Abstract
Background
Alcoholic steatohepatitis (ASH) is caused in part by the effects of ethanol on hepatic methionine metabolism.
Methods
To investigate the phenotypic and epigenetic consequences of altered methionine metabolism in this disease, we studied the effects of 4-wk intragastric ethanol feeding with and without the methyl donor betaine in cystathionine beta synthase (CβS) heterozygous C57BL/6J mice.
Results
The histopathology of early ASH was induced by ethanol feeding and prevented by betaine supplementation, while ethanol feeding reduced and betaine supplementation maintained the hepatic methylation ratio of the universal methyl donor S-adenosylmethionine (SAM) to the methyltransferase inhibitor S-adenosylhomocysteine (SAH). MethylC-Seq genomic sequencing of heterozygous liver samples from each diet group found 2–4% reduced methylation in gene bodies but not promoter regions of all autosomes of ethanol fed mice, each of which were normalized in samples from mice fed the betaine supplemented diet. The transcript levels of inducible nitric oxide synthase (Nos2) and DNA methyltransferase 1 (Dnmt1) were increased, while those of peroxisome proliferator receptor-a (Pparα) were reduced in ethanol fed mice, and each was normalized in mice fed the betaine supplemented diet. DNA pyrosequencing of CβS heterozygous samples found reduced methylation in a gene body of Nos2 by ethanol feeding that was restored by betaine supplementation, and was correlated inversely with its expression and positively with SAM: SAH ratios.
Conclusions
The present studies have demonstrated relationships among ethanol induction of ASH with aberrant methionine metabolism that was associated with gene body DNA hypomethylation in all autosomes and was prevented by betaine supplementation. The data imply that ethanol-induced changes in selected gene transcript levels and hypomethylation in gene bodies during the induction of ASH is a result of altered methionine metabolism that can be reversed through dietary supplementation of methyl donors.
Keywords: Alcohol, S-adenosylmethionine, S-adenosylhomocysteine, cystathionine beta synthase, DNA methylation
Introduction
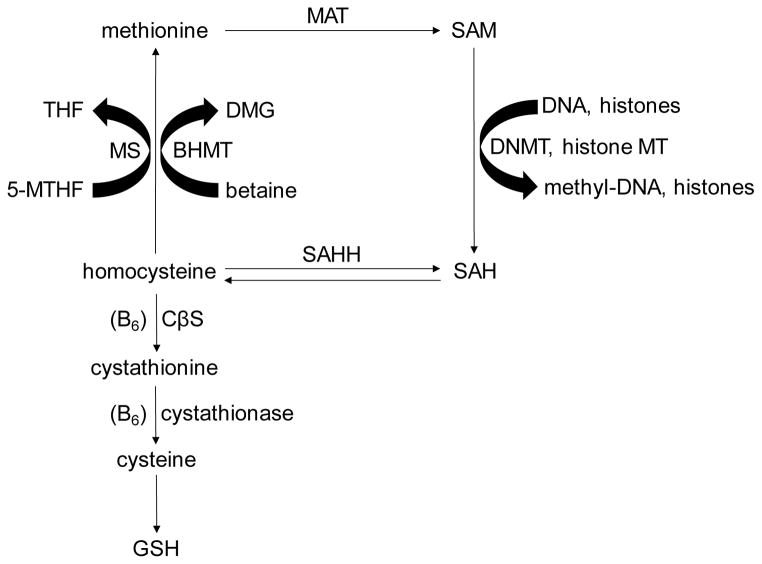
Chronic exposure to ethanol has profound effects on hepatic methionine metabolism which secondarily are involved in the regulation of DNA methylation and consequently the expressions of genes relevant to the pathogenesis of alcoholic steatohepatitis (ASH) (Halsted and Medici, 2011; Kharbanda, 2009; Mato and Lu, 2007). In particular, methionine metabolism is implicated in the production of the methyl donor, S-adenosylmethionine (SAM), which is generated from homocysteine by the enzymes methionine synthase and methionine adenosyltransferase and is the substrate for DNA and histone methyltransferases (Fig. 1). The alternative pathway leading to the production of methionine uses the alternate methyl donor betaine as a substrate and is catalyzed by the enzyme betaine homocysteine methyltransferase. Ethanol exposure decreases the gene expressions and activities of both methionine synthase and methionine adenosyltransferase, thereby reducing the production of SAM (Lu et al., 2000; Villanueva and Halsted, 2004). Since cystathionine beta synthase (CβS) is integral to the homocysteine degradation pathway (Fig. 1), the CβS heterozygous mouse was developed as a model for elevated homocysteine and, through the reverse pathway of SAH hydrolase, increased levels of the methyltransferase inhibitor S-adenosylhomocysteine (SAH) (Watanabe et al., 1995). Since SAM is the principal methyl donor and SAH is the principal methyltransferase inhibitor, the SAM:SAH ratio can be considered a reasonable index of methylation capacity (Clarke and Banfield, 2001). Previously, we showed reduced levels of the histone residue H3K9me3 together with up-regulation of several genes involved in the endoplasmic reticulum stress pathway after intragastric ethanol feeding of the CβS heterozygous mouse (Esfandiari et al., 2010).
