Summary
Bacteroides fragilis causes the majority of anaerobic infections in humans. The presence of a pathogenicity island in the genome discriminates pathogenic and commensal B. fragilis strains. The island encodes metalloproteinase II (MPII), a potential virulence protein, and one of three homologous fragilysin isozymes (FRA; also termed B. fragilis toxin or BFT). Here, we report biochemical data on the structural-functional characteristics of the B. fragilis pathogenicity island proteases by reporting the crystal structure of MPII at 2.13 Å resolution combined with detailed characterization of the cleavage preferences of MPII and FRA3 (as a representative of the FRA isoforms) identified using a high-throughput peptide cleavage assay with 18,583 substrate peptides. We suggest that the evolution of the MPII catalytic domain can be traced to human and archaebacterial proteinases, while the prodomain fold is a feature specific to MPII and FRA. We conclude that the catalytic domain of both MPII and FRA3 evolved differently relative to the prodomain, and that the prodomain evolved specifically to fit the B. fragilis pathogenicity. Overall, our data provide insights into the evolution of cleavage specificity and activation mechanisms in the virulent metalloproteinases.
Introduction
The gram-negative, anaerobic Bacteroides is one of the most prominent genera of the human microbiome. Commensal B. fragilis strains are critical to systemic and mucosal immunity and host nutrition [1]. However, pathogenic B. fragilis strains cause over 80% of anaerobic infections [2]. The presence of the 6-kb pathogenicity “islet” discriminates enterotoxigenic from commensal B. fragilis strains. There is a consensus that secretory metalloproteinases are virulence factors in B. fragilis [3]. In B. fragilis, a single pathogenicity island contains two distinct metalloproteinase genes coding for MPII and FRA (also called B. fragilis toxin or BFT) [4–6]. FRAs exist in three homologous isoforms (FRA1, 2 and 3) with sequence identities of over 95%, while the sequence identity between FRAs and MPII is ~25%. Both FRA and MPII are secretory metalloproteinases with a zinc-binding HEXXHXXGXXH motif and a characteristic Met-turn [7].
Currently, there is valuable structural and functional information for FRAs [6, 8–11]. The crystal structure of FRA3 was recently reported [12]. The FRA3 catalytic domain has a classic “metzincin” fold typical of eukaryotic matrix metalloproteases (MMPs) [7, 12], but the large N-terminal prodomain of FRA3 is unrelated to any known folds. The only in vivo demonstrated substrate of FRAs is E-cadherin, a key component of cell-cell contacts [13]. By cleaving E-cadherin, FRAs (either alone or in collaboration with other proteinases) ultimately weaken cell-to-cell contacts, enabling B. fragilis to penetrate the intestinal epithelium, where the bacterium’s polysaccharide capsule causes abscesses and inflammation within the tissue. The structural-functional characteristics of MPII, on the other hand, are barely known [14]. This information would aid in the identification of cleavage targets, thus helping to decipher the molecular effects of MPII and FRA proteolysis on host cells. Furthermore, this knowledge will help to establish how B. fragilis infection sets off multiple gastrointestinal pathologies and increases cancer risk, and how to design a therapeutic means to fight the disease.
This study provides biochemical data on the structural-functional characteristics of the B. fragilis pathogenicity island proteases by reporting the crystal structure of MPII at 2.13 Å resolution combined with detailed characterization of the cleavage preferences of MPII and FRA3.
Results
Crystal structure of MPII
The crystal structure of FRA3 was recently reported [12]. Here, we solved the crystal structure of the catalytically inactive E352A MPII zymogen mutant. In the mutant, Ala substituted for the catalytically essential Glu-352 active site residue. To facilitate protein crystallization, we purified the catalytically inactive MPII-E352A-T construct with the N-terminal Hisx6 tag followed by the thrombin cleavage sequence. The N-terminal tag was then cleaved by thrombin proteolysis. The mutant MPII-E352A protein was separated from the tag using size-exclusion chromatography (Figure 1).
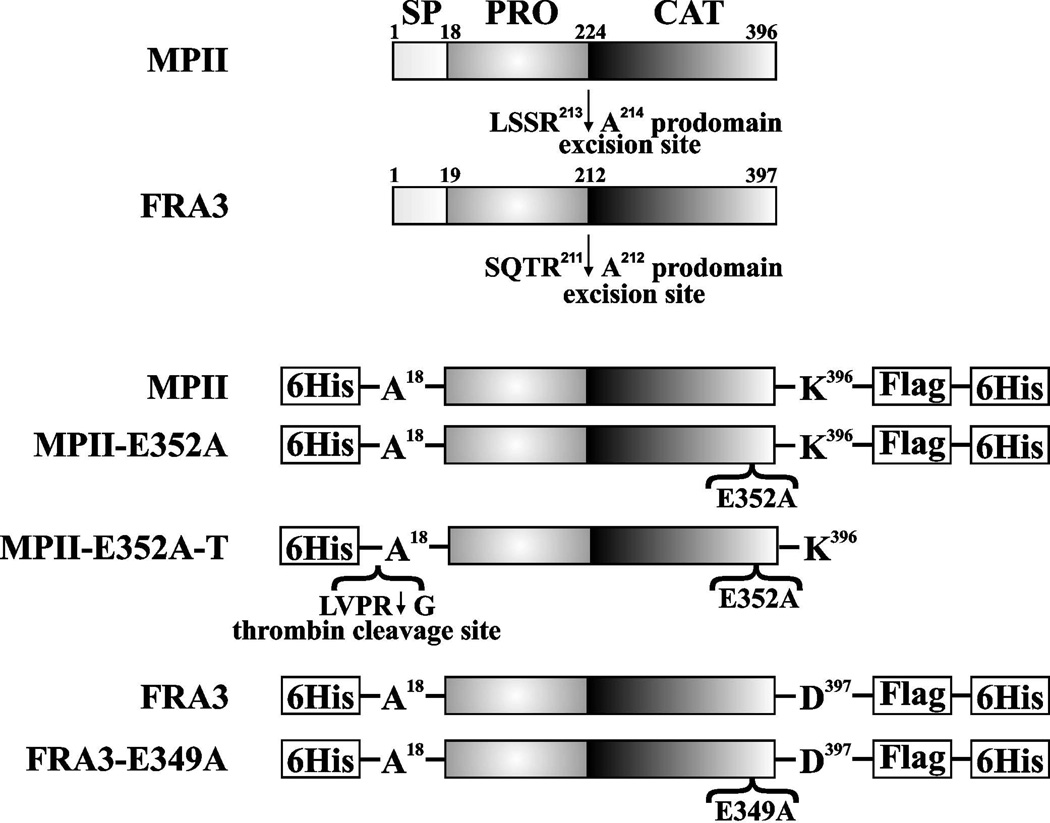
Figure 1. MPII and FRA3 constructs.
Top, wild-type MPII and FRA3. SP, PRO and CAT indicate the signal peptide, prodomain, and catalytic domain, respectively. The arrows indicate the cleavages at the prodomain excision sites. These cleavages release the prodomain and the mature protease. Bottom, the recombinant MPII and FRA3 constructs were tagged with the Hisx6 and FLAG tags. MPII-E352A and FRA3-E349A, the catalytically inactive mutants of MPII and FRA3, respectively. MPII-E352-T, the catalytically inert construct with the N-terminal Hisx6 tag followed by the thrombin cleavage sequence, was used in our crystallization studies.
The structure of MPII was solved using the data collected at 2.13 Å on the SYBYLS beamline at the Advanced Light Source, Berkeley. The data analysis revealed highly similar spatial structures of MPII and FRA3 despite relatively low (~25%) sequence identity between the two proteinases (Figures 2 and 3). The core 213 Cα atoms in the full-length proenzymes of MPII and FRA3 align with RMSD = 1.35 Å. The MPII zymogen consists of the N-terminal 32–184 prodomain and the C-terminal 217–396 catalytic domain connected by a 185–216 linker. The 186–195 residues of the linker run in the active site in a reverse orientation relative to the peptide substrate. This orientation of the linker prevents its cleavage by the preformed protease active site. The disordered 198–213 linker region had insufficient electron density for structural determination. Low electron density was also observed in the corresponding region of the FRA3 crystals [12]. Because both mature MPII and FRA3 commence at the N-terminal Ala-212, it is likely that the disordered linker state is a prerequisite for its efficient cleavage by an additional proteinase leading to the removal of the inhibitory prodomain. There were three additional regions in MPII with limited electron density (Figure 2). These regions include the G17ACADDLLHVEETAS31 sequence (in which Gly-Ala18 was the C-terminal portion of the thrombin cleavage site and Ala-18 was the N-terminus of the prodomain), and the disordered loops between α2 and β9, and β9 and β10 in the prodomain.
