA vs. sibling match wildtype controls at baseline and high workload conditions. n.s.= not significant. MVO2=myocardial oxygen consumption. p<0.05, unless otherwise noted in the panel.
MuRF1 Tg+ cardiac mitochondria have a substrate-depended decreased flux through the Krebs cycle
Mitochondria from MuRF1−/− and MuRF1 cardiac-specific Tg+ mice were assayed at basal and maximal ADP-stimulated rates of mO2 supported by pyruvate, malate, glutamate, and succinate in permeabilized cardiac myofibers. While cardiac mitochondria from MuRF1−/− mice did not differ from sibling-matched controls, MuRF1 Tg+ cardiac mitochondria consumed significantly less O2 (Figure 4A). Importantly, no significant differences in the respiratory control ratio, an indicator of mitochondrial coupling, were identified between MuRF1−/−, MuRF1 Tg+, and their respective WT control mice. (Figure 4B). Basal and maximal cardiac mitochondrial ADP-stimulated mO2 consumption supported by palmitoyl-L-carnitine and malate did not differ in the MuRF1−/− and MuRF1 Tg+ and their respective WT controls (Figure 4C), suggesting that MuRF1 expression does not affect fatty acid oxidation at the level of the mitochondria. Previous studies have identified that an increased sensitivity of the permeability transition pore, regulating Ca2+ uptake in the presence of Pi, is related to the rate of electron flow through complex I in muscle [32]. Since we identified significantly decreased oxygen consumption in MuRF1 Tg+ mitochondria when using Complex I substrates (Figure 4A), we then challenged isolated mitochondria from MuRF1 Tg+ hearts with increasing exogenous Ca2+ to determine the mCa2+ retention capacity before the opening of the mPTP (Figure 4D). Mitochondria from MuRF1 Tg+ hearts demonstrated no differences in mCa2+ retention capacity (i.e. sensitivity or mPTP to Ca2+) when supported by pyruvate + malate or succinate compared to WT controls, indicating that MuRF1 does not play a role in the modulating cell death regulated by the mitochondrial permeability transition apparatus.
Figure 4. Increasing MuRF1 expression inhibits Krebs cycle flux in permeabilized cardiac muscle fibers.