Figure 1. Methionine metabolism pathway.
Methionine is the precursor of the principal methyl donor SAM which is the substrate for several methylation reactions catalyzed by DNA and histone methyltranferases. Conversely, SAH, which is the product of both methyltransferases and the reverse activity of SAHH, inhibits methylation reactions. Homocysteine can be metabolized through the transsulfuration pathway, which includes the enzyme CβS, or through the transsmethylation pathway, which includes the enzymes MS and BHMT. The methyl donor betaine is the substrate of BHMT. SAM: S-adenosylmethionine; DNMT: DNA methyltransferase; SAH: S-adenosylhomocysteine; SAHH: S-adenosylhomocysteine hydrolase; BHMT: betaine-homocysteine methyltransferase; DMG: dimethylglycine; MS: methionine synthase; THF: tetrahydrofolate; MTHF: 5-methyltetrahydrofolate; MAT: methionine adenosyl transferase; CβS: cystathionine beta synthase; GSH: glutathione.
The effects of chronic alcoholism and experimental ethanol exposure on methionine metabolism have been extensively described in humans and in animal models of ASH (Halsted and Medici, 2011; Kharbanda, 2009; Watanabe et al., 1995), whereas a prior study that used the same intragastric ethanol feeding model in wild type mice demonstrated the reversal of histopathology and gene expressions relevant to the endoplasmic reticulum stress pathway by supplementation with betaine (Ji and Kaplowitz, 2003). Others found that supplemental betaine restored histopathology, the gene expression of nitric oxide synthase (Nos2), and hepatic SAM levels, while reducing SAH and increasing the SAM:SAH methylation ratio in an ethanol fed rat model of this disease (Kharbanda et al., 2012).
The present study is the first demonstration of the effects of ethanol exposure with and without betaine on global and genomic DNA methylation and their potential consequences for regulation of specific genes relevant to liver damage. The goals of the present study were to define the effects of ethanol-mediated aberrant methionine metabolism in wild type and heterozygous CβS-deficient mice on global, genomic, and gene-specific DNA methylation and activation of selected genes involved in the induction of ASH and the preventive effects of the methyl donor betaine on each of these processes. The criteria for methylation regulation of gene activation included significant changes in each parameter in response to ethanol treatment that were prevented by betaine supplementation.