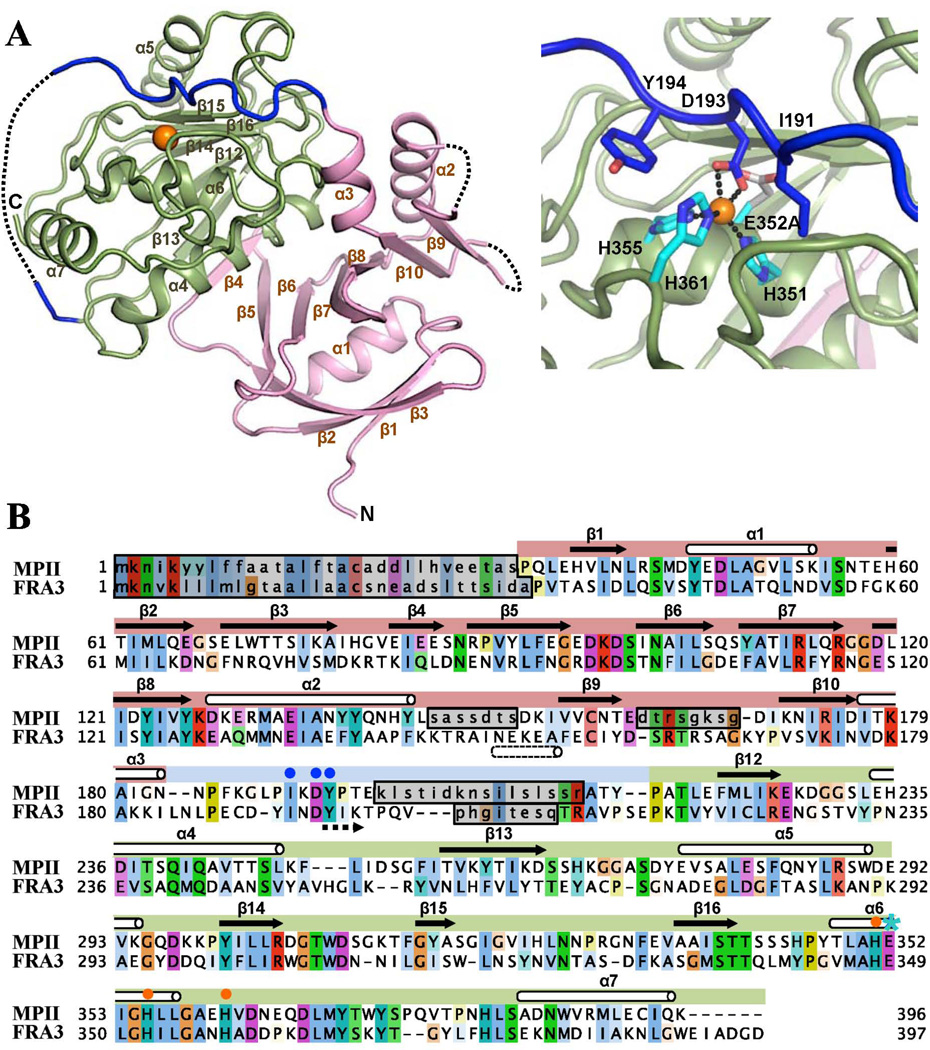
Figure 2. Structural organization of MPII.
(A) Right and left, cartoon presentation of MPII and its active site, respectively. The catalytic domain and the prodomain are colored green and magenta, respectively. The connecting linker is shown in dark blue and the Zn2+ ion as an orange ball. Regions with the missing electron density are shown as dotted lines. In the right panel active site histidines and the mutant E352A residue are shown as light blue sticks, and Ile-191, Asp-193 and Tyr-194, which discriminate MPII and FRA3, are pictured as dark blue sticks. The secondary structure elements are numbered as in FRA3 (PDB 3P24). (B) Sequence alignment of MPII and FRA3. Regions with the missing electron density are marked by grey boxes. The mutant E352A residue is marked by a blue star, the active site histidines by orange dots, and Ile-191, Asp-193 and Tyr-194 by blue dots. Residues are colored according to the Clustal X color scheme. Dotted arrow indicates antiparallel orientation of the prodomain relative to substrate.
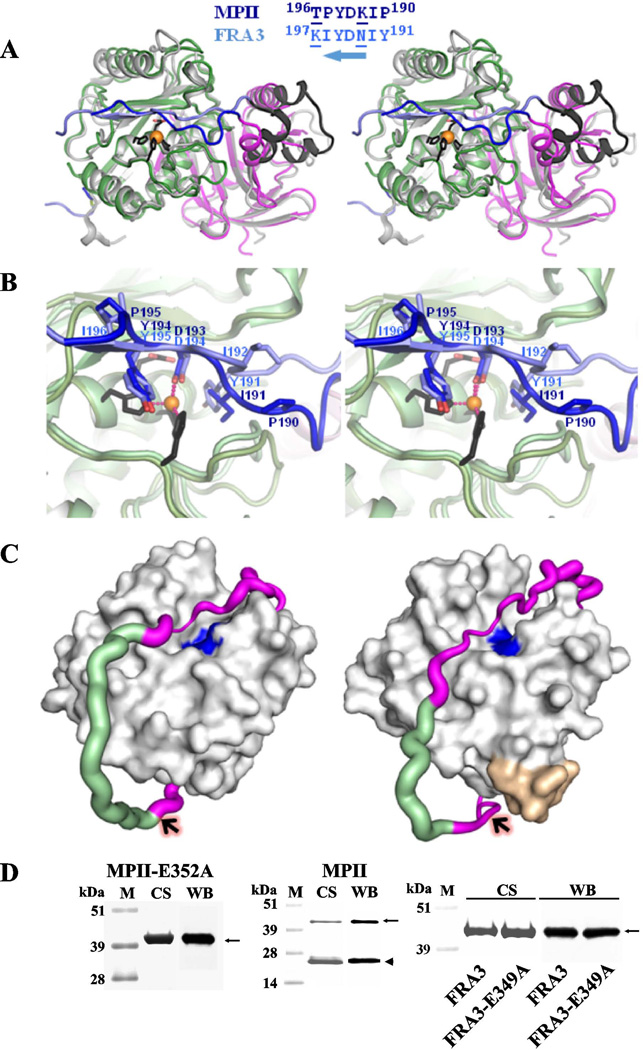
Figure 3. Structural differences between MPII and FRA3.
(A) Wall-eyed stereo view of the superimposed FRA3 and MPII structures. FRA3 is shown in grey and black. The prodomain and the catalytic domain of MPII are magenta and green, respectively, and the interdomain linker is blue. The C-terminal portion of the FRA3 prodomain is black. The active site Zn2+ ion is shown as an orange ball and the three histidines as black sticks. The alignment of the C-terminal linker is shown on the top. Substitutions of Tyr191, Asn193 and Ile196 (in FRA3) for Pro190, Lys192 and Pro195 (in MPII), respectively, result in a different fold of this linker region. Solvent-exposed residues are underlined. An arrow indicates that the prodomain runs in the antiparallel orientation in the active site relative to the substrate. (B) Wall-eyed stereo view of the active site in MPII and FRA3. The color scheme is as in A. (C) The linker is stabilized by the C-end segment of the catalytic domain in FRA3 (right) but not in MPII (left). The catalytic domain is represented as a grey surface. The active site is shown in blue. The C-terminal Glu-Ile-Ala-Asp-Gly-Asp397residue buckle (yellow) is present in FRA3, but not in MPII. The linker is shown as a magenta/green tube the width of which is proportional to the B-factor (a measure of mobility of a particular part of a protein) that varied from 20 to 80 A2. The residues with the missing electron density are modeled and colored in green. Arrows indicate the scissile bond between the linker and the catalytic domain in MPII and FRA3. (D) SDS-electrophoresis of the purified constructs. M, molecular weight markers. CS and WB, Coomassie staining and Western blotting with an anti-FLAG tag antibody, respectively. Solid arrows and arrowheads point to the proenzyme and active enzyme, respectively.
The prodomain structure and self-activation of MPII and FRA3
The MPII prodomain exhibits an unconventional fold that is similar to that of FRA3. This fold is a unique feature of the pro-metalloproteinases encoded by the pathogenicity island of B. fragilis [12]. We did not identify any homologues of this prodomain in an extensive search using the available sequence and structural homologies software. The level of sequence and structural homology between the prodomains of MPII and FRA3 is higher than between their catalytic domains. Thus, the RMSD value of the MPII versus FRA3 individual prodomain core (~80% Cα atoms) is 0.93 Å.
The crystal structure of MPII suggests that the prodomain includes the mixed parallel β1 and β2 strands linked by an α1 helix and followed by antiparallel β3 strand. The latter is followed by strand β4 that is a part of the domain core, which further consists of a large twisted antiparallel β-sheet (strands β5–β10). A long α2 helix connects strands β8 and β9, while strand β10 is followed by the C-terminal helix α3. The post-α3 linker region adopts an extended conformation, runs alongside the catalytic groove and directly interacts with the catalytic domain active site, thereby, maintaining the proenzyme state of the MPII zymogen (Figures 2 and 3).
There is, however, a significant difference in the orientation of the α3 helix between MPII and FRA3 (Figure 3A, B). In FRA3, the α3 helix directly binds the loop connecting the α2 helix with the β9 β-strand. There is a 30° rotation of α3 helix in MPII relative to FRA3. Because of this α3 rotation, there is no direct contact of α3 with the loop region between α2 and β9 and, as a result, this loop is disordered in MPII. Furthermore, because the C-terminus of the α3 helix is linked to the 185–216 interdomain linker, this helix rotation significantly changes the conformation of the linker in MPII versus FRA3. Thus, the 186–191 N-terminal sequence of the MPII linker moves away from the β15 strand, which directly binds the corresponding linker region in FRA3. However, in MPII the downstream Lys-Asp-Tyr 192–194 residues assume the positions of Asn-Asp-Tyr 193–195 observed in FRA3. The Asp-193 side chain of the MPII prodomain (Asp-194 of FRA3) interacts directly with the active site zinc and prevents its access to the substrate. In FRA3, the downstream Ile-Lys-Thr residues run as a parallel β-hairpin with β15 of the catalytic domain enhancing the prodomain binding to the active site. This fold allows Tyr-191 (Ile-191 in MPII) of the FRA3 prodomain to occupy the S1’ sub-site proximal to the catalytic Zn ion.
In turn, the presence of Pro-190 and Pro-195 in MPII abrogates several hydrogen bonds that associate the linker with β15, moves Ile-191 away from the substrate sub-site, and weakens the association of the linker with the active site. Additionally, in FRA3 the linker is stabilized by the C-terminal Glu-Ile-Ala-Asp-Gly-Asp397residue fragment. This C-terminal fragment is absent in the MPII peptide sequence (Figure 3C). The Arg-Ala scissile bond between the prodomain linker and the N-end of the catalytic domain better matches the cleavage preferences of MPII rather than FRA3 (see below “Multiplex peptide cleavage assay”). These differences, especially when combined, explain a looser association of the prodomain with the catalytic domain in MPII relative to FRA3 and provide structural requirements for the efficient self-activation of the MPII proenzyme [14]. Thus, the recombinant MPII proenzyme self-activates in the course of its purification from E. coli cells. In contrast, the rate of intermolecular self-activation of the recombinant FRA3 proenzyme is limited and the proenzyme only partially converts to the mature FRA3 protease during storage (Figure 3D).