A. Quantified rates of pyruvate/malate-supported O2 consumption (JO2) in permeabilized left ventricle myofibers from MuRF Tg+, MuRF −/−, and their respective WT controls. B. No differences in mitochondrial coupling were detected as indicated by RCR, represents the ratio of state 3 (phosphorylating) respiration to state 4 respiration. C. Quantified rates of palmitoyl-L-carnitine -supported O2 consumption in permeabilized left ventricle myofibers from MuRF Tg+, MuRF −/−, and their respective WT controls. D. Quantification of calcium retention capacity in isolated cardiac mitochondria from MuRF Tg+, MuRF −/−, and their respective WT controls. Data are expressed as mean ± SD and are representative of 3–5 mice per group. *p<0.05 by Student’s t-test vs. sibling-matched wildtype controls.
Mitochondria from MuRF1 Tg+ hearts generate significantly less ROS
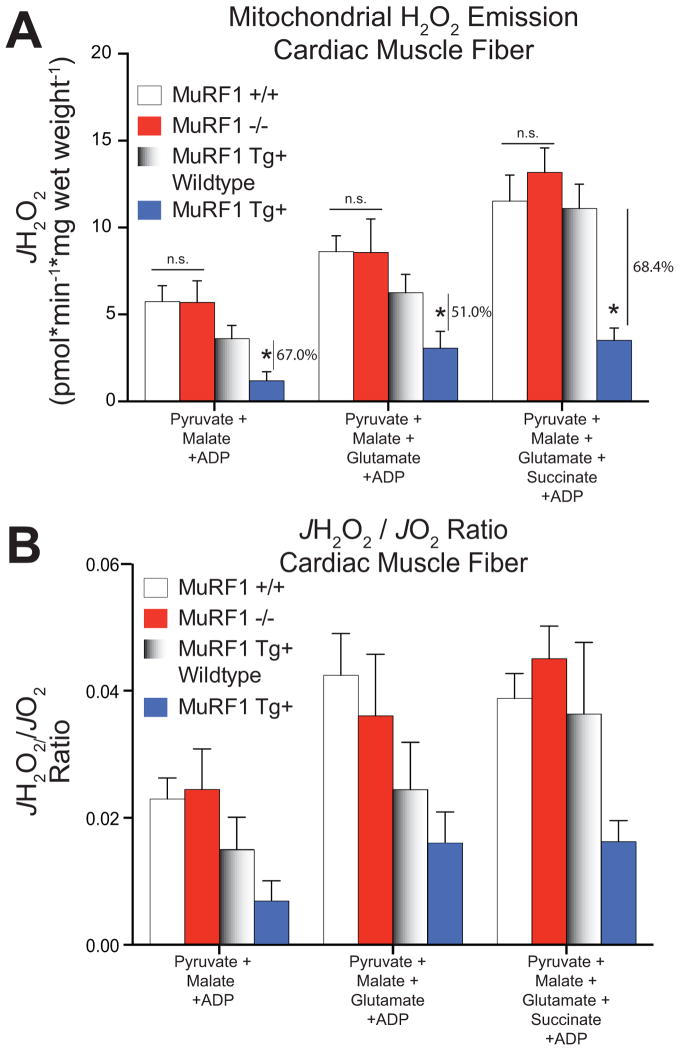
We next determined the rate of mH2O2 emission as a surrogate for reactive oxygen species formation from cardiac mitochondria using permeabilized myofibers in MuRF1−/− and MuRF1 Tg+ mice. While MuRF1−/− hearts did not differ from sibling-matched controls, surprisingly, MuRF1 Tg+ mitochondria supported by Pyruvate/Malate, Pyruvate/Malate/Glutamate, and Pyruvate/Malate/Glutamate/Succinate had a 67.0%, 51.0%, and 68.4% decrease in H2O2 emission compared to sibling-matched WT controls. This indicates a significant reduction in the mitochondria’s ability to form reactive oxygen species (ROS) in mice with increased cardiomyocyte MuRF1 expression (Figure 5A). When the JH2O2 is normalized to the oxygen consumption rate in the presence of Pyruvate/Malate, Pyruvate/Malate/Glutamate, and Pyruvate/Malate/Glutamate/Succinate (see Figure 4A), the MuRF1 Tg+ mitochondria JH2O2 (JH2O2/JO2, Figure 5B) is decreased, although no longer significantly less than the wildtype controls. Several potential sites of superoxide and H2O2 production have been reported in the Krebs cycle and electron transport chain in the mitochondria, including moieties on complex I [33] and complex III [34,35]. Recent studies on skeletal muscle mitochondria have found that complex II can generate superoxide or H2O2 [36], although complex II generation of significant ROS in vivo has mainly been demonstrated with mutations in the succinate dehydrogenase complex and flavoprotein subunit [37–39].
Figure 5. Mitochondrial H2O2 emission is inhibited in permeabilized cardiac muscle fibers from MuRF1 Tg+ hearts.
A. Quantified rates of mH2O2 emission (JH2O2) and B. the JH2O2/JO2 ratio from permeabilized muscle fibers obtained from MuRF Tg+, MuRF−/−, and their respective controls. Data are expressed as mean ± SD and are representative of 3–5 mice per group. *p<0.05 by Student’s t-test vs. sibling-matched wildtype controls.