Materials and Methods
Animals and diets
CβS wild-type and heterozygous littermates were raised by S. Dayal at the University of Iowa breeding colony. Mice heterozygous for disruption of the Cβs gene were crossbred to C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) for at least 20 generations and genotyped as described (Dayal et al., 2006; Watanabe et al., 1995). The CβS deficient mouse breeding protocol and subsequent care were approved by the Animal Care and Use Committee of the University of Iowa. Mice were shipped at 4 wk age to the Animal Core facility of the University of Southern California Research Center for Alcoholic Liver and Pancreatic Diseases for subsequent intragastric feeding of experimental diets. At age 5 months, the mice were grouped according to genotype and diet and intragastric feeding tubes were placed under intraperitoneal anesthesia with ketamine and xylazine as described (Tsukamoto et al., 2008) and each of three diets were infused after one week adaptation to the control diet. The total energy intake was set at 568 kcal/kg/day, with caloric percentages of dextrose, protein, and fat (corn oil) at 40%, 25%, and 35% in the control group (C), substituting ethanol for dextrose up to 37% in the ethanol group (E), and the addition of betaine at 1.5% volume in the supplemented group (B). Each diet contained all essential vitamins and minerals (Dyets Inc, Bethlehem, PA) as recommended by the Committee on Animal Nutrition of the National Research Council. After 4 weeks of intragastric feeding, the mice were anesthetized with intraperitoneal ketamine and xylazine and then killed by exsanguination, following which plasma and livers were removed and treated as described previously (Esfandiari et al., 2010). The care and experimental procedures were approved by the Animal Care and Use Committee of the University of Southern California according to the Guide for the Care and Use of Laboratory Animals of the National Research Council.
Liver methionine metabolite measurements
Liver SAM and SAH and plasma homocysteine were measured by high performance liquid chromatography by K. Kharbanda (Kharbanda et al., 2005).
Liver histopathology
Liver histopathology was evaluated independently by S.W. French by quantitative scoring of appropriately stained slides in blinded fashion using computerized software for 4 grades for parameters of lipid accumulation, lobular inflammation, necrosis, and fibrosis. Intrahepatic fat was further quantified as % per total pixels using the 10x objective. (French et al., 1993).
Analysis of global DNA methylation levels by dot blot analyses
Global DNA methylation was measured in mouse livers from each genotype and diet group using dot blot analysis as previously described (Woods et al., 2012). Data are expressed according to signal intensities of 5-methylcytosine.
Analysis of genomic DNA methylation levels by MethylC-seq
Genomic DNA methylation according to each autosome was analyzed in three representative heterozygous liver samples from each of the three feeding groups of heterozygous mice by MethylC-seq as described (Lister et al., 2009; Schroeder et al., 2011). MethylC-seq provides a genome-wide quantitative measurement of DNA methylation that is unbiased to genomic location of CpG sites (Lister et al., 2009). Briefly, genomic DNA from each sample was sheared, annealed to methylated sequencing adapters, and bisulfite converted. DNA methylation genomic sequencing of each prepared sample was performed at the University of California Berkeley Vincent J. Coates Genomics Sequencing Laboratory. Reads were mapped to the mm9 version of the mouse genome using BS Seeker (Chen et al., 2010). CpG site methylation data in promoter and gene body regions of genes from each of 19 autosomes were combined from both DNA strands, and multiple clonal reads with the same start site were removed. Custom Perl scripts were used to convert BS Seeker output to BED files of percent methylation per CpG site. The BED files were used for data visualization on the UCSC Human Genome Browser and for further data analysis. The data were analyzed using custom Perl scripts and R programs (Schroeder et al., 2013; Schroeder et al., 2011).
Quantitative PCR
Liver samples from all mice were analyzed using the Vet-For-All kit and BioSprint magnetic bead extraction instrument (QIAGEN, Valencia, CA) (Osman et al., 2012). We selected a total of 21 genes involved in alcoholic liver injury and methionine metabolism pathways for expression analysis by qPCR (Bustin et al., 2009), using the geometric mean of three reference genes Hprt1, B2m, and Gapdh for normalization. Primers for each gene were designed using Primer Express and are shown in Supplemental Table 1. Data were expressed as fold changes in each feeding group of each genotype compared to the mean gene expression values in control diet samples from mice of each genotype.