The catalytic domain structure
The overall structure model of MPII shows the classical metzincin architecture with the “upper N-terminal” and lower C-terminal” sub-domains with respect to the active site cleft and with the catalytic zinc ion located at the bottom of the cleft. The N-terminal sub-domain features a five-stranded β-sheet, in which β15 is antiparallel to the roughly parallel β12, β13, β14 and β16 β-strands, and two α-helices, α4 and α5. The active site α6 helix of the lower C-terminal sub-domain exhibits the first two zinc-binding histidines, His351 and His355, while the third zinc-binding histidine, His361, followed by the typical methionine (Met)-turn (Asp-Leu-Met369-Tyr), is localized in the extended conformation region downstream (Figure 2).
There are multiple homologues of the MPII catalytic domain fold, including the proteinases of archaebacteria and bacteria and, especially, MMP and ADAM metzincins (Figure 4). There is an unconventional α5 helix in the upper sub-domain. This helix was recorded only in the ADAM sub-family of the metzincins and in the achaebacterial metalloproteinase (Methanopyrus kandleri archaemetzincin).
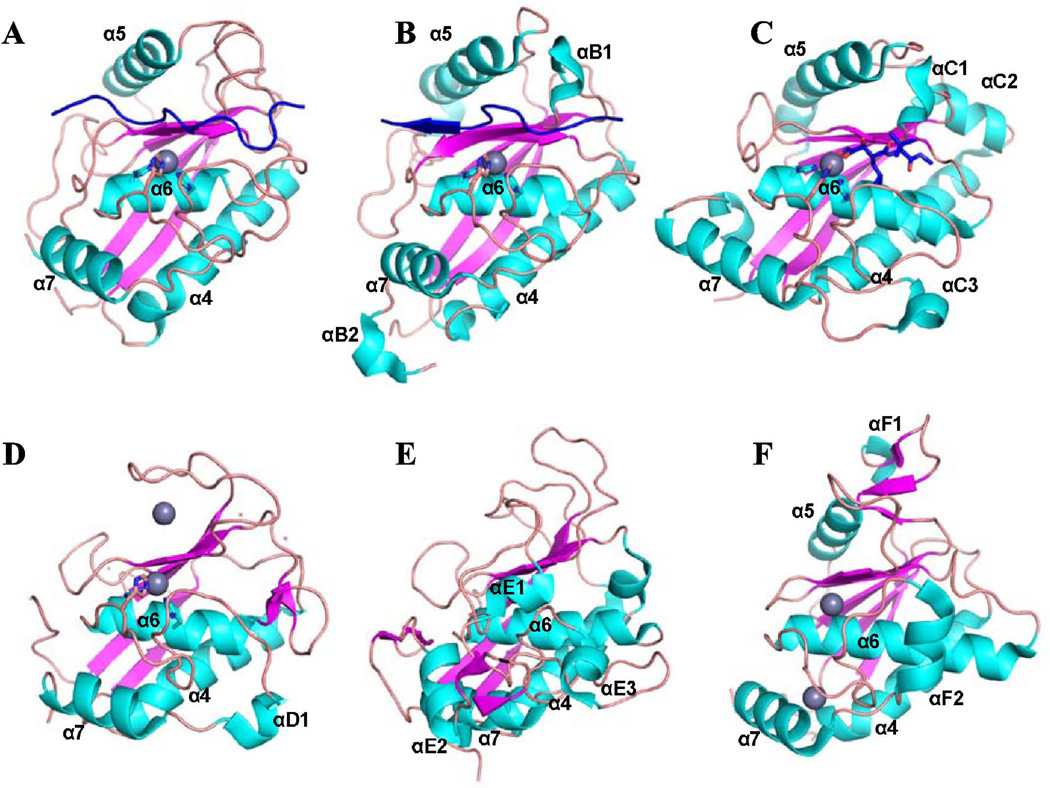
Figure 4. Structural homologues of the MPII catalytic domain.
(A) MPII (PDB 4ON1), (B) FRA3 (PDB 3P24), (C) human ADAM-33 (PDB 1R55), (D) human MMP-12 (PDB 1Y93), (E) Protease C from Erwinia chrysanthemi (PDB 1K7I) and (F) Archaemetzincin from Methanopyrus kandleri (PDB 2X7M). Helices, cyan; β-sheets, magenta; loops, orange; prodomain (in MPII and FRA3) and a hydroxamate inhibitor (in ADAM-33), blue; Zn ion, grey sphere. The homologues were identified in the PDB database using FFAS03 software (http://ffas.sanfordburnham.org). Structural elements are labeled as in the respective original publications.
Multiplex peptide cleavage assay
We used a high-throughput peptide-cDNA fusion-based cleavage profiling technology that has been employed previously to determine the cleavage preferences for several proteinases [14–17].
Here, we designed the peptide-cDNA fusions to contain a 10-residue peptide substrate flanked by N- and C-terminal common sequences. Statements below regarding peptide sequences refer only to the variable 10-mer portions of the peptides and disregard the flanking constant portions. We prepared a set of 18,583 peptides for elucidating cleavage sequence preferences of MPII and FRA3. The set included 1,643 peptides containing the dibasic furin cleavage motif [16] as our earlier studies [14] revealed that this motif is efficiently cleaved by MPII. We also included 1,369 peptides that were identified as the most efficient substrates of MMPs using phage display methods [18]. Additionally, we designed, in silico, 13,631 peptide sequences that we expected to be cleaved efficiently by various metalloproteinases. The design rules were derived from our bioinformatic analysis of the 1,369 experimentally verified MMP substrates mentioned above. We used these validated substrate sequences to identify amino acid residue types that are accepted by the following MMPs: MMP-2, MMP-9, -MMP-14, MMP-15, MMP-16, MMP-17, MMP-24 and MMP-25 at each of the individual P5-P5’ positions. We then used a random number generator to create multiple peptide sequences using random combinations of the accepted residues at each of the individual P5-P5’ positions, followed by filtering out redundant sequences. We also used our statistical substrate specificity profiling software [19] to exclude peptides that were predicted to be inefficient MMP substrates, resulting in a set of 13,631 sequences. Based on the specificity profiling software predictions and the crystal structures from the Protein Data Bank (PDB) database, we included 598 peptide sequences that were potential MMP cleavage substrates selected from secretory proteins. Finally, we included 1,342 positive and negative controls. The positive controls included known peptide substrates for thrombin, enterokinase, caspase-3, and West Nile and Dengue virus proteinases. The negative controls included randomly generated sequences and other peptides that were not cleaved by MMPs in our previous experiments.