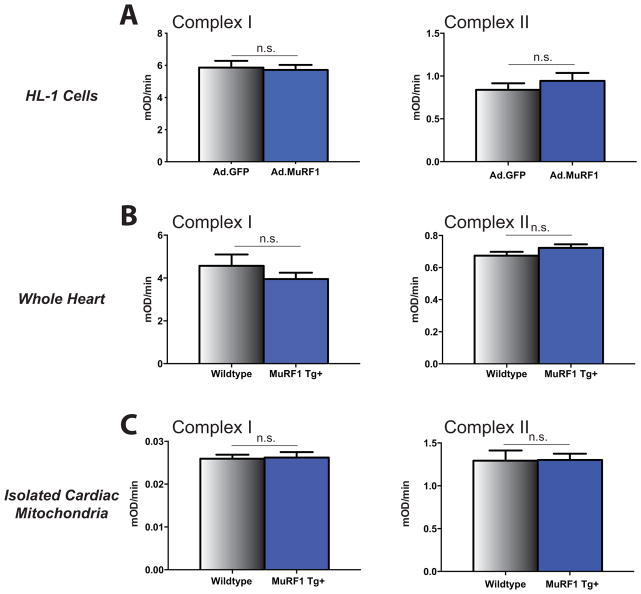
Complex I and II activity with increased MuRF1 expression
Using MuRF1 as bait, previous studies have identified that MuRF1 interacts with multiple substrates related to energy metabolism and mitochondria experimentally [5]. In complex I, NADH dehydrogenase (ubiquinone) and NADH-ubiquinone oxidoreductase were identified and in complex II Ubiquinol-cytochrome c reductase core protein I was found. This, in addition to the identification of reduced reactive oxygen species (ROS) production in the MuRF1 Tg+ mitochondria led us to investigate mitochondrial complex I and complex II activity in a cardiac-derived cell line, whole heart lysates, and isolated cardiac mitochondria. Despite having activity in mitochondria and binding to specific complex I and II substrates, no differences in complex I or complex II activity were identified in biochemical assays (Figure 6A). Similarly, using whole cardiac ventricular lysates from MuRF1 Tg+ hearts, we did not identify differences from WT control hearts in either complex I or complex II activity (Figure 6B). Lastly, we isolated mitochondria from MuRF1 Tg+ hearts and assayed complex I and complex II activity (Figure 6C). No changes from sibling WT control hearts were seen. Despite the previous identification of MuRF1 substrates in complex I and complex II (both implicated in ROS formation), increased MuRF1 expression did not affect complex I or complex II activity either in vitro or in vivo.
Figure 6. Mitochondrial Complex I/Complex II Activity with increased MuRF1 expression in HL-1 cells, MuRF1 Tg+ cardiac ventricular lysates and MuRF1 Tg+ isolated cardiac mitochondria.
Microplate activity assays measuring mitochondrial complex I activity (oxidation of NADH to NAD+) and complex II activity (reduction of ubiquinone to ubiquinol) in MuRF1 overexpressing HL-1 cells, MuRF1 Tg+ cardiac ventricular lysates and isolated cardiac mitochondria from MuRF1 Tg+ mice demonstrate no significant difference in complex I or complex II activity as a consequence of MuRF1 overexpression. (a) Mitochondrial complex I and complex II activity in total protein lysates isolated from HL-1 cells transduced with adenovirus expressing CMV GFP control or CMV GFP-tagged myc-MURF1. (b) Mitochondrial complex I and complex II activity in whole cardiac ventricular lysates from WT and MuRF1 Tg+ mice. (c) Mitochondrial complex I and complex II activity in mitochondria isolated from WT and MuRF1 Tg+ mouse hearts. n.s.= not significant by Student’s t-test vs. sibling-matched wildtype controls.
A consistent and critical finding in these studies was that mitochondrial function from MuRF1−/− hearts did not differ from WT hearts. We propose that this represents a redundancy of MuRF1 with other MuRF family proteins. Evidence for this redundancy of MuRF1 and MuRF2 in mitochondria may be seen in yeast two hybrid studies using p27 MuRF2 as bait (Table 1). We found that MuRF2 binds to muscle creatine kinase, aldolase A, NADH dehydrogenase (ubiquinone) 1a, and Aldo-keto reductase family 7 (Table 1), in addition to these redundant interacting proteins described earlier that MuRF1 interacts with [5], The redundancy in mitochondrial binding partners of MuRF1 and MuRF2 is consistent with our present studies, which did not detect any differences in MuRF1−/− cardiac mitochondrial function, but did detect differences when cardiac MuRF1 expression was increased. We believe this illustrates the redundancy that exists between MuRF1 and other MuRF family members, notably MuRF2, in the heart [40,41,5,7].
Table 1.
Comparison of new MuRF2 (p27) two-hybrid interacting proteins involved in energy metabolism compared to previously published MuRF1 interacting protein results.