Immunohistochemical staining
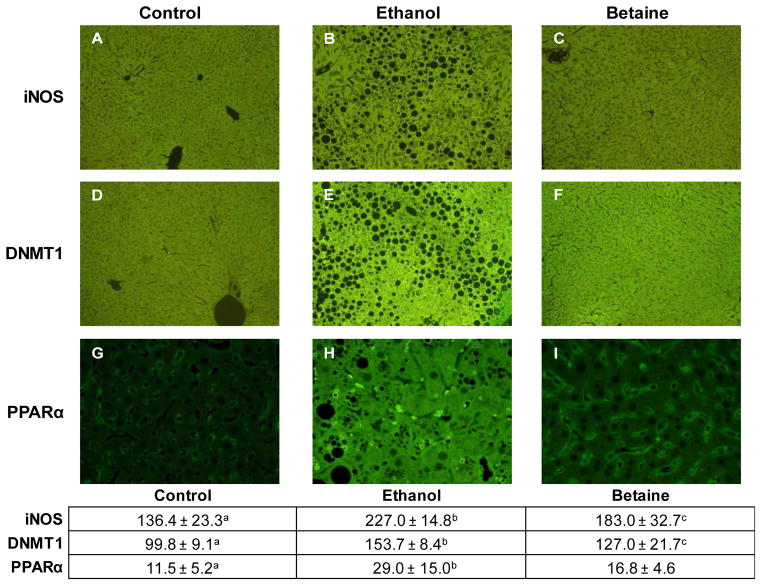
Liver tissues from heterozygous mice were fixed in neutral-buffered formalin, embedded in paraffin, cut into 4 μm thick sections, and stained with antibodies to inducible nitric oxide synthase (iNOS), DNMT1 and PPARα, each at 1/100 titer (Epitomics, Burlington, CA), followed by Donkey fluorescein isothiocyanate (FITC) labeled antibody 1/100 titer (Jackson ImmunoReaserch Labs Inc., Westgrove, PA). The expression of each of these genes had been identified by qPCR analysis as significantly altered in samples from ethanol fed mice and normalized in samples from betaine supplemented mice. Cellular fluorescence intensity was quantified in each slide in blinded fashion using a fluorescein isothiocyanate filter, Nikon morphometric software, and a Nikon 400 fluorescent microscope 40x objective with the same sensitivity setting throughout (Bardag-Gorce et al., 2008).
Pyrosequencing analysis of gene specific DNA methylation
Based on results from the qPCR analysis of gene expressions, we selected Nos2, Ppar-α, and Dnmt1 for pyrosequencing analyses of potential methylation differences in liver samples from each of the three heterozygous feeding groups, and to correlate these findings with their respective gene expressions and with liver SAM and SAM: SAH ratios. Regions of interest and primer selections were identified according to the prior MethylC-seq genomic analyses and were further defined by PyroMark Assay Design 2.0 and the UCSC Genome Browser July 2007 version. Bisulfite treatment of genomic DNA was performed using the EZ DNA Methylation-Direct Kit (Zymo Research, Irvine, CA). Triplicate PCR amplifications were performed using the recommended protocol for Pyromark CpG Assay (QIAGEN, Valencia, CA). Samples were sequenced on a Pyromark Q24 Pyrosequencer using Pyromark Gold Q24 Reagents (QIAGEN, Valencia, CA) and methylation levels were analyzed using Pyromark Q24 Software. An internal bisulfite conversion control was used in the pyrosequencing assay which measured methylation at selected CpG sites for each assay.
Statistical analyses
Outcome variables were assessed for conformance to the normal distribution and transformed if needed. Means were compared between diets and genotypes with 2-factor ANOVA and Tukey-Kramer tests were used for post-hoc pairwise comparisons. Spearman correlations were used to assess the relationships of Nos2 DNA methylation values in each heterozygous feeding group to its respective gene expression values. Analyses were performed with SAS for Windows Release 9.3 (Cary, NC). Group differences in MethylC-Seq analyses were determined separately by the Student’s t-test.