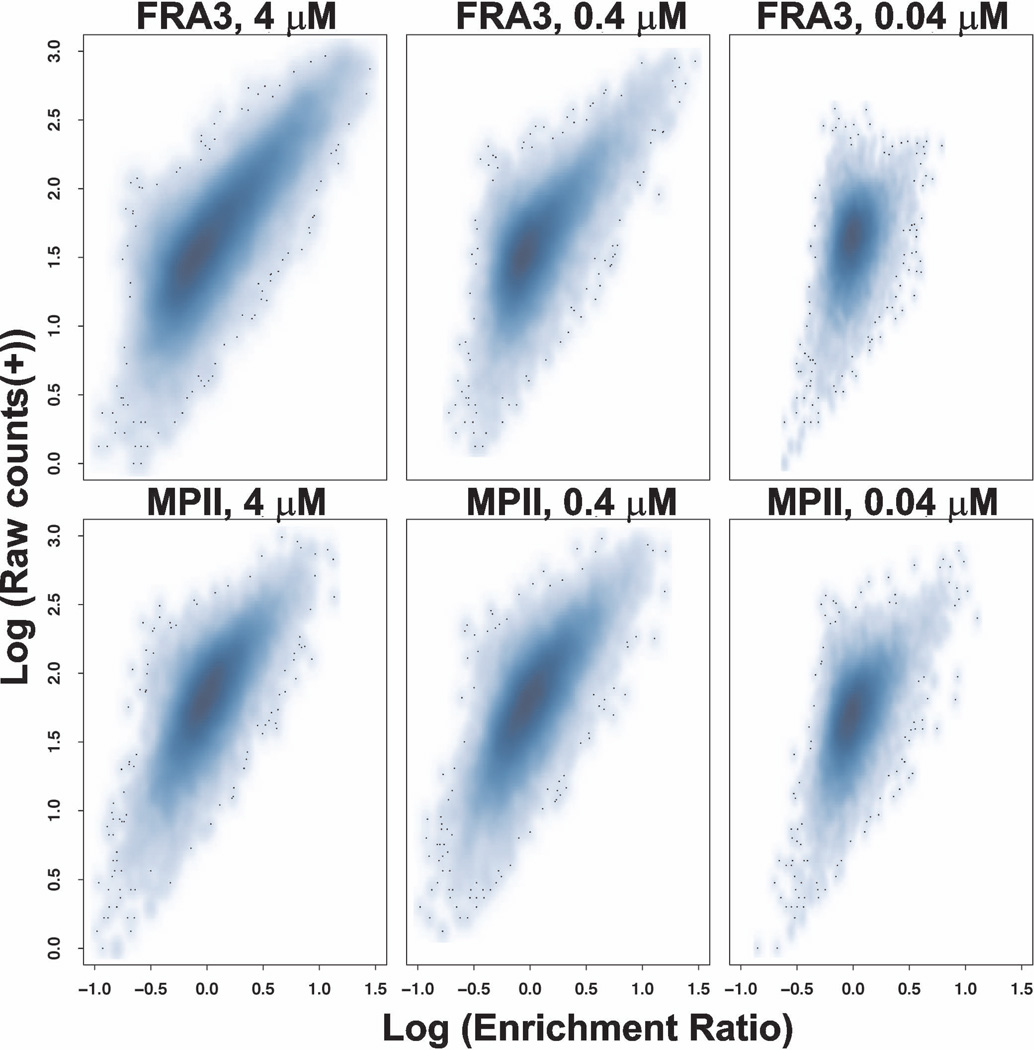
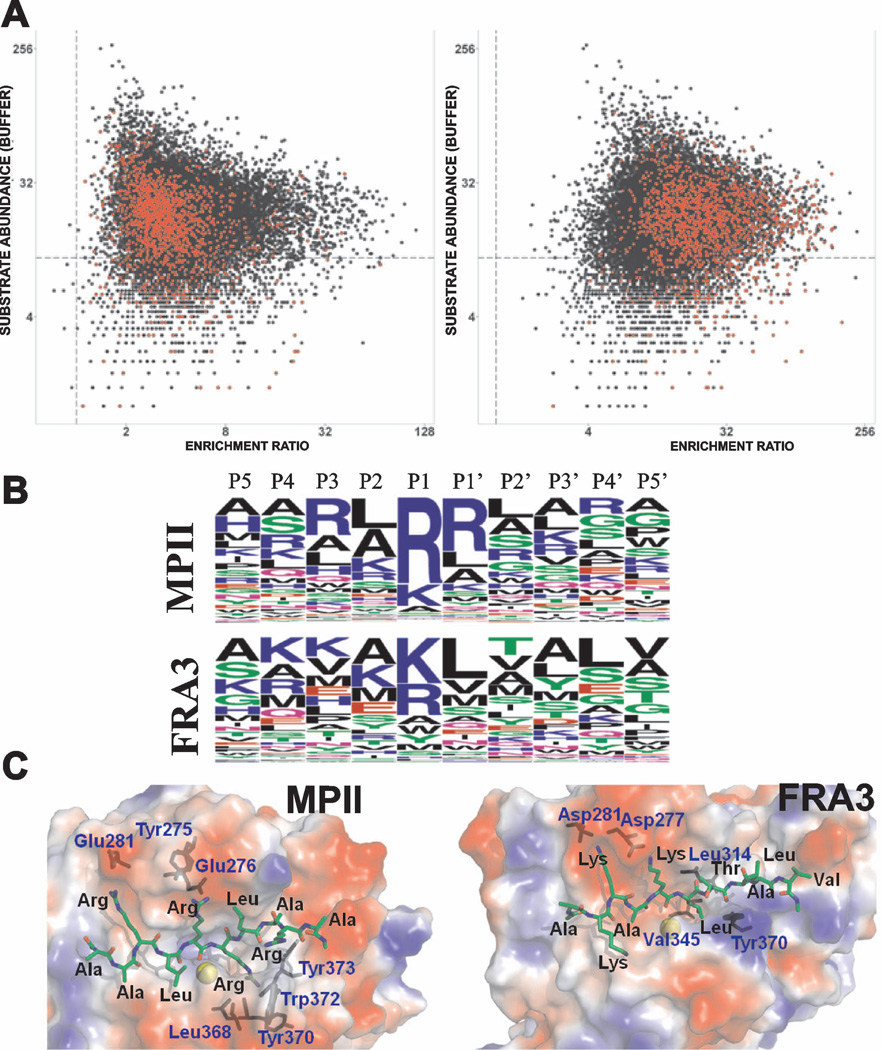
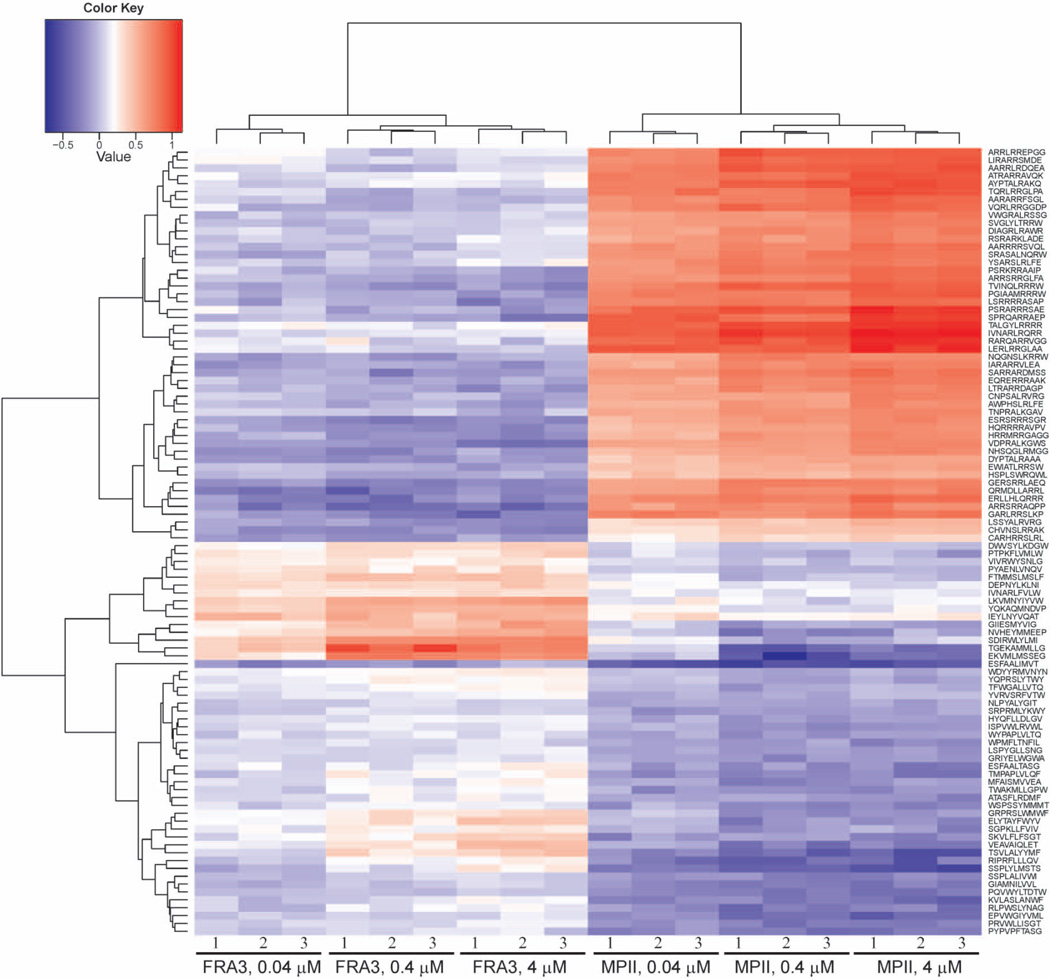
We evaluated cleavage of the peptide set described above by FRA3 and MPII at concentrations of 0.04, 0.4 and 4 µM. The Pearson correlation coefficient of technical replicates was >0.9 indicating good reproducibility at all three concentrations. Higher levels of non-specific cleavage were observed at 4 µM concentration indicating a loss of specificity, while assay sensitivity was diminished at 0.04 µM concentration (Figure 5). Consequently, we relied mainly on the 0.4 µM proteinase cleavage data for comparing the substrate specificity of MPII versus FRA3. The most specific substrates of MPII and FRA3, however, were identified using the data obtained at the 0.04 µM proteinase concentration. The dibasic peptides were more susceptible to cleavage by MPII than by FRA3 (Figure 6A). Overall, 300 and 93 substrate sequences for MPII and FRA3 (with the median normalized enrichment ratio values >3), respectively, were considered to be cleaved most efficiently. We used this subset of peptides to identify the preferred cleavage sequences for both enzymes. A direct relationship between the cleavage efficiency of MPII and FRA3 and the amino acid sequences of their peptide substrates are illustrated in the form of sequence logos (Figure 6B). Evidently, there is a partial overlap in the cleavage preferences of MPII and FRA3. Our data demonstrate that dibasic residues at the P1 and P1’ positions dominate the cleavage preference of MPII while other residue positions are much less selective. In contrast, dibasic motifs are not a prerequisite for the efficient cleavage of the peptides by FRA3. In general, our in vitro cleavage results indicate that the cleavage preferences of FRA3 are not highly selective and that FRA3 accepts hydrophobic, hydrophilic and charged residue types in multiple substrate positions. We selected 100 of the most specific substrates (50 for each proteinase) and performed a supervised clustering to demonstrate that the cleavage preferences were largely irrespective of protease concentration (Figure 7).
Figure 5. Scatter density plot of FRA3 and MPII at three concentrations (0.04, 0.4 and 4 µM each).
Density plot of cleavage enrichment ratios of 18,583 peptides. X-axis denotes the log10 -transformed enrichment ratios where the median enrichment ratio is normalized to 1, while Y-axis shows log10 raw DNA counts in a digest sample. Darker blue represents a higher density of substrates, with outlying substrates are represented as dots. This facilitates comparison of the three different concentrations, where specificity can be ascertained from the distance between the outlying high-ratio substrates and the high-density region of the plot representing non-specific cleavage. The plots suggests that the assay is more sensitive and less specific at the high, 4 µM, concentration both FRA3 and MPII. In turn, at the low, 0.04 µM, concentration of FRA3 and MPII the assay is less sensitive, but more specific.
Figure 6. Proteolysis of peptides by FRA3 and MPII.
