Abstract
Objective
To determine and compare the frequency of cancer-associated genetic abnormalities in esophageal metaplasia biopsies with and without goblet cells.
Background
Barrett’s esophagus (BE) is associated with increased risk of esophageal adenocarcinoma (EAC) but the appropriate histologic definition of BE is debated. Intestinal metaplasia (IM) is defined by the presence of goblet cells while non-goblet cell metaplasia (NGM) lacks goblet cells. Both have been implicated in EAC risk but this is controversial. While IM is known to harbor genetic changes associated with EAC, little is known about NGM. We hypothesized that if NGM and IM infer similar EAC risk then they would harbor similar genetic aberrations in genes associated with EAC.
Methods
Ninety frozen NGM, IM, and normal tissues from 45 subjects were studied. DNA copy number abnormalities (CNA’s) were identified using microarrays and fluorescence in situ hybridization (FISH). Targeted sequencing of all exons from twenty EAC-associated genes was performed on metaplasia biopsies using Ion AmpliSeq™ DNA sequencing.
Results
Frequent CNA’s targeting cancer-associated genes were found in IM whereas no such changes were observed in NGM. In one subject, FISH confirmed loss of CDKN2A and amplification of chromosome 8 in IM but not in a nearby NGM biopsy. Targeted sequencing revealed 11 non-synonymous mutations in 16 IM samples and 2 mutations in 19 NGM samples.
Conclusions
This study reports the largest and most comprehensive comparison of DNA aberrations in IM and NGM genomes. Our results show that IM has a much higher frequency of cancer-associated mutations than NGM.
INTRODUCTION
Once a rare disease, the incidence of esophageal adenocarcinoma (EAC) has increased 300-500% in the United States and other Western Countries over the past 30-40 years1, 2. Paralleling the marked increase in incidence, death from EAC also continues to rise rapidly. EAC is known to develop through a series of metaplastic, dysplastic and neoplastic cellular changes that are largely initiated by chronic gastroesophageal reflux disease (GERD)3. GERD precipitates a metaplastic change of the esophageal epithelium from the normal squamous to a more acid-resistant columnar epithelium known as Barrett’s esophagus (BE). BE is estimated to be present in approximately 1.6%4 of adults in Western populations (up to 4 million Americans)5, 6 and progression to EAC is estimated to occur in 0.12-0.5% of patients per year5, 7-9. Consequently, individuals with BE are recommended to undergo regular endoscopic surveillance with multiple biopsies to identify signs of dysplasia or EAC10. The appropriate definition of BE however is currently a matter of considerable debate. In the United States, specialized intestinal metaplasia (IM) containing histologically confirmed goblet cells is required for a diagnosis of BE11 while in the United Kingdom, the presence of goblet cells is not currently required for the diagnosis of BE12.
Historically, the presence of IM has been associated with EAC risk but recent studies have indicated that metaplasia without goblet cells (NGM) may also be pre-malignant and may carry a neoplastic progression risk similar to that of IM13, 14. If correct patients with NGM would also be candidates for BE surveillance programs, the cost-effectiveness of which is often criticized given the low rate of progression from BE to cancer15, 16. Unfortunately, the cancer-risk of NGM relative to IM remains controversial17, 18. One thing that is known however, is that EAC arises through accumulation of multiple DNA alterations (mutation, DNA copy number abnormality, methylation), many of which are also observed in early dysplastic lesions19-21. Furthermore, IM contains frequent alterations in known cancer-associated genes (TP53, CDKN2A, FHIT, WWOX)20, 22, 23 and these DNA alterations are presumably the underlying cause for the increased cancer risk associated with IM24. On the other hand, very little is known about DNA alterations in NGM with ploidy changes being the only possible aberrations noted to date25. Thus, the goal of this study was to determine and compare the spectrum and frequency of DNA copy number alterations and cancer-associated point mutations in NGM and IM. This was performed on fresh-frozen esophageal biopsy specimens using a combination of Affymetrix SNP 6.0 microarrays, targeted re-sequencing of cancer-associated genes and fluorescence in situ hybridization.
MATERIALS & METHODS
Subject cohort
Fresh frozen esophageal mucosa specimens including non-goblet metaplasia (NGM), intestinal metaplasia (IM), and normal squamous epithelium (NSE) from 45 subjects were used in this study. Biopsies were collected from 41 subjects who underwent upper endoscopy at the University of Rochester Medical Center between 2004-2010 and mucosal tissue pieces were obtained from four subjects who underwent esophagectomies at the University of Pittsburgh Medical Center between 2003-2008. For each subject, the most advanced disease state (EAC > dysplasia > IM > NGM) prior to, or at the time of tissue collection was noted (Table 1). All subjects at both institutions provided informed consent and research was approved by appropriate IRB’s. Additional information on subjects and specimens is provided in Table 1.
Table 1. Subject cohort demographics.
Fifty-seven tissues (indicated by specimen ID’s; SIDs) from 45 subjects (indicated by patient ID’s; PIDs) were included in this study. Gender, subject’s highest disease state, specimen location, and consensus tissue type for the metaplastic tissues are listed. Specimens included in the SNP array and Ion AmpliSeq re-sequencing specimens are indicated along with goblet-cell (GC) score (0= none, 1= rare, 2 = common, 3 = numerous) for each tissue and the NGM sub-types. PID= patient ID, SID= Specimen ID; GEJ= Gastro-esophageal junction; Eso= Esophagus; NGM= non-goblet metaplasia; IM= intestinal metaplasia; CM= cardiac mucosa; OCM= oxynto-cardiac mucosa; CM-OCM= Mixture of CM and OCM type.