Results
Baseline characteristics and methionine metabolites
Mouse body weights were similar among all six groups after 4 weeks of feeding (Table 1). Ethanol feeding increased liver weights and consequently liver/body weight ratios in both wild-type and heterozygous mice, and this ratio was restored to control levels by betaine supplementation in heterozygous mice. Blood ethanol levels were appropriately elevated in the ethanol groups and were unchanged by betaine supplementation of the ethanol diets. Plasma alanine aminotransferase (ALT) levels were increased 3–4 fold by ethanol feeding in each genotype but were not normalized by betaine supplementation of the ethanol diet, with wide variation of individual results. SAM levels were reduced by about one half in each genotype by ethanol feeding and were amplified by ~50-fold by supplementation of the SAM precursor betaine to the ethanol diet. SAH levels were increased in both genotypes of ethanol-fed mice and were further amplified by betaine treatment in response to the extreme elevation of its precursor SAM. As a result of these changes, the SAM:SAH ratio in each genotype was reduced by ethanol feeding and was increased by betaine supplementation. Mean plasma homocysteine levels were increased more than 6- fold in the ethanol-fed heterozygotes and were significantly lowered to control levels by betaine supplementation. There were no genotype effects or interactions on these parameters except for genotype effects on plasma homocysteine levels when comparing results from the two ethanol groups (p<0.003).
Table 1.
Effects of diets in CβS wild-type and heterozygous mice.
| Wild-type Control (6) | Wild-type Ethanol (4) | Wild-type Betaine (6) | Hetero Control (7) | Hetero Ethanol (8) | Hetero Betaine (8) | |
|---|---|---|---|---|---|---|
| Body weight (grams) | 28±1.2 | 28.9±3.4 | 26.8±5.3 | 30±1.3 | 29.1±4.6 | 26.2±4 |
| Liver weight (grams) | 1.5±0.1a | 2.5±0.4b | 2±0.43 | 1.6±0.1a | 2.7±0.7b | 1.9±0.4a |
| Liver/body weight | 0.05±0.01a | 0.08±0.01b | 0.07±0.01b | 0.05±0.01a | 0.09±0.01b | 0.07±0.01c |
| Blood ethanol (mg/mL) | 5.3±1.5a | 286±167b | 249±110b | 5.1±2.1a | 156±121b | 185±97b |
| ALT (U/L) | 19.6±6.6a | 93.5±20.4b | 113.5±109b | 30.8±28.9a | 107.3±48b | 121±99b |
| Liver SAM (nmol/g) | 57.4±5a | 27.6±16.0b | 2862±1113c | 69.5±14.4a | 32.0±16.2b | 3087±2123c |
| Liver SAH (nmol/g) | 31.6±10.7a | 57.2±25.0b | 100.8±46.7b | 59.6±19.7a | 74.6±21.4a | 131.2±56.2b |
| SAM:SAH | 2±0.6a | 0.65±0.6b | 39.1±35.8c | 1.31±0.5a | 0.48±0.3b | 39.5±56.0c |
| Plasma Homocysteine (nmol/mL) | 4.3±2.6a | 14.0±6.8 b | 10.4±8.2ab | 12.2±6.2a | 80.6±52.8b | 12.6±9.1a |
Values are expressed as mean ± standard deviation. The number of mice in each group is reported in parentheses. Values with different letter symbol are significantly different (p<0.05) from each other within each genotype. There were no genotype effects except for plasma homocysteine (p<0.003) in the two different ethanol fed groups. SAM: S-adenosylmethionine; SAH: S-adenosylhomocysteine.