(A) Cleavage of the peptides with the dibasic furin cleavage motif by MPII and FRA3. Peptide cleavage was evaluated after treatment of 18,583 peptides by FRA3 or MPII (0.4 µM each). The peptide cleavage levels are defined as the number of sequencing reads of the corresponding DNA templates in the samples. Each black dot represents a unique peptide substrate. Enrichment ratios were calculated for proteinase-treated peptides versus their respective “buffer alone” controls. Because the abundance of the individual peptides varied in the synthesized pool, we plot peptide abundance versus enrichment ratio. The data confirm that the enrichment ratio is independent of the abundance of the individual peptides in the synthesized pool. Red dots mark peptides with the dibasic cleavage motif, which were efficiently cleaved by MPII (right) relative to FRA3 (left). The vertical and horizontal dotted lines represent unity ratio (=1) and 10 DNA sequencing counts, respectively. (B) Frequency plot of the cleavage sequences of MPII and FRA3 in an IceLogo format. The height of a character is proportional to the frequency of the amino acid residue at the individual position of the cleaved peptide. In MPII and FRA3, the scissile bond is Arg-Arg and Lys-Leu, respectively. (C) Structural modeling of MPII (PDB 4ON1) and FRA3 (PDB 3P24) with the Ace-Ala-Ala-Arg-Leu-Arg-Arg-Leu-Ala-Arg-Ala-NMe and Ace-Ala-Lys-Lys-Ala-Lys-Leu-Thr-Ala-Leu-Val-NMe peptide substrates, respectively. Substrates and the relevant protease residues are shown as sticks and labeled using black and blue letters, respectively. Zinc ions are shown as yellow spheres. The molecular surface of MPII and FRA3 is colored according to electrostatic potential (red, blue and white represent negative, positive, and neutral electrostatic potential values, respectively).
Figure 7. Supervised clustering of the 100 most selective peptide substrates of FRA3 and MPII in a heatmap format.
P-values of substrates (50 for FRA3 and 50 for MPII) are < 0.001 each after Bonferroni multiple testing correction. X-axis shows clustering of FRA3 and MPII samples in triplicate at three concentrations (0.04, 0.4 and 4 µM each). Y-axis shows clustering of peptide substrates based on their enrichment ratios normalized to the mean enrichment for the respective protease concentration. Red, white and blue colors designate high, medium and low enrichment ratios, respectively.
Structural requirements for specificity
To elucidate structural elements that guide the cleavage preferences of MPII and FRA3, we modeled the hypothetical optimal substrates (Ace-Ala-Ala-Arg-Leu-Arg-Arg-Leu-Ala-Arg-Ala-NMe and Ace-Ala-Lys-Lys-Ala-Lys-Leu-Thr-Ala-Leu-Val-NMe) bound to the MPII and FRA3 crystal structure, respectively (Figure 6C). The long, positively charged side-chain of Arg fits well into the tunnel-like sub-sites of MPII. In MPII, the guanidinium group of the substrate P1’ Arg interacts with both the Leu-368 and Trp-372 carbonyls and the nitrogen atom from the peptide bond of Tyr-373. In turn, the hydrophobic side chains of Leu-314, Val-345 and Tyr-370 form the S1’ subsite in FRA3, which prefers Leu. The presence of Tyr-275, Glu-276 and Glu-281 in MPII versus Asn-275, Asp-277 and Asp-281 in FRA3 explain the preference of MPII for Arg versus the preference of FRA3 for Lys at both P1 and P3. An additional difference between the substrate binding sites is that the S2'-S5’ subsites are positively charged (or neutral) in FRA3 and negatively charged (or neutral) in MPII. This difference explains why MPII with Trp-372 accepts Arg and Lys at the P3’-P4’ positions while FRA3 with Lys-369 prefers hydrophobic and negatively charged residues.
Exemplary substrate validation
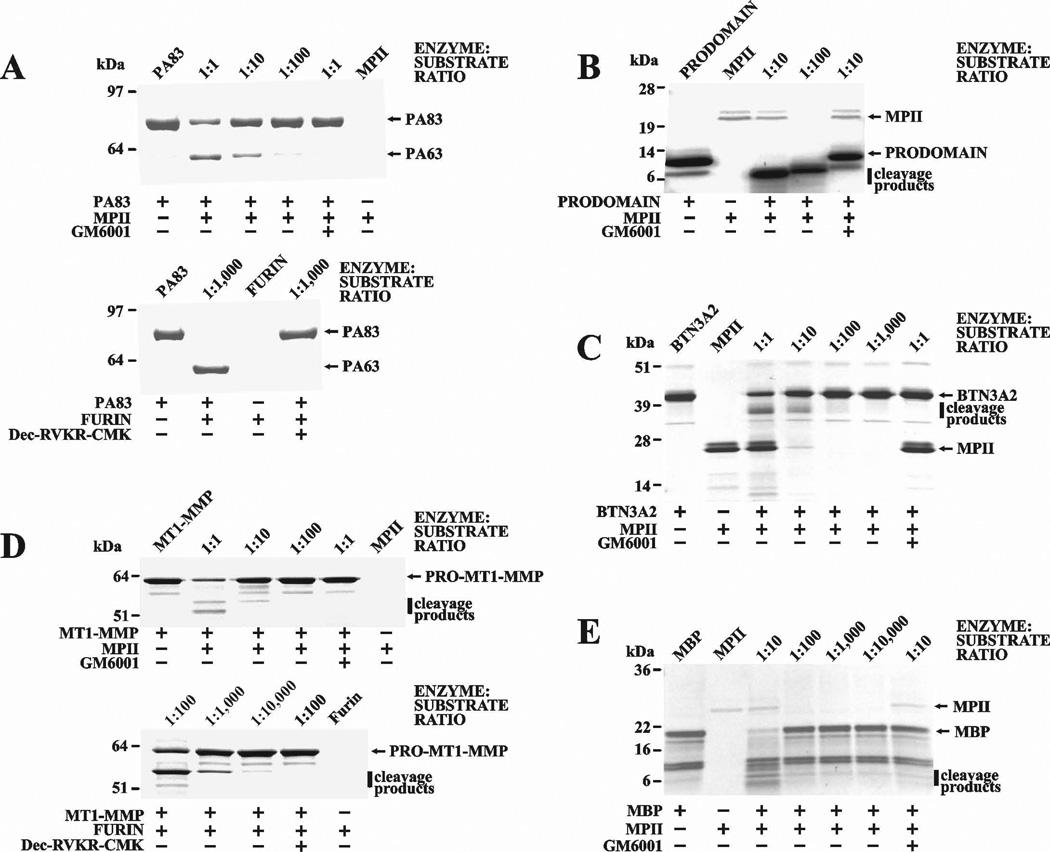
As a result of our studies, we are now able to elucidate the cleavage motifs of MPII. The peptide cleavage data suggest that the dibasic cleavage motif of MPII is most similar to that of furin. To test this suggestion, we co-incubated several commercially available proteins with the dibasic motif [anthrax protective antigen-83 (PA83), the membrane type-1 matrix metalloproteinase (MT1-MMP) proenzyme, the individual furin prodomain, butyrophilin A2 (BTN3A2) and myelin basic protein (MBP)] with MPII. Where indicated, a hydroxamate inhibitor (GM6001) was added to the reactions to block MPII activity. In agreement with our suggestion, anthrax PA83 treated with MPII appeared to be cleaved at the Arg193-Lys-Lys-Arg196 furin cleavage motif, resulting in the C-terminal PA63 and the N-terminal PA20 fragments (Figure 8A). The furin 27–107 prodomain contains two basic motifs (Val-Thr-Lys-Arg75↓Ser76-Leu-Ser-Pro and Arg-Leu-Gln-Arg89↓Glu90-Pro-Gln-Val) [20]. Similar to anthrax PA83, MPII readily cleaved the furin prodomain, generating at least two digest fragments (Figure 8B). As expected, MPII also cleaved the MT1-MMP proenzyme. As a result of the cleavage of the Arg108-Arg-Lys-Arg111↓Tyr112 site, where Tyr112 is the N-terminal residue of the MT1-MMP enzyme, the mature MT1-MMP was generated. MPII also cleaved human MBP and BTN3A2, both of which exhibit basic sequence motifs (Figure 8C–E).
Figure 8. MPII proteolysis of selected proteins.
Anthrax PA83 (A), the prodomain of human furin (B), human BTN3A2 (C), human MT1-MMP (D), and human MBP (E) were co-incubated with MPII at the indicated enzyme:substrate molar ratio. The protein samples in the A and D panels were also co-incubated with furin. Where indicated GM6001 and decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone were added to the reactions to inhibit MPII and furin activity, respectively.