| Patient Information | Specimen Information | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PID | Gender | Advanced Disease State |
SID | Location | Tissue type | SNP | Ion Torrent |
GC Score |
NGM Sub-type |
| 4 | M | EAC | UR109 | GEJ | NGM | Y | Y | 0 | CM |
| 12 | M | IM | UR28 | GEJ | Composite | Y | Y | 3,0 | CM |
| 18 | M | LGD | UR58 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 27 | M | EAC | UR550 | Eso | NGM | Y | - | 0 | OCM |
| 31 | M | LGD | UR84 | Eso | IM | Y | Y | 3 | N/A |
| 36 | M | HGD | UR99 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 37 | F | IM | UR103 | Eso | Composite | Y | Y | 3,0 | N/A |
| 39 | M | EAC | UR117 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 41 | M | LGD | UR122 | Eso | IM | Y | Y | 3 | N/A |
| 43 | M | HGD | UR194 | Eso | NGM | Y | Y | 0 | OCM |
| 44 | F | IM | UR309 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 45 | F | NGM | UR128 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| 50 | M | LGD | UR151 | GEJ | IM | Y | Y | 3 | N/A |
| UR152 | GEJ | NGM | Y | Y | 0 | CM-OCM | |||
| UR158 | GEJ | NGM | Y | Y | 0 | CM-OCM | |||
| 55 | M | LGD | UR159 | GEJ | Composite | Y | Y | 3,0 | CM |
| UR162 | Eso | IM | Y | Y | 3 | N/A | |||
| 56 | M | EAC | UR165 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| 59 | M | EAC | UR218 | Eso | IM | Y | Y | 3 | N/A |
| 61 | M | LGD | UR301 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 63 | M | IM | UR342 | Eso | NGM | Y | Y | 0 | OCM |
| 66 | M | EAC | UR182 | GEJ | NGM | Y | - | 0 | CM-OCM |
| 67 | M | EAC | UR199 | Eso | IM | Y | Y | 3 | N/A |
| UR221 | GEJ | Composite | Y | Y | 3,0 | CM | |||
| 68 | M | IM | UR222 | Eso | NGM | Y | Y | 0 | CM-OCM |
| UR223 | Eso | IM | Y | Y | 3 | N/A | |||
| 87 | F | IM | UR269 | Eso | IM | Y | Y | 3 | N/A |
| UR286 | Eso | Composite | Y | Y | 1,0 | CM | |||
| 89 | M | IM | UR289 | Eso | IM | Y | Y | 3 | N/A |
| 92 | M | HGD | UR649 | Eso | IM | Y | Y | 3 | N/A |
| UR274 | Eso | NGM | Y | Y | 0 | OCM | |||
| 94 | M | HGD | UR284 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| 95 | M | IM | UR291 | GEJ | NGM | Y | Y | 0 | CM |
| UR627 | Eso | Composite | Y | - | 3,0 | CM | |||
| 104 | M | HGD | UR771 | Eso | NGM | Y | - | 0 | CM-OCM |
| UR773 | Eso | IM | Y | - | 3 | N/A | |||
| 111 | F | LGD | UR482 | Eso | IM | Y | Y | 2 | N/A |
| 116 | M | HGD | UR369 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| 124 | F | HGD | UR402 | Eso | IM | Y | - | 3 | N/A |
| UR582 | Eso | NGM | Y | - | 0 | CM-OCM | |||
| 165 | M | HGD | UR762 | Eso | IM | Y | - | 2 | N/A |
| 169 | M | EAC | UR637 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| UR776 | Eso | IM | Y | Y | 3 | N/A | |||
| 172 | M | LGD | UR653 | Eso | IM | Y | - | 1 | N/A |
| 188 | M | HGD | UR727 | GEJ | NGM | Y | Y | 0 | CM-OCM |
| UR931 | Eso | IM | Y | Y | 3 | N/A | |||
| 209 | M | HGD | UR830 | Eso | NGM | Y | - | 0 | CM |
| 211 | M | IM | UR840 | Eso | IM | Y | - | 3 | N/A |
| 213 | M | EAC | UR242 | Eso | NGM | Y | Y | 0 | CM-OCM |
| 217 | M | HGD | UR881 | GEJ | IM | Y | - | 3 | N/A |
| 221 | M | LGD | UR937 | Eso | IM | Y | - | 2 | N/A |
| 222 | M | HGD | UR946 | Eso | IM | Y | - | 1 | N/A |
| 261 | M | LGD | UR1137 | Eso | IM | Y | - | 3 | N/A |
| UP1034 | M | EAC | 4938 | Eso | IM | Y | Y | 3 | N/A |
| UP1064 | M | EAC | 4936 | Eso | IM | Y | Y | 2 | N/A |
| UP1066 | M | EAC | 4937 | Eso | IM | Y | Y | 3 | N/A |
| UP163 | M | EAC | 1881 | Eso | IM | Y | Y | 1 | N/A |
Specimen Screening
The location of each biopsy/tissue piece was noted as either tubular esophagus or gastroesophageal junction (GEJ) at the time of endoscopy (or on the resection specimen). Specimens were immediately frozen in tissue cassettes containing optimal cutting temperature compound (OCT) to facilitate histologic review. Hematoxylin and eosin (H&E) stained sections of each specimen were independently screened by two pathologists (Z.Z, K.S). Tissues were classified as NSE, NGM or IM. IM was defined by the presence of any number of goblet cells (GCs) but goblet cell density was also recorded using a scale of 1-3 with 1= rare GCs, 2= common GCs and 3 = numerous GCs (Table 1). NGM was defined as columnar cell metaplasia without GCs in the entire specimen. NGM was further classified into two subgroups of mucosa: oxyntocardiac mucosa (OCM) and cardiac mucosa (CM). OCM consisted of both cardiac and oxyntic glands and CM consisted of only cardiac mucosa without parietal cells (Table 1).
In some cases, more than one biopsy from the same esophageal level (distance from the incisors) was included in a single OCT block. When the histology of these two biopsies was different (one IM and one NGM), the tissue block was referred to as being a composite sample (Supplemental Figure 1C). It is important to note that composite samples consist of two separate biopsies that were typically separated by 1-2cm in esophageal location and are thus independent tissue pieces.