Histopathology
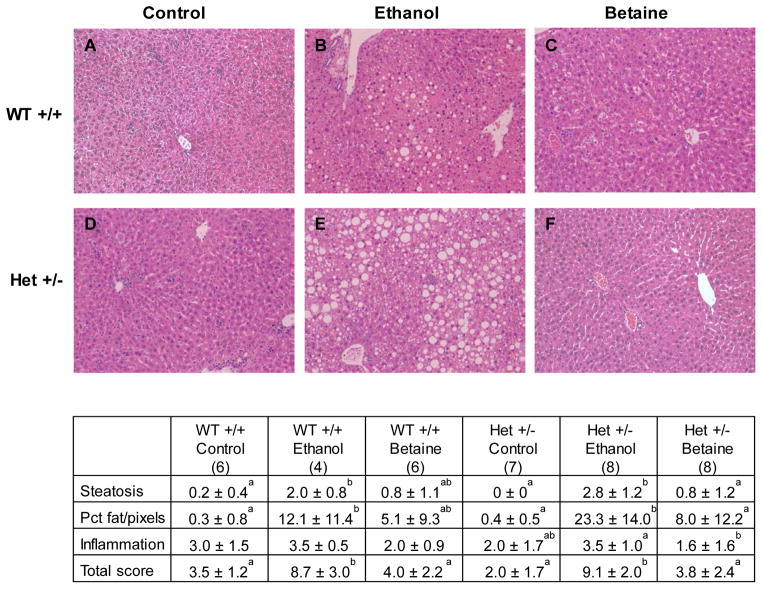
Scores of liver injury were influenced by diets with no independent or interactive genotype effects. As depicted (Fig. 2), total and steatosis scores were significantly increased by ethanol feeding within each genotype and were reduced by betaine supplementation in heterozygotes. There were no changes among the groups in inflammation scores, and there were minimal to no scores for necrosis or fibrosis among the groups (not shown).
Figure 2. Histopathology of mouse livers after 4 weeks of control (A, D), ethanol (B, E), and betaine-supplemented ethanol diet (C, F) (hematoxylin and eosin, 10x).
Selective images show typical changes in wild-type (top) and heterozygous mice (bottom) and followed by quantitative scores for each diet and genotype. Values in the table are expressed as mean ± standard deviation. The number of mice in each group is reported in parentheses. Values with different letter symbols are significantly different (p<0.05) from each other within each genotype.
Global DNA methylation by dot blot analysis
Global methylation levels were assessed by signal intensities of 5-methylcytosine in liver samples from each genotype and diet group. According to ANOVA of all results, there were no genotype effects or differences among the diet groups.
Genome MethylC-seq analysis
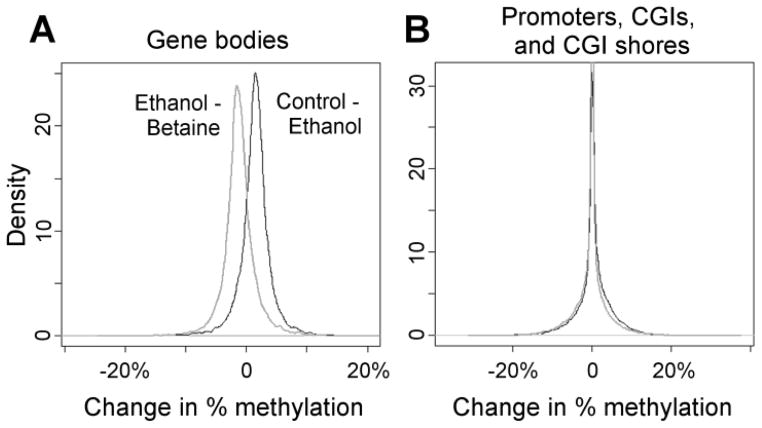
Each of 19 autosomes from heterozygous ethanol- fed mice had lower percent genomic methylation compared to values in control mice, whereas methylation in each autosome was restored to its respective control level by betaine supplementation (Table 2). Combining these data, the average methylation of all autosomes was 72.6% in control diet group samples and was decreased to 71.1% in ethanol group samples (p<0.03 v control samples) but was maintained at control diet levels by betaine supplementation with 72.4% average methylation (p<0.04 vs ethanol samples with p = 0.0182 overall diet effect). Evaluating regions of each genome in each autosome, the average methylation at CpG sites in gene bodies in the ethanol group was significantly less than that found in the control and betaine groups (Fig. 3a) However, methylation of the majority of CpG sites in promoter regions and in CpG island shores were unaffected by ethanol feeding and betaine treatment (Fig. 3b).
Table 2.