| MuRF-2 Two-hybrid interaction results (previously unpublished) | MuRF-1 Two-hybrid interaction results* (Accession Code) |
|---|---|
| Glycolysis | |
| Aldolase A Glycogen phosphorylase Phosphofructokinase, muscle |
Aldolase A (BC000367) Pyruvate kinase (NM182471) |
| Krebs cycle | |
| Malate dehydrogenase, mitochondrial (Mor1) Succinate-CoA ligase, GDP-forming, alpha subunit (Thiokinase) Isocitrate dehydrogenase |
Pyruvate dehydrogenase (lipoamide) beta (BC000439) |
| Respiratory chain, mitochondrial | |
| NADH dehydrogenase (ubiquinone) | NADH dehydrogenase (ubiquinone) (NM005005) NADH-ubiquinone oxidoreductase (AF067186) Ubiquinol-cytochrome c reductase core protein 1 (BC009586) |
| Fatty acid synthesis | |
| Malonyl-CoA decarboxylase | No substrates in this category identified |
| Fatty acid oxydation | |
| Enoyl coenzyme A hydratase 1, peroxisomal | No substrates in this category identified |
| ATP regeneration | |
| Creatine kinase, muscle (CKM) | Creatine kinase, muscle (CKM) (NM001824) Adenylate kinase 1 (AK1) (NM000476) |
| Lactic Acid Cycle | |
| No substrates in this category identified | Pyruvate dehydrogenase kinase (NM002612) |
| NADH regeneration | |
| No substrates in this category identified | 3-Hydroxyisobutyrate dehydrogenase (BC032324) |
| Sarcomeric proteins | |
| Myosin light chain (MLC) | Myosin light chain-2 (BC031006) Telethonin (Tcap) (NM003673) Myotilin (NM006790) Titin (A-band region D-zone) (X90568) Nebulin (X83957) Nebulin-related anchoring protein (NM006175) Troponin I (L21715) Troponin T1 (BC010963) Troponin T3 (BT019997) |
Witt et a., J Mol Biol. 2005 Jul 22;350(4):713–22.
Initial studies yeast two hybrid assays using MuRF1 as bait identified binding a number of binding partners with cellular activities throughout the cell, including pathways involved in ATP/energy production [5]. These studies identified myofibrillar, substrates, in addition to substrates involved in energy metabolism, mitochondria, and 10 of unknown (undescribed) function [5]. However, the significance of MuRF1’s interactions with muscle creatine kinase, Aldolase A, and NADH dehydrogenase (ubiquinone) 1a, for example, were unknown [5]. Moreover, these initial studies demonstrated MuRF1’s redundant interactions with muscle ring finger-2 (MuRF2) with respect to multiple sarcomeric proteins, troponin I (TnI), troponin T (TnT), telethonin, titin, nebulin, myotilin, and myosin light chain 2 (MLC2) [5] (see Table 1). However, prior to this current study, the significance of both MuRF1’s regulation of mitochondrial function and its potential redundancy had been proven. Together, the lack of differences in MuRF1−/− heart mitochondrial function and changes in MuRF1 Tg+ heart mitochondrial flux and ROS production are consistent with MuRF1’s redundant regulation of cardiac mitochondrial function.
Discussion
MuRF1 has previously been shown to be a muscle-specific ubiquitin ligase involved in muscle mass regulation presumably through it’s ability to target troponin I [6] and myosin heavy chain [7] for proteasome-dependent degradation. However, recent studies have extended MuRF1’s regulation of sarcomere degradation to include the regulation of signal transduction, through interactions with cytoplasmic (cJun) [16] and nuclear transcription factors (e.g. SRF and E2F1) [17,40] that are regulated by both proteasome dependent and proteasome-independent (non-degradative) mechanisms, respectively. With MuRF1’s expanding role in signaling transduction and recently metabolism through its regulation of creatine kinase activity [8,42], we investigated MuRF1’s role in cardiac mitochondrial function in vivo. We found significant decreases in mitochondrial flux and ROS production in MuRF1 Tg+ hearts, but not MuRF1−/− hearts, illustrating MuRF1’s potential redundant regulation of mitochondrial function.