Specimen Selection
Specimens were selected for study only if diagnostic consensus was reached between the two pathologists and if the sample consisted of >70% tissue of interest with <30% NSE or submucosa. Furthermore, OCM samples were only included if the endoscopic location was tubular esophagus. OCM samples from the GEJ were excluded to avoid the possibility that they were actually oxyntic mucosa from the stomach. CM (and IM) samples were included from both GEJ and tubular esophagus as the histologic distinction between CM and oxyntic mucosa from the stomach is more easily made. In all cases, both pathologists were confident that the biopsies included for study represented esophageal metaplasia.
Final Histologic Review and DNA isolation
Selected specimens were cut on a cryostat (20-30 × 5μM sections) for DNA isolation. The first, middle and final sections were placed on slides, H&E stained and reviewed by both pathologists. Genomic DNA was isolated from the intervening tissue sections using the QiaAmp DNA Mini Kit (Qiagen, CA). DNA concentration and purity was assessed by UV absorbance (NanoDrop 1000, Thermo Fisher Scientific, Waltham MA) and the DNA quality was assessed by gel electrophoresis.
Genome-wide Human SNP 6.0 Assay
Fifty-eight specimens of NGM, IM, or composite histology were analyzed for DNA copy number abnormalities using the Affymetrix SNP 6.0 GeneChip arrays (Affymetrix Inc, CA). DNA labeling, array hybridization, washing and staining was carried out according to the standard Genome-Wide Human SNP Assay Kit instructions (Affymetrix Inc. CA) at the SUNY Upstate Medical University microarray core facility (Syracuse, NY).
DNA Copy number analysis
Data analysis was performed in Nexus 6.0 Copy number analysis software (BioDiscovery, CA). All samples were normalized to a baseline reference file created using DNA from blood of 15 individuals from a similar subject population. The data was pre-processed and the log2 DNA copy number ratios for each sample were generated for each probeset on the array using the reference files. The SNP-Rank Segmentation algorithm (implemented in Nexus 6.0) was used to segment the data with a minimum requirement of 8 probesets and a significance threshold p-value of 10−6. Log2 copy number thresholds for gains and losses were set at +0.15 (~2.2 copies) and −0.2 (~1.7 copies) respectively while high level gains and homozygous loss were set at +0.5 (~2.8 copies) and −0.8 (~1.1 copies) respectively.
Fluorescence in situ hybridization (FISH)
For FISH analysis, frozen tissue samples were first formalin-fixed and paraffin-embedded and 4 μM sections were then cut and fixed on positively charged slides. H&E stained sections were reviewed by the pathologists to ensure that the reoriented tissue was the same histology as determined prior to the genomic studies. FISH for CDKN2A (9p21.3) loss and chromosome 8 gain was performed in the Department of Cytogenetics FISH Core Facility (University of Rochester Medical Center, NY) using a VP2000 processor (Abbott Molecular, IL) and standard protocols. Probes for CDKN2A with Spectrum Orange label in combination with an internal control centromere 9 (CEP9) with Spectrum Green label (Cat. No: 05J51-001) and centromere 8 (CEP8) with Spectrum Green label (Cat. No: 06J37-018) were purchased from Abbott Molecular (Des Plaines, IL).
FISH Analysis
Manual enumeration of the FISH signals was performed using a Nikon eclipse 80i microscope (Nikon Instruments Inc., NY) and Cytovision software (Leica Microsystems, IL) under 60x oil immersion lens. Signals from overlapping nuclei were not included and at least 100 nuclei per biopsy were counted. The number of CDKN2A and CEP9 signals or the number of CEP8 signals were recorded for each cell nucleus.
Targeted re-sequencing of genes frequently mutated in EAC
A subset of metaplasia samples including 19 NGM, 16 IM and 5 composite tissues (Table 1) along with matched NSE specimens were analyzed using targeted re-sequencing of 20 genes that are frequently somatically mutated in EAC. These 20 genes were identified from a preliminary analysis of whole exome sequencing data on 56 EAC tumor specimens (Dulak et al. Nature Genetics in press). Targeted re-sequencing libraries were generated using the Ion AmpliSeq™ Technology (Life Technologies, CA). Briefly, 1395 PCR amplicons (no longer than 140 bp) were designed to cover all exons from these 20 genes (>90kb of total DNA) using the Ion AmpliSeq Designer pipeline (Life Technologies, CA). Primers were pooled for two multiplex PCR pre-amplification reactions each with 10ng of input DNA using reagents provided by Ion Torrent. After 14-16 PCR cycles, the product was partially digested to polish the ends and adapter sequences were added for multiplexing. After an additional 5-cycle library amplification step, the libraries were combined with Ion Sphere™ particles for clonal amplification using emulsion PCR. The Ion Sphere™ particles, DNA polymerase and sequencing primers were then loaded into Ion AmpliSeq sequencing chips for sequencing on Ion Personal Genome Machine™ (PGM™) sequencer26. Raw sequence reads were then processed using the Ion AmpliSeq Software Suite and mapped to the hg19 reference sequence. Sequence data from metaplasia samples was compared with subject-matched normal DNA in order to filter sequence polymorphisms and identify variants. Sequence variants were identified using Ion AmpliSeq Variant caller software followed by manual review using the Integrated Genome Viewer (IGV) (http://www.broadinstitute.org/igv/).