Cellular effects of MPII and FRA3
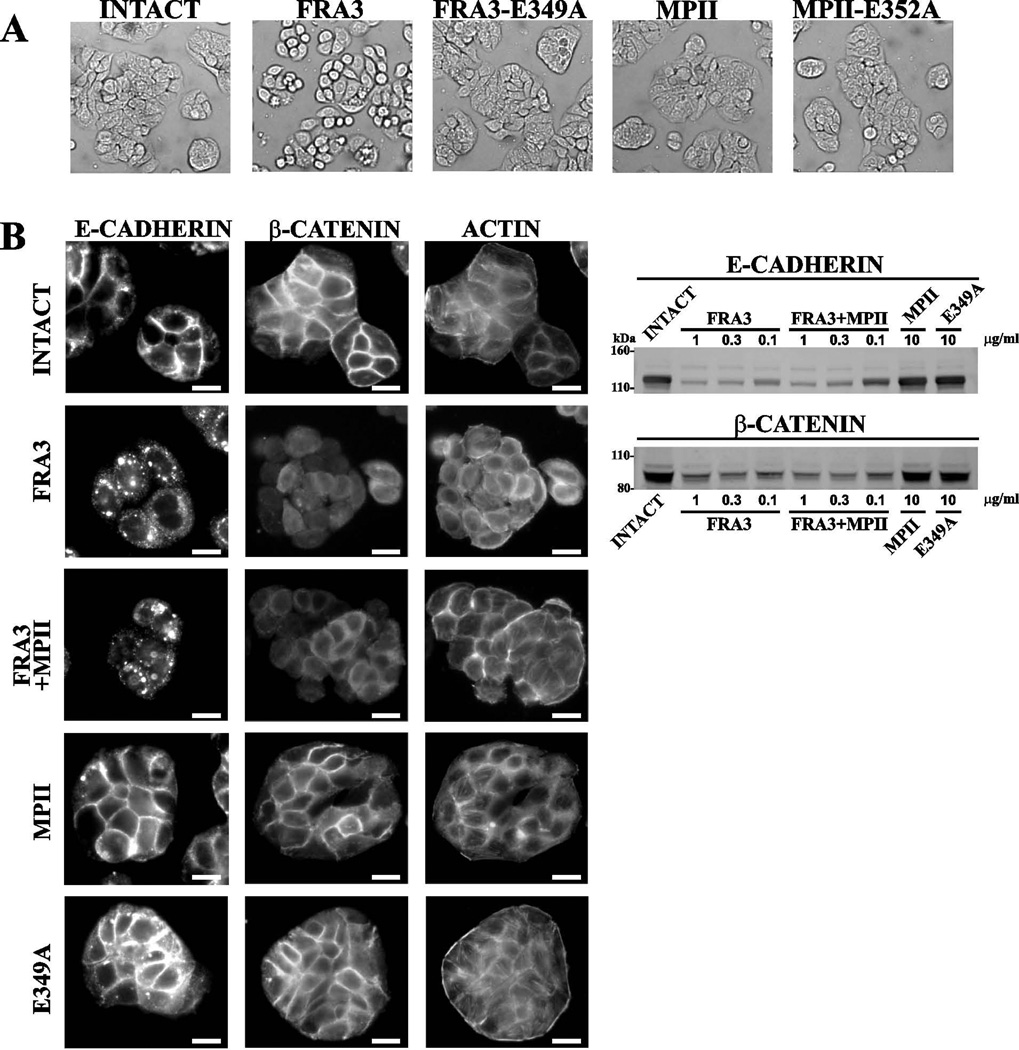
Here, we used colon carcinoma HT-29 cells, which are known to be responsive to B. fragilis proteinases [3, 21]. Cells were co-incubated with MPII, MPII-E352A, FRA3 and FRA3-E349A and then observed using a microscope. Cell rounding was readily observed in cells co-incubated with FRA3 alone but not in those co-incubated with FRA3-E349A, MPII-E352A, or active MPII (Figure 9). These observations correlate with the findings by others who recorded FRA3 proteolysis of cellular E-cadherin followed by changes in cell morphology [13]. To corroborate these observations, HT-29 cells were co-incubated with MPII, MPII-E352A, FRA3 and FRA3-E349A alone and in combination, and then stained with the E-cadherin and β–catenin antibodies. The membrane immunoreactivity of both E-cadherin and β-catenin was observed in the intact cells and in the samples treated with MPII and FRA3-E349A. In turn, the immunoreactivity of membrane E-cadherin and β–catenin was reduced in the cells co-incubated with FRA3. MPII did not enhance the FRA3-dependent cleavage of E-cadherin and reduction in the β–catenin levels. The results of Western blotting of the cell lysates directly supported our immunostaining results and suggested that FRA3 alone cleaved E-cadherin and that this cleavage decreased the level of cellular β–catenin. This decrease in the level of β–catenin that follows FRA3 proteolysis of cellular E-cadherin correlates with γ-secretase proteolysis of the E-cadherin cytoplasmic tail and the subsequent liberation and proteasomal degradation of the released β-catenin [8]. In agreement with the results of others [13, 22], we conclude that E-cadherin is susceptible to FRA3, but not to MPII, in HT-29 cells.
Figure 9. FRA3 proteolysis of cellular E-cadherin and β-catenin.
(A) Morphology of HT-29 cells. Cells were left untreated or co-incubated for 3 h at 37°C with FRA3 and FRA3-E349A (5 nM each), and MPII and MPII-E352A (50 nM each). Cell morphology was observed using a bright-field microscope. (B) Left, immunostaining of HT-29 cells. Cells were left intact or incubated for 3 h at 37°C with FRA3 (5 nM), MPII (50 nM) and E349A (5 nM) alone or in combination. Cells were then fixed, permeabilized and stained for E-cadherin β-catenin (middle) and phalloidin/F-actin (right). Scale bar, 20 µm. Right, Western blotting of HT-29 cells with the E-cadherin and β-catenin antibodies. Cells were left intact or co-incubated for 3 h with the indicated concentrations of FRA3, MPII and E349A. Where indicated, MPII (10 µg/ml, ~500 nM) was added jointly with FRA3. E349A, the E349A proteolytically inactive mutant of FRA3.
There are, however, multiple potential cleavage targets of MPII in the human secretome that are distinct from E-cadherin. Based on the peptide cleavage data, we selected over 80 soluble and membrane-associated human proteins that include an MPII cleavage site and could, therefore, represent potential cleavage targets of MPII (Supplemental Table S2). It is likely that the combined proteolytic action against multiple substrates, rather than the cleavage of a single cellular substrate, constitutes the virulent function of MPII.
Discussion
Enterotoxigenic B. fragilis is the most frequent diarrhea-causing anaerobe and a risk factor for colorectal cancer [3, 23–26]. It is likely that the role of enterotoxigenic B. fragilis in increasing colorectal cancer risk is similar to that of Helicobacter pylori in gastric cancer [27, 28]. The presence of a single pathogenicity island discriminates toxigenic from non-toxigenic B. fragilis strains [29, 30]. The island encodes two distinct secretory metalloproteinases, MPII and FRA [3]. The latter exists in three highly homologous enterotoxigenic isoforms (FRA1, FRA2, and FRA3), which differ by only a few substitutions [9, 10, 31, 32]. In turn, there is only ~25% peptide sequence identity between MPII and FRAs [14]. Whereas FRAs have been the focus of studies by multiple laboratories [reviewed by [3]], the characteristics of MPII were virtually unknown.
Here, we obtained substrate preference data for MPII and FRA3 by empirically testing 18,583 individual peptides in a highly parallel assay. Comparative analysis of our results indicates that dibasic residues at the P1 and P1’ positions dominate the cleavage preference of MPII. Other residue positions are less selective in this proteinase. The presence of a dibasic motif, however, is not required for the efficient peptide cleavage by FRA3. The presence of the P1 Arg or Lys followed by a P1’ hydrophobic residue, especially Leu, largely suffices for FRA3. In general, our cleavage results indicate that FRA3 is not a highly selective proteinase and that, similar to human MMPs, FRA3 tolerates the presence of multiple hydrophobic, hydrophilic and charged residue types in multiple substrate positions.
We further concluded that MPII is capable of cleaving the basic motifs in human cellular proteins. This aberrant proteolysis is likely to interfere with the normal ratio of those membrane and soluble precursor proteins, which are incompletely processed by cellular furin in normal gut epithelium. As a result, MPII proteolysis may cause an abnormal precursor/mature protein ratio in the multiple targets of furin, leading to pathological consequences. Multiple additional soluble and membrane proteins with the dibasic cleavage motif may be the cleavage targets of MPII in the gut environment. Our current observations support our earlier suggestion that MPII appears to be the first bacterial zinc metalloproteinase with a dibasic cleavage preference [14]. Metalloproteinases (antareases) with similar cleavage preferences exist in the venom of predatory arthropods such as scorpions [33, 34], thereby demonstrating a high level of versatility of metalloproteinase proteolysis.
To increase our understanding of the enterotoxiogenic B. fragilis proteinases, we determined the crystal structure of the MPII zymogen at 2.13 Å resolution. To avoid self-proteolysis, we used the catalytically inert E352A mutant in our crystallization studies. The MPII zymogen consists of the N-terminal ~150 residue prodomain and the C-terminal catalytic domain of ~180 residues connected by a 20 residue long linker. The fold of the catalytic domain in MPII is similar to that in FRA3. This fold is largely conserved in multiple archaebacterial, microbial and mammalian metalloproteinases. The common architecture of the catalytic domain, the presence of the Met-turn motif and the unconventional α5 helix in B. fragilis MPII and FRA3, archaebacterial archaemetzincin and human ADAMs allow us to hypothesize that the catalytic domain evolved to adopt a similar fold in Bacteroides, archaebacteria and mammals. This likely optimal fold has been independently refined by multiple metalloproteinases in multiple, evolutionally unrelated organisms. In sharp contrast to the catalytic domain, the MPII prodomain exhibits an unconventional fold that is a unique feature of the enzymes encoded by the pathogenicity island of B. fragilis. Overall, the data suggest that the catalytic domain of both MPII and FRA3 evolved separately from the prodomain.
The uniqueness of its sequence and fold suggests that the prodomain evolved specifically to fit the B. fragilis pathogenicity. In agreement with others [35], we speculate that the prodomain might be required for binding to specific receptor(s) in host cells and that this receptor is distinct from E-cadherin. Studies to identify possible receptors of MPII and FRA3 are currently in progress.
Materials and Methods
General reagents
The reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless indicated otherwise. 5-FAM-SLGRKIQIQK(QXL520)-NH2 substrate was acquired from AnaSpec (Fremont, CA, USA). GM6001 (a broad-specificity hydroxamate inhibitor of MMPs) was from EMD Millipore (Temecula, CA, USA). Anthrax PA83 was purchased from List Biological Laboratories (Campbell, CA, USA). Murine monoclonal antibodies to β-catenin and E-cadherin were from BD Transduction Laboratories (San Jose, CA, USA). Recombinant human BTN3A2 was from OriGene (Rockville, MD, USA). Alexa Fluor-594 goat anti-mouse antibody and Alexa Fluor-488 phalloidin were from Life Technologies (Santa Cruz, CA, USA). The horseradish peroxidase-conjugated donkey anti-mouse antibody was from Jackson ImmunoResearch (West Grove, PA, USA). Human colon carcinoma HT-29 cells were obtained from ATCC (Manassas, VA, USA) and routinely grown in McCoy’s 5A medium supplemented with 10% fetal bovine serum and gentamicin (10 µg/ml). The catalytically active MPII and FRA3 constructs and their catalytically inactive mutants (E352A and E349A, respectively) were obtained earlier [14]. Furin and its individual prodomain were isolated as described previously [16, 20]. MBP was from Biodesign (Saco, ME, USA). Decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone was obtained from Bachem (Torrance, CA, USA).