While ubiquitin ligases have rarely been reported in mitochondria, proteomic studies suggest that ubiquitin ligase activity in the heart is abundant. One recent proteomics study of the heart identified that most ubiquitinated proteins are found in the mitochondria [9]. Specifically, of all the ubiquitinated proteins found in the heart, 38% of all the ubiquitinated proteins identified were from mitochondria, contrast to the 27.3% from the cytosolic compartment, 13.2% in the Golgi/SR/Vesicle, 9.1% in the nucleus (the remaining 12.4% are of an unknown cellular localization) [9]. Previously, only two ubiquitin ligases have been described in mitochondria, including MARCH5 and MULAN/MAPL [15]. These ubiquitin ligases, incidentally identified in the heart as well, have been shown to regulate mitochondrial dynamics in non-cardiac cells [43,10,44,45,12,46], as recently reviewed [15]. Consistent with MuRF1’s role in mitochondria, most MuRF1 protein expression and activity determined by auto-ubiquitination in cardiomyocytes, could be found in the mitochondrial fraction of striated muscle cells, including cardiac-derived muscle cells transduced with MuRF1 (Figure 1). While MuRF1 was present in the cytosolic fraction, most ubiquitin ligase activity (auto-ubiquitination) could be seen in the mitochondrial fractions in the cardiac-derived cells (Figure 1B). These findings are consistent with both MuRF1’s previous interactions with mitochondrial proteins [5], and our present findings that MuRF1 regulates cardiomyocyte flux and ROS in MuRF1 Tg+ hearts. This makes MuRF1 the heart-specific ubiquitin ligase to be described in the mitochondria, as well as its functional consequences.
A key finding in this study is the remarkable reduction in reactive oxygen species production in the MuRF1 cardiomyocyte-specific transgenic muscle fibers (Figure 1). ROS production is a substantial cause of injury after an ischemia/reperfusion event [47]; thus, MuRF1’s reduction in ROS production would be predicted to be cardioprotective. One possible way that MuRF1 may be doing this is through its proposed association with complex I through NADH dehydrogenase, identified previously in yeast two hybrid studies as a MuRF1 interacting protein [5]. The significance of this association on electron transport chain complex I and II function was investigated in the current study. Complex I serves as a major source of ROS in the mitochondria. Therefore, we speculated that increased MuRF1 expression may target complex I for degradation, decreasing complex I dysfunction by increased turnover, and; therefore, decrease ROS production. However, in vitro and in vivo biochemical analysis revealed no functional difference (Figure 5). This implies that a reduction in Krebs cycle flux could be the mechanism of decreased respiration in MuRF1 transgenic mice.
How MuRF1 may be targeted to the mitochondria is unknown. MuRF1 has been reported in the cytosol, nucleus, and sarcomere; it does not contain an N-terminal mitochondrial targeting sequence that would transport it to the mitochondrial matrix (e.g. H2N-Met-Leu-Ser-Leu-Arg-Gln-Ser-Ile-Arg-Phe-Phe-Lys-Pro-Ala-Thr-Arg-Thr-Leu-Cys-Ser-Ser-Arg-Tyr-Leu-Leu-) [6,48,38]. Nor is MuRF1 strongly predicted to localize to mitochondria itself based on properties of proteins targeted to the nucleus (Mitoprot II, v1.101, probability of export to mitochondria 0.0013–0.0018) [49]. This does not preclude that MuRF1 is not found in the mitochondria. For example, the ubiquitin ligase MULAN is targeted to the mitochondria but does not contain an N-terminal mitochondrial targeting sequence. Interestingly, MULAN is targeted to the mitochondria by two novel transmembrane domains just identified (named TMD1 and TMD2) that are necessary for MULAN to translocate to the mitochondria [12]. The other ubiquitin ligase found to associate with mitochondria, Parkin, mediate the process of “mitophagy”. Parkin is directed to damaged mitochondria, where it interacts with phosphorylated PINK1 (with its own mitochondrial targeting sequence); it subsequently ubiquitinates PINK1 to direct downstream mitochondrial autophagy [50,51]. Both the MULAN and Parkin examples illustrate mechanisms by which ubiquitin ligases are targeted to the mitochondria independent of N-terminal mitochondrial targeting sequences. Given the paucity of ubiquitin ligases (zero) identified in the MitoCarta inventory of mammalian mitochondrial genes [52] and the fact that most ubiquitinated proteins in the heart are of mitochondrial origin (38%)[9], it would not be surprising that ubiquitin ligases end up in the mitochondria by novel mechanisms yet to be identified, which differ from traditionally mitochondrial targeting sequences. Given MuRF1’s wide array of substrate specificities, including as key regulators of metabolism [8,53,54] and mitochondrial oxidative phosphorylation (NADH dehydrogenase, aka ubiquinone, NADH ubiquinone oxidoreductase, and 3-hydroxyisobutyrate dehydrogenase), including the mitochondrial ATP synthase beta-subunit involved in the regeneration of ATP [5], MuRF1’s interaction of proteins targeted to the mitochondria is possible and warrants further investigation. Taken in context of the current studies, the evidence presented that MuRF1 activity is found in mitochondrial fractions is not conclusive evidence of MuRF1’s localization. Further studies are needed to elucidate MuRF1’s localization to the mitochondria and the mechanisms involved in light of the novel mechanisms found in the other two ubiquitin ligases identified in mitochondria to date.