PCR amplification and Sanger sequencing of CDKN2A exon 2
Exon 2 from CDKN2A was PCR amplified using forward (5′-GACGACAGCTCCGCAGAAGTT-3′) and reverse primers (5′-TCTGAGCTTTGGAAGCTCTCAGG-3′) flanking the 5′ and 3′ ends of the exon 2. The PCR reaction consisted of 1X GC buffer, 0.25mM each of dNTP mix, 2μM each of forward and reverse primers, 1.25X of Advantage® GC polymerase mix (Cat. No: 639114, Clontech Laboratories Inc., CA), and 50ng genomic DNA in a final volume of 20ul. PCR conditions included an initial hot start at 94°C for 3 minutes followed by 40 cycles of denaturation at 94°C for 30 seconds and annealing/renaturation at 70°C for 1 minute and with a final hold at 25°C. Gel electrophoresis was performed on a 2% agarose gel to check for amplified DNA products. PCR products were then purified using ExoSAP-IT (Affymetrix, CA) and sequencing was performed with the PCR primers at Bio Basic Inc. (Ontario, Canada) on an ABI 3730XL sequencer (Life Technologies, CA). Chromatograms were assembled in DNA Baser Sequence Assembler v2 (Heracle BioSoft SRL, http://www.DnaBaser.com) followed by multiple sequence alignment in ClustalW 2.0 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) using the RefSeq entry NM_000077.4 as the reference sequence.
RESULTS
Tissue Pathology
Ninety fresh frozen tissues (82 biopsies and 8 resection specimens) from 45 subjects met all criteria and were included in the study (73 from the tubular esophagus and 17 from the GEJ; Table 1). Pathology review classified the samples as IM (n=26), NGM (n=25), composite (n=6) and NSE (n=33). Within this tissue set, multiple specimens (n= 21) were obtained from 9 subjects (indicated in Table 1) while the remaining 69 samples were from independent subjects.
Subject Cohort Characteristics
The subject cohort was predominantly male (39 males vs 6 females) with an average age of 67 years. Prior to, or at the time of biopsy collection for this study, 13 subjects had a diagnosis of EAC, 22 had dysplasia, 9 had non-dysplastic IM and 1 subject had NGM only with no IM found (Table 1).
DNA Copy Number Aberrations in NGM and IM
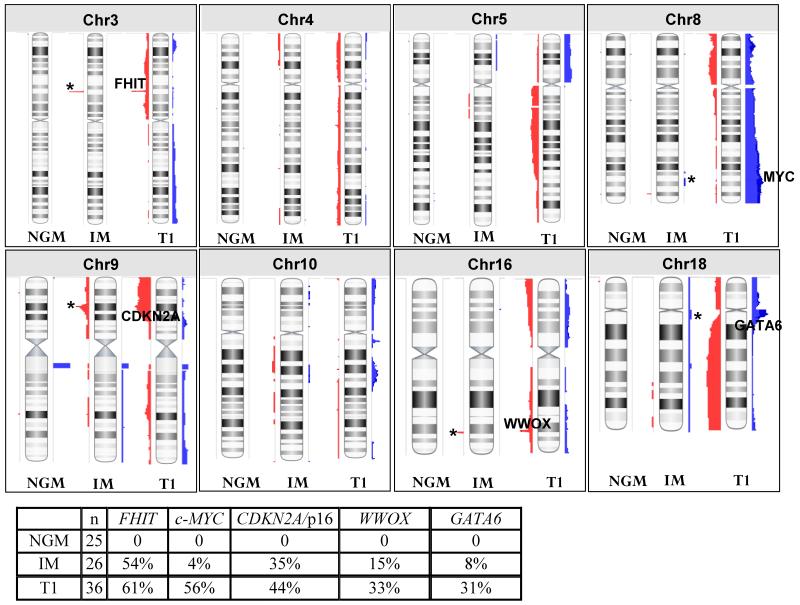
DNA copy number analysis was performed on 25 NGM, 26 IM and 6 composite specimens. For comparison, we also included previously analyzed data from 36 early stage (T1) esophageal adenocarcinoma specimens (Dulak et al. 201227; Supplemental Figure 2). The 26 pure IM samples harbored a total of 73 regions of copy number aberrations. The median length of these regions was 667035bp (range 29256-28588324bp). Sixty-four out of 73 regions contain a total of 1935 unique genes (Supplemental Table 1). Consistent with previous reports20, 28 recurrent aberrations were observed that target well known cancer-associated genes (Figure 1). These include deletions at 3p14 (FHIT; 54% of samples), 9p21.3 (CDKN2A; 35%) and 16q23.1 (WWOX; 15%) and amplifications at 8q24 (c-MYC; 4%) and 18q11.2 (GATA6; 8%). In addition, other aberrations were observed in IM samples that appear to match those occurring in T1 EAC samples although the specific genes driving these events are not well defined. These include regional deletions on 4p and 4q, amplification on 5q and deletion on 5p, deletion on 9q and amplifications on both 10p and 10q (Figure 1).
Figure 1. Comparison of copy number alterations between 25 non-goblet metaplasia (NGM), 26 intestinal metaplasia (IM), and 36 T1 adenocarcinoma genomes.
Copy number gains are represented in blue while losses are in red. Known cancer associated genes are indicated on the chromosomes for the T1 genomes and their alteration frequencies are indicated in the accompanying table. Presence of these changes in NGM and/or IM genomes is indicated by ‘*’. Complete genomes are provided in Supplemental Figure 2.
In the 25 NGM samples, 40 regions of copy number aberrations were observed with a median length of 218086bp (range 23691-3781221bp). Twenty-nine regions contain a total of 297 unique genes (Supplemental Table 2 and Supplemental Figure 2) none of which appear to have any previous association with EAC. Six recurrent regions were identified but all have 100% overlap with regions of known copy number variation (CNV) and therefore likely represent copy number polymorphism29 rather than true aberrations (Supplemental Table 2). All other changes observed occurred in only one sample each.
Comparison of NGM and IM from the same subjects
From 9 subjects, specimens with different histologies were available for study. This included 5 subjects (PID’s: 50, 92, 124, 169,188) with both IM and NGM, 1 subject (PID: 89) with both composite and IM and 3 subjects (PID’s: 55, 68 and 104) with composite, IM and NGM. In the 5 subjects with IM and NGM specimens, DNA copy losses were identified at the FHIT locus in 4 cases, at the CDKN2A locus in 3 cases and at the WWOX locus in 1 case. In all 5 subjects, these losses were observed in the IM sample only and not in the matched NGM sample (Supplemental Figure 3). In subject 89, losses at the FHIT and WWOX genes were observed in the IM sample in addition to losses on chromosomes 5 and 10, but no changes were seen in the composite sample (Supplemental Figure 3).