Enzyme cloning, expression and purification
The catalytically inactive MPII-E352A construct comprising the N-terminal Hisx6 tag followed by the thrombin cleavage sequence was constructed by PCR using the original MPII-E352A Hisx6–tagged construct as a template [14], and the 5’–CACCATGCACCATCACCATCACCATGGACTAGTACCGCGAGGAGCCTGTGCCGAT GACCTG–3’ (thrombin cleavage sequence is underlined) and 5’–TCACTTTTGGATGCACTCCAG–3’ oligonucleotides as the forward and reverse primers, respectively. The resulting PCR product was re-cloned into the pET101 vector (Invitrogen, Carlsbad, CA, USA) and used to transfect competent E.coli BL21 (DE3) Codon Plus cells (Stratagene, Santa Clara, CA, USA). Transformed cells were grown at 30°C in Luria–Bertani broth containing ampicillin (0.1 mg/ml). Cultures were induced with 0.6 mM IPTG and growth was continued for an additional 16 h at 18°C. The cells were then collected by centrifugation (5,000 × g; 15 min), re-suspended in Tris-HCl buffer, pH 8.0, containing 1 M NaCl, and disrupted by sonication. The pellet was removed by centrifugation (40,000 × g; 30 min). The MPII-E352A construct was then purified from the supernatant fraction using a Ni2+-chelating Sepharose Fast Flow column. The purified material (1.5 ml; 30 mg/ml) was co-incubated for 18 h at 4°C with thrombin (60 U). Thrombin was next inactivated using 0.25 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride. The resulting untagged MPII-E352A construct was purified using a Superdex S-75 High Resolution 10/30 column, equilibrated in 25 mM HEPES buffer, pH 7.5, containing 0.1 M NaCl, and the purified sample was concentrated using an Ultracel-10K concentrator (Millipore, Billerica, MA, USA) to reach a 20 mg/ml concentration. The final MPII-E352A sample was used for crystallization.
Crystallization and structure solution
Initial screens for the identification of crystallization conditions were performed using a 96 reagent crystallization screen Index (Hampton Research, Aliso Viejo, CA, USA) and a Phoenix robot (Art Robbins Instruments, Sunnyvale, CA, USA). MPII-E352A was crystallized using the sitting drop vapor diffusion method. Drops (0.4 µl each) contained 0.2 µl 25 mM HEPES buffer, pH 7.5, 100 mM NaCl, 4 mM CaCl2 and MPII-E352A (20 mg/ml), and 0.2 µl crystallization buffer (20% PEG3330-0.2M MgCl2). Needle-like crystals (0.3×0.04×0.04 mm3 in size) were observed in 10–14 days. Crystals were flash-frozen in liquid nitrogen using 20% v/v glycerol as a cryoprotectant. Diffraction data were collected at 2.13 Å resolution from the cryo-cooled crystals at 100°K on the SYBYLS beamline at the Advance Light Source (Berkeley, CA) and processed using the HKL2000 and ccp4i program suites [36, 37]. The structures were solved using the Phaser molecular replacement program [38] and PDB 3P24 as a template. The model rebuilding and refinement were done with COOT [39] and the Phenix program suite [40]. The data collection and refinement statistics are presented in Table 1.
Table 1. Data collection and refinement statistics.
Residue count includes the 18-residue signal peptide.
| Space Group | P212121 | ||
| Unit Cell a, b, c /Å | 44.4 - 114.4 - 140.2 | ||
| Resolution/Å (outer shell) | 50-2.13 (2.17-2.13) | ||
| Unique reflections collected | 41324 | ||
| Completeness (%) | 99.8 (99.7) | ||
| Average Redundancy | 12.4 (10.8) | ||
| <I/σ(I)> | 21 (4.8) | ||
| Rmerge | 0.135 (0.58) | ||
| Refinement statistics: | |||
| Resolution range (Å) | 44-2.13 | ||
| No reflections work set (RFREE set) | 41241 (2064) | ||
| RWORK (RFREE) | 0.16 (0.20) | ||
| RMS Deviations | |||
| bond lengths (Å) | 0.005 | ||
| bond angles (Å) | 0.92 | ||
| Ramachandran plot (%) | |||
| Favored by MolProbity (%) | 98.0 | ||
| Outliers by MolProbity (%) | 0.31 | ||
| Coordinate errors, estimated by Phenix (Å) | 0.3 | ||
| Residues in chain A observed/present | 334/396# | ||
| Residues in chain B observed/present | 333/396# | ||
| Ligands per a chain (Zn) | 1 | ||
| Water molecules (total) | 429 | ||
| Temperature factors, ADP (Å2) | |||
| average | 32.0 | ||
| protein | 31.5 | ||
| ligands (Zn2+) | 19.5 | ||
| solvent | 37.6 | ||
| Wilson B | 27 |
Protease activity assays with FRET peptide
MPII cleavage activity assays were performed in triplicate in wells of a 96 well plate in 0.2 ml reactions containing 50 mM HEPES, pH 8.0, 1 mM CaCl2, 0.5 mM MgCl2 and 10 µM ZnCl2. The 5-FAM-SLGRKIQIQK(QXL520)-NH2 substrate (AnaSpec) and enzyme concentrations were 10 µM and 50 nM, respectively. Initial reaction velocities were monitored continuously at λex=488 nm and λem=520 nm on a Spectramax Gemini EM fluorescence spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
MPII proteolysis of proteins in vitro
Anthrax PA83 (2 µg; ~1 µM) and the following purified human proteins [the furin 27–107 prodomain (1.8 µg, ~9 µM) [20], the MT1-MMP proenzyme (2 µg; ~2 µM), MBP (2 µg; ~5 µM) and BTN3A2 (2 µg; ~2.5 µM) were each co-incubated for 1 h at 37°C with increasing concentrations of MPII in 50 mM HEPES, pH 8.0, containing 1 mM CaCl2, 0.5 mM MgCl2 and 10 µM ZnCl2. As a control, PA83 and the MT1-MMP proenzyme were co-incubated for 1 h at 37°C with furin (at the 1:100–1:10,000 enzyme:substrate molar ratio) in 20 µl reactions containing 100 mM HEPES, pH 7.5, and 1 mM CaCl2. Where indicated, GM6001 (10 µM) and decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone (50 µM) were added to the reaction to inhibit MPII and furin, respectively. The reactions were stopped by adding a 5×SDS sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis in either 4–12% or 4–20% gradient polyacrylamide gels.
Protease treatment and immunostaining of the cells
HT-29 cells were grown on 15-mm glass coverslips. Where indicated, cells were co-incubated for 3 h at 37°C with MPII (1 µg/ml, ~50 nM), FRA3 (0.1 µg/ml, ~5 nM), FRA3-E349A (0.2 µg/ml, ~5 nM) or MPII-E352A (2 µg/ml, ~50 nM) individually or in combination in serum-free McCoy’s 5A medium supplemented with 20 mM HEPES, pH 7.0 and 0.2% BSA. Cells were then fixed for 16 min with 4% paraformaldehyde, permeabilized for 4 min using 0.1% Triton X-100 in 4% paraformaldehyde, blocked for 1 h in 10% BSA, and stained for 16–18 h at 4°C using the murine E-cadherin and β-catenin monoclonal antibodies (dilution 1:100 and 1:50, respectively) followed by the Alexa Fluor 594-conjugated goat anti-mouse antibody (dilution 1:200). Cellular actin was stained using Alexa Fluor 488-conjugated phalloidin (1 h; dilution 1:500). The slides were mounted in Vectashield DAPI-containing medium and analyzed using an Olympus BX51 fluorescence microscope equipped with a MagnaFire digital camera. Where indicated, cells were observed using a bright-field microscope.
Western blotting
HT-29 cells were grown in wells of a 12-well plate and co-incubated for 3 h at 37°C with FRA3 (0.1–1 µg/ml, ~5–50 nM), MPII (10 µg/ml, ~500 nM) or FRA3-E349A (10 µg/ml, ~250 nM) individually or in combination in serum-free McCoy’s 5A medium. Cells were then lysed for 1 h at 4°C using 50 mM octyl-β-D-glucopyranoside in TBS, pH 7.4, containing 1 mM phenylmethylsulfonyl fluoride, 25 µM GM6001 and a protease inhibitor mixture set III. Insoluble material was removed by centrifugation (14,000×g, 15 min). The supernatant aliquots (10 µg total protein) were analyzed by Western blotting using the murine E-cadherin and β-catenin monoclonal antibodies (dilution 1:2,000) followed by the horseradish peroxidase-conjugated donkey anti-mouse antibody (dilution 1:5,000) and a TMB/M substrate (SurModics, Eden Prairie, MN, USA).
Multiplex peptide cleavage assay and data analysis
Biotinylated peptide-cDNA pool preparation was performed as reported earlier [15]. The template DNAs encoded 18,583 10-mer peptide substrates flanked by N-terminal (Biotin-linker-Gly-Ala) and C-terminal (Gly-Asn-Ala-Ser-Ala-Ser-Ala-Ala-Gly-Ala-linker-DNA) common sequences. Six biotinylated oligonucleotides were spiked into the peptide-cDNA pool as internal standards for normalization after PCR reactions. Streptavidin-coated magnetic beads with the immobilized peptide-cDNA pool (2 pmol total, ~0.1 fmol/peptide) were co-incubated with MPII or FRA3 at 37°C for 30 min, in 3 µl reactions containing 50 mM HEPES, pH 6.8, 10 mM CaCl2 and 10 µM ZnCl2. Reactions without the proteinases were used as controls. To identify cleaved peptide substrates, the cDNA molecules released by the proteinase treatment were collected, DNA adapters required for sequencing were installed by PCR, and the obtained DNA constructs were sequenced using a MiSeq sequencing instrument (Illumina, San Diego, CA, USA).