These studies identify the first ubiquitin ligase found in cardiomyocytes that are found in the mitochondria and regulates flux through the Krebs cycle while also reducing ROS generation. Interestingly, increased expression of MuRF1, but not deletion of MuRF1, results in changes in ROS generation compared to WT controls. Since these effects are not seen in the MuRF1−/− hearts, we hypothesized that MuRF1 and MuRF2 redundantly regulated mitochondrial function. Consistent with this, yeast two hybrid using MuRF2 as bait identified interacting mitochondrial proteins redundant with previous MuRF1 yeast two hybrid substrates (Table 1). By reducing cardiomyocyte mitochondrial ROS production capacity, increased MuRF1 expression may be an additional mechanism by which cardiac MuRF1 is cardioprotective against ischemia/reperfusion injury both in vivo and ex vivo by its regulation of complex I and II of the electron transport chain activity, the primary sources of cardiac ROS.
Supplementary Material
Normalized oxygen consumption and heart rate of MuRF1−/− and MuRF1 Tg+ hearts compared to strain matched wildtype controls. Hearts were isolated and immediate perfused in the working mode at basal and high workloads. Values are expressed as mean ± SEM and represent 5–7 mice per group. Groups were analyzed by a One Way ANOVA vs. sibling match wildtype controls at baseline and high workload conditions. n.s.= not significant. p<0.05, unless otherwise noted in the panel.
Acknowledgments
The authors wish to thank Dr. William Claycomb for the generous gift of the HL-1 cells used in these studies. This work is supported by the National Institutes of Health (R01HL104129 to M.W.), the Fondation Leducq (to M.W.), and the Jefferson-Pilot Fellowship in Academic Medicine (to M.W.).
Non-standard abbreviations
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- α-MHC
α-myosin heavy chain
- JNK
c-Jun N-terminal protein kinases
- mCa2+
mitochondrial Ca2+
- mH2O2
mitochondrial H2O2
- MITOL/MARCHV
membrane-associated ring finger 5
- mPTP
mitochondrial permeability transition pore
- ROS
reactive oxygen species
- MULAN
mitochondrial ubiquitin ligase activator of NF-κB
- MuRF1
Muscle Ring Finger-1
- MuRF2
Muscle Ring Finger 2
- KO
knockout
- MuRF3
muscle ring finger 3
- NADH
reduced form of nicotinamide adenine dinucleotide
- PDH
pyruvate dehydrogenase complex
- SERCA
sarcoplasmic/endoplasmic reticulum Ca2+-ATPase
- Tg
transgenic
- WT
wildtype
- mO2
mitochondrial O2
Footnotes
Disclosures
There are no conflicts of interest to disclose.