In the 3 subjects with IM, NGM and composite samples multiple DNA copy losses and gains were identified in IM and composite samples but not in the NGM samples. For subject 68, loss was observed at the CDKN2A locus in the IM and composite samples but not in the NGM sample. Based on the boundaries of the deleted region, the loss in these two tissues appears to be clonally related. However, the composite sample also has an amplification event on chromosome 15 that is not observed in the IM or NGM samples. Similarly, for subject 104, clonally appearing losses of FHIT, chromosome 9 (including CDKN2A but also in other regions) and chromosome 18 were found in the IM and composite samples but not in the NGM sample. Chromosome 10 on the other hand had multiple regions of loss in the IM sample but regions of gain in the composite indicating some clonal divergence. Finally, subject 55 had a loss of CDKN2A in both IM and composite samples in addition to a gain of whole chromosome 8 and FHIT loss only in the composite sample (Supplemental Figure 3).
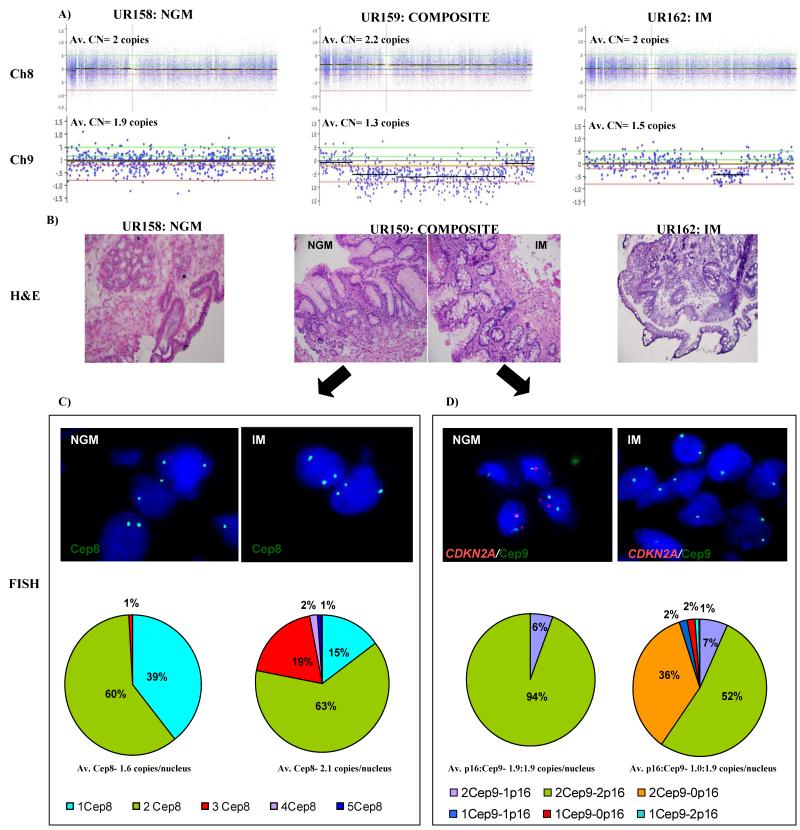
For tissues from subject 55, we used fluorescence in situ hybridization (FISH) to determine if the alterations observed in the composite sample (UR159) were present in the IM or the NGM biopsy. The SNP array results from the composite specimen (UR159) of subject 55 indicates a gain of chromosome 8 and loss at the CDKN2A locus on chromosome 9 (Figure 2A). The FISH results clearly show that both of these events are present only in the IM biopsy of the composite sample. For the chromosome 8 amplification, 60% of nuclei in the NGM portion had two copies of CEP8, 39% had 1 copy and 1% had 3 copies. This is almost identical to the control NSE sample (Supplemental Figure 4). However, in the IM biopsy 63% of nuclei had two copies, 15% had one copy while 22% had three or more copies of CEP8. For CDKN2A/CEP9, 94% of nuclei in the NGM biopsy had two copies of both CDKN2A and CEP9 while 6% had two copies of CEP9 and one copy of CDKN2A. In the IM biopsy 52% of nuclei had two copies each of CDKN2A and CEP9, 7% had two copies of CEP9 and one copy of CDKN2A while 36% had two copies of CEP9 but no copies (homozygous deletion) of CDKN2A (Figure 2).
Figure 2. Validation of genomic changes using fluorescence in situ hybridization (FISH) in composite sample (UR159) from subject 55.