We treated the peptide-cDNA pool with FRA3 and MPII proteinases at 0.04, 0.4 and 4 µM each, in triplicate. Sequencing counts for each sample were normalized using the spiked-in internal standards. The enrichment ratio (Ratio) for each substrate in a sample was then calculated by taking the ratio of average sequencing counts of three replicates in proteinase samples over average counts in negative samples (buffer alone). A list of preferred peptide substrates for each of FRA3 and MPII was determined by taking the 50 most selective peptides with respect to the buffer controls by Student t-test (1-way) on log-transformed median-normalized enrichment ratio data for each comparison. Following Bonferroni correction all 100 selected peptides had p-value<0.001.
Molecular modeling
Molecular modeling of the peptide Ala-Ala-Arg-Leu-Arg-Arg-Leu-Ala-Arg-Ala complex with MPII was initiated from the crystal structure of MPII (PDB 4ON1). Molecular modeling of the peptide Ala-Lys-Lys-Ala-Lys-Leu-Thr-Ala-Leu-Val was performed using the FRA3 crystal structure (PDB 3P24). In these structures, the C-terminal portion of the prodomain directly fits into the active site of MPII and FRA3. and runs in the opposite orientation relative to the substrate. To model the peptide substrate, the Asn184-Glu197 and Leu186-Val201 segments of the prodomain were deleted in silico from the MPII and FRA3 structure, respectively. The substrates were then modeled by placing the substrate's side chain heavy atoms at the positions of the heavy atoms in the prodomain. Further optimization of the main and side chain atoms of the substrate was performed using molecular mechanical minimization and short molecular dynamics simulations via the Amber12 modeling package and FF99SB force field, and generalized-Born method for representing solution environment implicitly [41–44]. In the our optimization procedure, the proper orientation of the carbonyl group of the scissile bond relative to the active site Zn ion was maintained, and acetyl and N-methyl groups for capping the N- and C-termini of the modeled substrate were used. The final models were displayed using PyMol (www.pymol.org).
Supplementary Material
Acknowledgements
Our work was supported by R01CA83017, R01CA157328 and R01DE022757 (AYS), R44GM085884 (IAK), and R01GM098835 (PC) grants from the NIH. We thank Andy Arvai and John Tainer (The Scripps Research Institute) for their assistance in data collection. We would also like to thank all employees of Prognosys Biosciences for their help. In particular, we are thankful to Mark Chee for useful comments on the manuscript, Petr Capek for preparation of modified oligonucleotides, Erina He for assistance with DNA sequencing, and Wayne Delport for help with sequencing data analysis.
Abbreviations
- BTN3A2
butyrophilin A2
- FRA
fragilysin
- MBP
myelin basic protein
- MMP
matrix metalloproteinase
- MPII
metalloproteinase II
- PA83
protective antigen-83
Footnotes
Author contribution
SAS performed protein purification, crystallization and biochemical experiments; AEA and RCL did protein crystallization and X-ray analysis; NM contributed to experimental design and performed cleavage assay experiments; MK and DAR contributed to the cleavage assay experimental design and analyzed the data; AGR did cellular experiments; PC analyzed the cleavage and structural data and did modeling; IAK and DAR designed the cleavage assay experiments and analyzed the data; AYS analyzed the data and drafted the paper. All authors discussed the results and contributed to writing of the manuscript.
References
- 1.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dejea C, Wick E, Sears CL. Bacterial oncogenesis in the colon. Future Microbiol. 2013;8:445–460. doi: 10.2217/fmb.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco AA, Cheng RK, Chung GT, Wu S, Oh HB, Sears CL. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J Bacteriol. 1999;181:6623–6633. doi: 10.1128/jb.181.21.6623-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obiso RJ, Jr, Lyerly DM, Van Tassell RL, Wilkins TD. Proteolytic activity of the Bacteroides fragilis enterotoxin causes fluid secretion and intestinal damage in vivo. Infect Immun. 1995;63:3820–3826. doi: 10.1128/iai.63.10.3820-3826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moncrief JS, Obiso R, Jr, Barroso LA, Kling JJ, Wright RL, Van Tassell RL, Lyerly DM, Wilkins TD. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect Immun. 1995;63:175–181. doi: 10.1128/iai.63.1.175-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomis-Ruth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotechnol. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- 8.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120:1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung GT, Franco AA, Wu S, Rhie GE, Cheng R, Oh HB, Sears CL. Identification of a third metalloprotease toxin gene in extraintestinal isolates of Bacteroides fragilis. Infect Immun. 1999;67:4945–4949. doi: 10.1128/iai.67.9.4945-4949.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kling JJ, Wright RL, Moncrief JS, Wilkins TD. Cloning and characterization of the gene for the metalloprotease enterotoxin of Bacteroides fragilis. FEMS Microbiol Lett. 1997;146:279–284. doi: 10.1016/s0378-1097(96)00488-0. [DOI] [PubMed] [Google Scholar]
- 11.Riegler M, Lotz M, Sears C, Pothoulakis C, Castagliuolo I, Wang CC, Sedivy R, Sogukoglu T, Cosentini E, Bischof G, et al. Bacteroides fragilis toxin 2 damages human colonic mucosa in vitro. Gut. 1999;44:504–510. doi: 10.1136/gut.44.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulas T, Arolas JL, Gomis-Ruth FX. Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog. Proc Natl Acad Sci U S A. 2011;108:1856–1861. doi: 10.1073/pnas.1012173108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci U S A. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiryaev SA, Remacle AG, Chernov AV, Golubkov VS, Motamedchaboki K, Muranaka N, Dambacher CM, Capek P, Kukreja M, Kozlov IA, et al. Substrate Cleavage Profiling Suggests a Distinct Function of Bacteroides fragilis Metalloproteinases (Fragilysin and Metalloproteinase II) at the Microbiome-Inflammation-Cancer Interface. J Biol Chem. 2013;288:34956–34967. doi: 10.1074/jbc.M113.516153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlov IA, Thomsen ER, Munchel SE, Villegas P, Capek P, Gower AJ, Pond SJ, Chudin E, Chee MS. A highly scalable peptide-based assay system for proteomics. PLoS One. 2012;7:e37441. doi: 10.1371/journal.pone.0037441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiryaev SA, Chernov AV, Golubkov VS, Thomsen ER, Chudin E, Chee MS, Kozlov IA, Strongin AY, Cieplak P. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PLoS One. 2013;8:e54290. doi: 10.1371/journal.pone.0054290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiryaev SA, Thomsen ER, Cieplak P, Chudin E, Cheltsov AV, Chee MS, Kozlov IA, Strongin AY. New details of HCV NS3/4A proteinase functionality revealed by a high-throughput cleavage assay. PLoS One. 2012;7:e35759. doi: 10.1371/journal.pone.0035759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratnikov BI, Cieplak P, Gramatikoff K, Pierce J, Eroshkin A, Igarashi Y, Sun Q, Godzik A, Osterman AL, Stec B, et al. Basis for substrate recognition and distinction by matrix metalloproteinases. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1406134111. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratnikov B, Cieplak P, Smith JW. High throughput substrate phage display for protease profiling. Methods Mol Biol. 2009;539:93–114. doi: 10.1007/978-1-60327-003-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gawlik K, Shiryaev SA, Zhu W, Motamedchaboki K, Desjardins R, Day R, Remacle AG, Stec B, Strongin AY. Autocatalytic activation of the furin zymogen requires removal of the emerging enzyme's N-terminus from the active site. PLoS One. 2009;4:e5031. doi: 10.1371/journal.pone.0005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Tassell RL, Lyerly DM, Wilkins TD. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect Immun. 1992;60:1343–1350. doi: 10.1128/iai.60.4.1343-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sears CL, Buckwold SL, Shin JW, Franco AA. The C-terminal region of Bacteroides fragilis toxin is essential to its biological activity. Infect Immun. 2006;74:5595–5601. doi: 10.1128/IAI.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holton J. Enterotoxigenic Bacteroides fragilis. Curr Infect Dis Rep. 2008;10:99–104. doi: 10.1007/s11908-008-0018-7. [DOI] [PubMed] [Google Scholar]
- 24.Wick EC, Sears CL. Bacteroides spp. and diarrhea. Curr Opin Infect Dis. 2010;23:470–474. doi: 10.1097/QCO.0b013e32833da1eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 27.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salama NR, Hartung ML, Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claros MC, Claros ZC, Hecht DW, Citron DM, Goldstein EJ, Silva J, Jr, Tang-Feldman Y, Rodloff AC. Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates. Anaerobe. 2006;12:17–22. doi: 10.1016/j.anaerobe.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Franco AA. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J Bacteriol. 2004;186:6077–6092. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco AA, Mundy LM, Trucksis M, Wu S, Kaper JB, Sears CL. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect Immun. 1997;65:1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kharlampieva DD, Manuvera VA, Podgorny OV, Kovalchuk SI, Pobeguts OV, Altukhov IA, Alexeev DG, Lazarev VN, Govorun VM. Purification and characterisation of recombinant Bacteroides fragilis toxin-2. Biochimie. 2013;95:2123–2131. doi: 10.1016/j.biochi.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Fletcher PL, Jr, Fletcher MD, Weninger K, Anderson TE, Martin BM. Vesicle-associated membrane protein (VAMP) cleavage by a new metalloprotease from the Brazilian scorpion Tityus serrulatus. J Biol Chem. 2010;285:7405–7416. doi: 10.1074/jbc.M109.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz E, Rendon-Anaya M, Rego SC, Schwartz EF, Possani LD. Antarease-like Zn-metalloproteases are ubiquitous in the venom of different scorpion genera. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagen.2013.12.012. in press. [DOI] [PubMed] [Google Scholar]
- 35.Wu S, Shin J, Zhang G, Cohen M, Franco A, Sears CL. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect Immun. 2006;74:5382–5390. doi: 10.1128/IAI.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative, Computational, Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, et al. Amber 12. University of California, San Francisco. 2012 [Google Scholar]
- 43.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.