References
- 1.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. 1065874 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296(4):H997–H1006. doi: 10.1152/ajpheart.00660.2008. 00660.2008 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] J Mol Biol. 2001;306(4):717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 4.Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, et al. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167(6):1147–1159. doi: 10.1083/jcb.200402033. jcb.200402033 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol. 2005;350(4):713–722. doi: 10.1016/j.jmb.2005.05.021. S0022-2836(05)00552-8 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] Proc Natl Acad Sci U S A. 2004;101(52):18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117(9):2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willis MS, Schisler JC, Li L, Rodriguez JE, Hilliard EG, Charles PC, et al. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ Res. 2009;105(1):80–88. doi: 10.1161/CIRCRESAHA.109.194928. CIRCRESAHA.109.194928 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, et al. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem Biophys Res Commun. 2007;357(3):731–736. doi: 10.1016/j.bbrc.2007.04.015. S0006-291X(07)00706-1 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178(1):71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonashiro R, Kimijima Y, Shimura T, Kawaguchi K, Fukuda T, Inatome R, et al. Mitochondrial ubiquitin ligase MITOL blocks S-nitrosylated MAP1B-light chain 1-mediated mitochondrial dysfunction and neuronal cell death. [Research Support, Non-U.S. Gov’t] Proc Natl Acad Sci U S A. 2012;109(7):2382–2387. doi: 10.1073/pnas.1114985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3(1):e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang F, Wang B, Li N, Wu Y, Jia J, Suo T, et al. RNF185, a Novel Mitochondrial Ubiquitin E3 Ligase, Regulates Autophagy through Interaction with BNIP1. PLoS One. 2011;6(9):e24367. doi: 10.1371/journal.pone.0024367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livnat-Levanon N, Glickman MH. Ubiquitin-proteasome system and mitochondria - reciprocity. [Research Support, Non-U.S. Gov’t Review] Biochim Biophys Acta. 2011;1809(2):80–87. doi: 10.1016/j.bbagrm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Zungu M, Schisler J, Willis MS. All the little pieces. -Regulation of mitochondrial fusion and fission by ubiquitin and small ubiquitin-like modifer and their potential relevance in the heart. [Research Support, N.I.H., Extramural Review] Circ J. 2011;75(11):2513–2521. doi: 10.1253/circj.cj-11-0967. [DOI] [PubMed] [Google Scholar]
- 16.Li HH, Du J, Fan YN, Zhang ML, Liu DP, Li L, et al. The Ubiquitin Ligase MuRF1 Protects Against Cardiac Ischemia/Reperfusion Injury by Its Proteasome-Dependent Degradation of Phospho-c-Jun. Am J Pathol. 2011;178(3):1043–1058. doi: 10.1016/j.ajpath.2010.11.049. S0002-9440(10)00174-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100(4):456–459. doi: 10.1161/01.RES.0000259559.48597.32. 01.RES.0000259559.48597.32 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saks VA, Veksler VI, Kuznetsov AV, Kay L, Sikk P, Tiivel T, et al. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. [Research Support, Non-U.S. Gov’t] Mol Cell Biochem. 1998;184(1–2):81–100. [PubMed] [Google Scholar]
- 19.Anderson EJ, Yamazaki H, Neufer PD. Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. [Comparative Study] J Biol Chem. 2007;282(43):31257–31266. doi: 10.1074/jbc.M706129200. [DOI] [PubMed] [Google Scholar]
- 20.Anderson EJ, Thayne K, Harris M, Carraway K, Shaikh SR. Aldehyde stress and up-regulation of Nrf2-mediated antioxidant systems accompany functional adaptations in cardiac mitochondria from mice fed n-3 polyunsaturated fatty acids. [Research Support, N.I.H., Extramural] Biochem J. 2012;441(1):359–366. doi: 10.1042/BJ20110626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294(2):H1036–1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- 22.Sloan RC, Moukdar F, Frasier CR, Patel HD, Bostian PA, Lust RM, et al. Mitochondrial permeability transition in the diabetic heart: contributions of thiol redox state and mitochondrial calcium to augmented reperfusion injury. [In Vitro Research Support, Non-U.S. Gov’t] J Mol Cell Cardiol. 2012;52(5):1009–1018. doi: 10.1016/j.yjmcc.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Kamimura M, Matsumoto K, Koshiba-Takeuchi K, Ogura T. Vertebrate crossveinless 2 is secreted and acts as an extracellular modulator of the BMP signaling cascade. [Research Support, Non-U.S. Gov’t] Dev Dyn. 2004;230(3):434–445. doi: 10.1002/dvdy.20069. [DOI] [PubMed] [Google Scholar]
- 24.Anderson EJ, Rodriguez E, Anderson CA, Thayne K, Chitwood WR, Kypson AP. Increased propensity for cell death in diabetic human heart is mediated by mitochondrial-dependent pathways. [Research Support, N.I.H., Extramural] Am J Physiol Heart Circ Physiol. 2011;300(1):H118–124. doi: 10.1152/ajpheart.00932.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsien RY. Fluorescent indicators of ion concentrations. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. Review] Methods Cell Biol. 1989;30:127–156. doi: 10.1016/s0091-679x(08)60978-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, et al. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. [Comparative Study Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Proteomics. 2008;8(8):1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. [Research Support, Non-U.S. Gov’t] J Biol Chem. 1990;265(13):7248–7256. [PubMed] [Google Scholar]