(A) Probe level copy number data showing the gain of entire chromosome 8 in the composite specimen (UR159) and loss of CDK2NA loci in composite (UR159) and intestinal metaplasia (IM; UR162) specimens compared to the NGM specimen (UR158). Y-axis represents the log2 signal ratio where log20 = 2 copies (baseline). Average copy number per locus is indicated above each plot. The two green and red lines above and below the baseline respectively indicate the gain/high copy gain and loss/homozygous loss thresholds. (B) Representative H & E images for NGM (UR158), composite (UR159), and IM (UR162) taken at 20X magnification. (C) & (D) Representative fluorescence in situ hybridization (FISH) images of the NGM and IM portions of the composite tissue (UR159). Pie-charts summarize the result into different categories of normal (green) and abnormal signals (other colors as indicated) observed in each biopsy for gain of chromosome 8 (C) and loss of CDKN2A (D). Av.= average; CN= copy number; Cep8= centromere8; Cep9= centromere 9; p16= CDKN2A
Targeted re-sequencing of frequently mutated EAC genes in NGM and IM
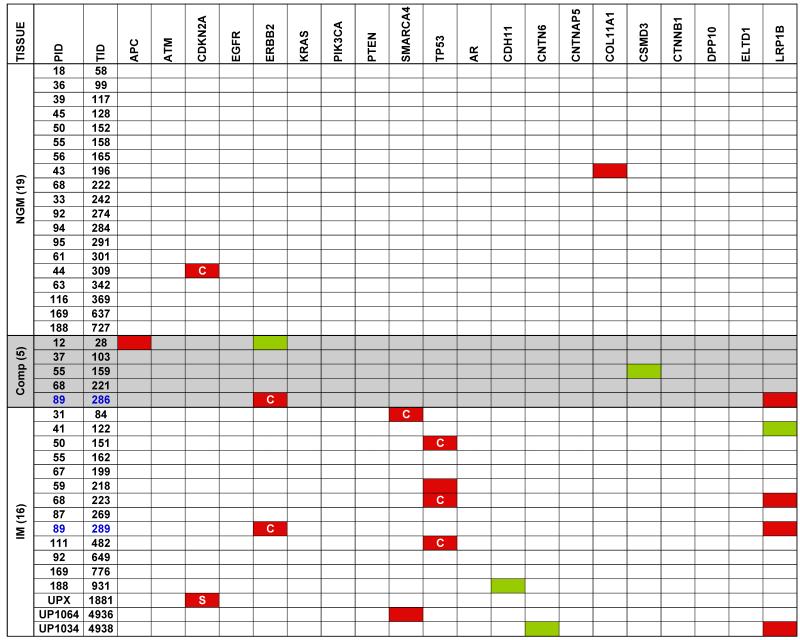
In order to identify cancer-associated point mutations in NGM and IM, we used a highly multiplexed PCR amplification approach to re-sequence 20 genes that are frequently mutated in EAC based upon an interim analysis of an emerging study of the spectrum of mutations in this disease. This focused sequencing was performed for 19 NGM, 16 IM, and 5 composite samples along with their matched normal samples. This analysis revealed a total of 16 non-synonymous (amino acid modifying) and 5 synonymous (no change to the amino acid) mutations (indicated in Figure 3). Two of these changes were observed in independent NGM samples (2/19; 10.5%) and both were non-synonymous mutations (in CDKN2A and COL11A1). A total of 14 mutations were observed in the IM samples. Eleven of these were non-synonymous changes that occurred in 9 (53%) independent samples with two samples each having mutations in two different genes. The genes with non-synonymous mutations in the IM samples were TP53 (n=4; 25%), SMARCA4 (n=2; 13%), LRP1B (n=3; 19%), ERBB2 (n=1; 6%) and CDKN2A (n=1, identified by Sanger sequencing; 6%). Finally, three non-synonymous mutations were identified in the composite samples including a mutation in APC in one sample and mutations in ERBB2 and LRPB1 in another sample (sample UR286 from subject 89) (Figure 3). Interestingly, the IM sample (UR289) from the same subject had identical ERBB2 and LRPB1 mutations indicating clonality. Furthermore, the mutant fraction observed in the IM sample was almost exactly double that seen in the composite for both mutations.
Figure 3. Targeted re-sequencing of 20 frequently mutated EAC genes from pre-neoplastic biopsies.
Nineteen NGM, 5 composites, and 16 IM specimens from 36 subjects were analyzed along with matched normal squamous mucosa. Mutations in the coding regions within the specific genes are indicated in red (non-synonymous mutation) and green (silent substitutions) while ‘C’ indicates the mutation is reported in the COSMIC database and ‘S’ indicates the point mutation was identified by Sanger sequencing.
Mutation Load Discriminates Between Metaplasia and EAC
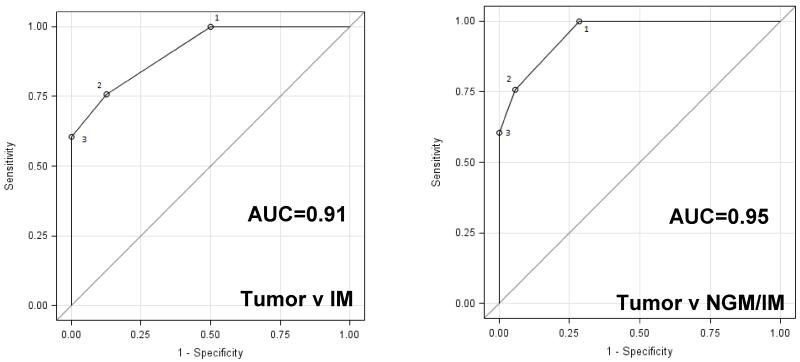
Detecting progression of metaplasia to EAC (or dysplasia) is the goal of surveillance in subjects with BE. Therefore, we evaluated the discriminatory capacity of mutation load (number of mutations per sample in the 20-gene panel) to detect this event. Data from whole exome sequencing of 66 EAC samples was used for this analysis along with the targeted re-sequencing data from metaplasia samples. A pairwise Receiver-Operator Characteristic (ROC) analysis resulted in an area under the curve (AUC) of 0.954 for any metaplasia (NGM or IM) versus EAC and 0.915 for IM alone versus EAC (Figure 4). Thus, mutation load appears to have good potential for detecting disease progression in subjects with BE and this approach may be applied to esophageal cytology samples as well as biopsies.
Figure 4. Power of mutation load to discriminate between metaplasia and EAC.
Pairwise Receiver-Operator Characteristics (ROC) curves analysis was performed for Tumor v IM and Tumor v any metaplasia (NGM/IM) using the 20-gene AmpliSeq mutation data from NGM and IM samples along with mutation data from 66 EAC tumor samples obtained from whole exome sequencing.
DISCUSSION
These data report the largest and most comprehensive comparison of cancer-associated genetic changes in NGM and IM. We have carefully controlled the collection, preparation and histologic characterization of the specimens. The location of biopsies with respect to the GEJ was carefully noted, tissues were snap frozen in OCT to facilitate sectioning and histologic analysis and multiple sections of the same specimen used for DNA isolation were examined independently by two pathologists. Using high resolution DNA copy number analysis of the whole genome we identified numerous genetic changes in IM epithelium. These findings are in agreement with previous studies and reveal that the changes frequently target well known cancer-associated genes and regions which are similarly altered in early stage EAC. In contrast, we found far fewer DNA copy number changes in NGM. When present in NGM, abnormalities were commonly in regions with established copy number polymorphisms, were smaller than those in IM and often did not contain any known genes. Furthermore, when analyzing multiple tissues from the same subject, IM samples were found to have copy number changes while the matched NGM samples did not. This was true for at least one subject where NGM and IM biopsies from the same level (distance from the incisors) were analyzed using FISH. In these tissues, chromosome 8 gain and CDKN2A loss were found only in the IM biopsy and not in the adjacent NGM biopsy.
We further refined the findings with targeted re-sequencing of the entire coding region of 20 cancer-associated genes frequently mutated in EAC. This was achieved using novel technology (AmpliSeq™) capable of assembling customized next-generation sequencing libraries starting with very low DNA input. Eleven non-synonymous mutations in 16 IM samples were identified including mutations in the TP53 and CDKN2A genes as previously reported. We also identified non-synonymous mutations in other cancer-associated genes not previously reported, including ERBB2, SMARCA4 and LRP1B. Many of the observed mutations in IM tissues are present in the COSMIC database, indicating that they may be drivers of cancer risk. As with DNA copy number changes, point mutations in NGM were much less common than in IM with only 2/19 samples each having a single mutation. Interestingly one of these mutations was in the CDKN2A gene and this specific mutation is also in the COSMIC database. Taken together these data show that NGM harbors far fewer cancer-associated genetic aberrations than IM. This was true even though the NGM samples were collected from an extremely “high-risk” cohort of subjects with most having coincident dysplasia or EAC.
Since the recognition of the columnar lined esophagus (CLE) in the 1950’s and subsequent studies drawing a link between metaplastic esophageal epithelium and EAC, the diagnosis of BE has become synonymous with risk of developing EAC. It is now recognized that esophageal columnar metaplasia is not a simple or single entity but can be classified into a variety of sub-types depending on the type and distribution of specific cell types (e.g. parietal cells, chief cells, mucous cells, goblet cells, Paneth cells and pancreatic acinar cells) and/or the presence and structure of sub-mucosal glands. Among these, the presence or absence of goblet cells has emerged as the most clinically relevant as it identifies intestinal metaplasia (IM), currently required for the definition of BE in the United States and many other countries. The diagnosis of BE triggers recommendations to enter endoscopic surveillance programs aimed at detecting progression to dysplasia or EAC. Of significance, the requirement for presence of goblet cells in the definition has recently been challenged as a result of studies implicating non-goblet cell metaplasia (NGM) in the development of EAC (reviewed in Riddell 200918). The requirement of the goblet cell to define BE, and its inferred cancer risk, remains a highly controversial topic and one that carries major clinical and economic implications.
Cancer develops as a result of genetic abnormalities that drive transformation and progression. Numerous studies have also shown the presence of genetic changes (mutation, DNA copy number changes, methylation etc.) in samples of IM20, 22, 28. These include changes in well-known cancer genes including TP53, CDKN2A/p16, APC, SMAD4, FHIT and WWOX. Many of these changes have also been associated with a markedly increased risk of progression to dysplasia or EAC19, 30, 31. Specifically, loss or mutation of TP53 and CDKN2A, aneuploidy and the extent of clonal diversity observed in IM are all generally accepted biomarkers for increased risk of progression3, 19, 24. In contrast, only one study has specifically analyzed DNA changes in NGM25. DNA content abnormalities, DNA heterogeneity index, DNA exceeding 5N and aneuploidy were reported to occur with equal frequency and extent in NGM and BE. In another study, Chaves and colleagues reported that gains of chromosome 7 and 18 are more common in goblet cells than in columnar cells32. Although this study that has been quoted as evidence for genetic abnormalities in NGM18, it should be noted that all specimens used in the Chaves study were clearly described as being IM. As such this study does not globally identify changes in NGM but rather shows that at the cellular level within intestinalized epithelia (IM), the frequency of genetic changes is lower in the goblet cells than in the surrounding columnar cells. These reports represent what is very limited data on the genetic abnormalities present in NGM epithelia.
Genomic analyses of the scale performed here are currently not economically feasible for clinical testing, particularly considering the need for multiple biopsies performed every one-two years, nor do they address the issue of sampling error. It is feasible however that adding genetic analysis to the recently reported use of a Cytosponge33, 34 to collect esophageal cytology specimens may prove clinically useful. For example, given the identification of IM via a screening cytology analysis, a paired mutation analysis could provide a baseline estimate of progression risk (regardless of IM or NGM status). Abnormal cytology or a high incidence of mutations could also trigger endoscopy. Repeat cytology samples could be collected and mutation analysis used to detect/monitor (not predict) progression from baseline and triage subjects to endoscopy. Even with the limited set of 20 genes analyzed, our data clearly show that mutation load can differentiate metaplasia from EAC. Additional studies would be required to identify an optimal gene panel, evaluate mutation load in dysplasia and develop appropriate classification algorithms. Next generation sequencing technologies are perfect for this type of test as they are capable of detecting very rare mutation events in the large background of normal cell DNA present in cytology specimens. Furthermore, the 20-gene assay presented here was designed (and has been tested) to work on highly fragmented DNA such as that obtained from routinely fixed and processed clinical specimens. Perhaps the question to ask when determining effective management strategies for subjects with esophageal columnar metaplasia should no longer be the presence or absence of goblet cells but rather the spectrum and frequency of mutations present in each individual case.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the support provided by the Department of Cytogenetics FISH Core Facility at the University of Rochester Medical Center and by Michael Thornton, Rob Bennett, Ben Kong, Iris Casuga and David Joun at Life Technologies.