- 28.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimes BW, Brandt BL. Properties of a clonal muscle cell line from rat heart. Exp Cell Res. 1976;98(2):367–381. doi: 10.1016/0014-4827(76)90447-x. [DOI] [PubMed] [Google Scholar]
- 30.Willis MS, Li HH, Rodriguez JE, Li L, Rojas M, Lockyer P, et al. MuRF1 Inhibits JNK Signaling in Cardiac Ischemia Reperfusion injury by Degrading Phosphorylated cJun. FASEB J. 2008;22:751–711. [Google Scholar]
- 31.Wadosky KM, Rodriguez JE, Hite RL, Min JN, Walton B, Willis MS. Muscle RING Finger-1 Attenuates IGF-1-dependent cardiomyocyte hypertrophy by Inhibiting JNK Signaling. Am J Physiol Endocrinol Metab. 2014 doi: 10.1152/ajpendo.00326.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontaine E, Eriksson O, Ichas F, Bernardi P. Regulation of the permeability transition pore in skeletal muscle mitochondria. Modulation By electron flow through the respiratory chain complex i. J Biol Chem. 1998;273(20):12662–12668. doi: 10.1074/jbc.273.20.12662. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem. 2002;80(5):780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- 34.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191(2):421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cadenas E, Boveris A, Ragan CI, Stoppani AO. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys. 1977;180(2):248–257. doi: 10.1016/0003-9861(77)90035-2. [DOI] [PubMed] [Google Scholar]
- 36.Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem. 2012;287(32):27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Mol Cell Biol. 2008;28(2):718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Yu L, Yu CA. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] J Biol Chem. 1998;273(51):33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- 39.Agarwala GC. Effect of intracisternal injection of ACTH on blood glucose and hepatic glycogen in dogs. Indian J Physiol Pharmacol. 1979;23(4):297–304. [PubMed] [Google Scholar]
- 40.Willis MS, Wadosky KM, Rodriguez JE, Schisler JC, Lockyer P, Hilliard EG, et al. Muscle ring finger 1 and muscle ring finger 2 are necessary but functionally redundant during developmental cardiac growth and regulate E2F1-mediated gene expression in vivo. Cell Biochem Funct. 2013 doi: 10.1002/cbf.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27(2):350–360. doi: 10.1038/sj.emboj.7601952. 7601952 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willis MS, Zungu M, Patterson C. Cardiac muscle ring finger-1--friend or foe? Trends Cardiovasc Med. 2010;20(1):12–16. doi: 10.1016/j.tcm.2010.03.001. S1050-1738(10)00032-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21(3):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7(10):1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25(15):3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18(2):102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 47.Paradies G, Petrosillo G, Pistolese M, Di Venosa N, Federici A, Ruggiero FM. Decrease in mitochondrial complex I activity in ischemic/reperfused rat heart: involvement of reactive oxygen species and cardiolipin. Circ Res. 2004;94(1):53–59. doi: 10.1161/01.RES.0000109416.56608.64. [DOI] [PubMed] [Google Scholar]
- 48.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] J Cell Biol. 2002;157(1):125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. [Comparative Study Research Support, Non-U.S. Gov’t] Eur J Biochem. 1996;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. [Research Support, N.I.H., Extramural] Science. 2013;340(6131):471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. [Research Support, N.I.H., Intramural Review] Antioxid Redox Signal. 2011;14(10):1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao TJ, Yan YB, Liu Y, Zhou HM. The generation of the oxidized form of creatine kinase is a negative regulation on muscle creatine kinase. J Biol Chem. 2007;282(16):12022–12029. doi: 10.1074/jbc.M610363200. M610363200 [pii] [DOI] [PubMed] [Google Scholar]
- 54.Koyama S, Hata S, Witt CC, Ono Y, Lerche S, Ojima K, et al. Muscle RING-finger protein-1 (MuRF1) as a connector of muscle energy metabolism and protein synthesis. J Mol Biol. 2008;376(5):1224–1236. doi: 10.1016/j.jmb.2007.11.049. S0022-2836(07)01533-1 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalized oxygen consumption and heart rate of MuRF1−/− and MuRF1 Tg+ hearts compared to strain matched wildtype controls. Hearts were isolated and immediate perfused in the working mode at basal and high workloads. Values are expressed as mean ± SEM and represent 5–7 mice per group. Groups were analyzed by a One Way ANOVA vs. sibling match wildtype controls at baseline and high workload conditions. n.s.= not significant. p<0.05, unless otherwise noted in the panel.