GRANT SUPPORT: This work was supported in part by NIH grant R01 CA130853 (TEG).
Footnotes
CONFLICTS OF INTEREST: DR, S-MC and C-YL are employed by Life Technologies and helped develop the Ion AmpliSeq™ technology. All other authors have no conflicts.
References
- 1.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–7. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid BJ, Li X, Galipeau PC, et al. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–31. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 5.Jankowski J, Barr H, Wang K, et al. Diagnosis and management of Barrett’s oesophagus. BMJ. 2010;341:c4551. doi: 10.1136/bmj.c4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankowski JA, Harrison RF, Perry I, et al. Barrett’s metaplasia. Lancet. 2000;356:2079–85. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 7.Hvid-Jensen F, Pedersen L, Drewes AM, et al. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–83. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 8.Bhat S, Coleman HG, Yousef F, et al. Risk of malignant progression in Barrett’s esophagus patients: results from a large population-based study. J Natl Cancer Inst. 2011;103:1049–57. doi: 10.1093/jnci/djr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai TK, Krishnan K, Samala N, et al. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett’s oesophagus: a meta-analysis. Gut. 2012;61:970–6. doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 10.American Gastroenterological A. Spechler SJ, Sharma P, et al. American Gastroenterological Association medical position statement on the management of Barrett’s esophagus. Gastroenterology. 2011;140:1084–91. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 11.Wang KK, Sampliner RE. Practice Parameters Committee of the American College of G. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett’s esophagus. Am J Gastroenterol. 2008;103:788–97. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 12.Playford RJ. New British Society of Gastroenterology (BSG) guidelines for the diagnosis and management of Barrett’s oesophagus. Gut. 2006;55:442. doi: 10.1136/gut.2005.083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelty CJ, Gough MD, Van Wyk Q, et al. Barrett’s oesophagus: intestinal metaplasia is not essential for cancer risk. Scand J Gastroenterol. 2007;42:1271–4. doi: 10.1080/00365520701420735. [DOI] [PubMed] [Google Scholar]
- 14.Gatenby PA, Ramus JR, Caygill CP, et al. Relevance of the detection of intestinal metaplasia in non-dysplastic columnar-lined oesophagus. Scand J Gastroenterol. 2008;43:524–30. doi: 10.1080/00365520701879831. [DOI] [PubMed] [Google Scholar]
- 15.Inadomi JM. Surveillance in Barrett’s esophagus: a failed premise. Keio J Med. 2009;58:12–8. doi: 10.2302/kjm.58.12. [DOI] [PubMed] [Google Scholar]
- 16.Inadomi JM, Sampliner R, Lagergren J, et al. Screening and surveillance for Barrett esophagus in high-risk groups: a cost-utility analysis. Ann Intern Med. 2003;138:176–86. doi: 10.7326/0003-4819-138-3-200302040-00009. [DOI] [PubMed] [Google Scholar]
- 17.Chandrasoma P, Wickramasinghe K, Ma Y, et al. Is intestinal metaplasia a necessary precursor lesion for adenocarcinomas of the distal esophagus, gastroesophageal junction and gastric cardia? Dis Esophagus. 2007;20:36–41. doi: 10.1111/j.1442-2050.2007.00638.x. [DOI] [PubMed] [Google Scholar]
- 18.Riddell RH, Odze RD. Definition of Barrett’s esophagus: time for a rethink--is intestinal metaplasia dead? Am J Gastroenterol. 2009;104:2588–94. doi: 10.1038/ajg.2009.390. [DOI] [PubMed] [Google Scholar]
- 19.Ong CA, Lao-Sirieix P, Fitzgerald RC. Biomarkers in Barrett’s esophagus and esophageal adenocarcinoma: predictors of progression and prognosis. World J Gastroenterol. 2010;16:5669–81. doi: 10.3748/wjg.v16.i45.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Galipeau PC, Sanchez CA, et al. Single nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett’s esophagus neoplastic progression. Cancer Prev Res (Phila) 2008;1:413–23. doi: 10.1158/1940-6207.CAPR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai LA, Paulson TG, Li X, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007;46:532–42. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 22.Paulson TG, Maley CC, Li X, et al. Chromosomal instability and copy number alterations in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–14. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulson TG, Galipeau PC, Xu L, et al. p16 mutation spectrum in the premalignant condition Barrett’s esophagus. PLoS One. 2008;3:e3809. doi: 10.1371/journal.pone.0003809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid BJ. Early events during neoplastic progression in Barrett’s esophagus. Cancer Biomark. 2010;9:307–24. doi: 10.3233/CBM-2011-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Hahn H, Odze RD, et al. Metaplastic esophageal columnar epithelium without goblet cells shows DNA content abnormalities similar to goblet cell-containing epithelium. Am J Gastroenterol. 2009;104:816–24. doi: 10.1038/ajg.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 27.Dulak AM, Schumacher S, van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai LA, Kostadinov R, Barrett MT, et al. Deletion at fragile sites is a common and early event in Barrett’s esophagus. Mol Cancer Res. 2010;8:1084–94. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 30.Merlo LM, Shah NA, Li X, et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–97. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 32.Chaves P, Crespo M, Ribeiro C, et al. Chromosomal analysis of Barrett’s cells: demonstration of instability and detection of the metaplastic lineage involved. Mod Pathol. 2007;20:788–96. doi: 10.1038/modpathol.3800787. [DOI] [PubMed] [Google Scholar]
- 33.Kadri S, Lao-Sirieix P, Fitzgerald RC. Developing a nonendoscopic screening test for Barrett’s esophagus. Biomark Med. 2011;5:397–404. doi: 10.2217/bmm.11.40. [DOI] [PubMed] [Google Scholar]
- 34.Kadri SR, Lao-Sirieix P, O’Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett’s oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. doi: 10.1136/bmj.c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.