Methylation of autosomes by MethylC-seq genome analyses.
| Chromosome | Control | Ethanol | Betaine | p-value Ethanol v Control | p-value Betaine v Ethanol |
|---|---|---|---|---|---|
| 1 | 73.95±0.41 | 72.51±0.57 | 73.84±0.51 | .0282 | .0410 |
| 2 | 71.84±0.43 | 70.41±0.51 | 71.60±0.40 | .0219 | .0312 |
| 3 | 74.02±0.44 | 72.52±0.61 | 73.90±0.53 | .0318 | .0451 |
| 4 | 71.39±0.45 | 69.99±0.55 | 71.29±0.47 | .0292 | .0376 |
| 5 | 72.05±0.46 | 70.59±0.52 | 71.89±0.41 | .0233 | .0312 |
| 6 | 73.56±0.39 | 72.07±0.58 | 73.35±0.44 | .0274 | .0432 |
| 7 | 70.00±0.43 | 68.64±0.52 | 69.87±0.40 | .0273 | .0352 |
| 8 | 72.38±0.44 | 70.95±0.53 | 72.25±0.39 | .0245 | .0306 |
| 9 | 72.35±0.43 | 70.88±0.57 | 72.21±0.45 | .0270 | .0372 |
| 10 | 73.87±0.40 | 72.46±0.53 | 73.72±0.39 | .0253 | .0346 |
| 11 | 69.23±0.44 | 67.88±0.49 | 69.03±0.29 | .0248 | .0364 |
| 12 | 73.26±0.50 | 71.72±0.61 | 73.07±0.45 | .0311 | .0435 |
| 13 | 74.20±0.45 | 72.80±0.57 | 74.03±0.44 | .0323 | .0468 |
| 14 | 73.91±0.43 | 72.37±0.58 | 73.77±0.50 | .0253 | .0358 |
| 15 | 72.51±0.36 | 71.08±0.52 | 72.31±0.40 | .0215 | .0349 |
| 16 | 74.31±0.42 | 72.90±0.59 | 74.16±0.43 | .0325 | .0457 |
| 17 | 71.77±0.37 | 70.44±0.46 | 71.61±0.37 | .0191 | .0291 |
| 18 | 74.43±0.37 | 72.93±0.53 | 74.21±0.42 | .0206 | .0346 |
| 19 | 70.91±0.40 | 69.58±0.46 | 70.74±0.28 | .0208 | .0291 |
| X | 73.48±0.57 | 71.93±0.71 | 73.13±0.58 | .0461 | .0914 |
| All autosomes | 72.55±0.43 | 71.11±0.54 | 72.39±0.42 | .0254 | .0358 |
Figure 3. Genome-wide changes in methylation in (A) gene bodies and (B) promoters, CpG islands (CGIs), and CGI shores.
The average methylation was computed for each gene/promoter/CGI and CGI shore within each of the three groups, and differences among groups were plotted as density histograms. As shown in the left box for gene bodies, ethanol diet samples had on average 1.2% less methylation than betaine supplemented diet samples, whereas control diet samples had on average 1.5% greater methylation than ethanol diet samples. As shown in the right box, ethanol and betaine treatments did not change methylation levels in promoters, CG Islands (CGI), or CGI shores.
Effects of ethanol and betaine on transcript levels of Dnmt1, Nos2, and Pparα
We selected a total of 21 genes in pathways of inflammation, lipogenesis, endoplasmic reticulum stress, and methionine metabolism for expression analysis by qPCR. Specific primer sequences and qPCR results for each gene according to the two genotypes are listed in Supplemental Material, Tables 1 and 2. Among these 21 genes, only Dnmt1, Nos2, and Pparα met criteria for changes in expression being altered by ethanol feeding and corrected by betaine supplementation. Since the expressions of Nos2 and Pparα were unaffected by genotype, each of their dietary effects were analyzed by pooling data from both genotypes. On the other hand, Dnmt1 expression was influenced by both genotype and diet, and dietary effects were only significant in heterozygotes. As shown in Table 3, the mean expression of Nos2 in pooled data from both genotypes was increased 7.6 -fold by ethanol treatment and was reduced significantly to control levels by betaine supplementation. Whereas ethanol feeding reduced Pparα expression in pooled results from both genotypes, it increased Dnmt1 expression among heterozygotes, and values for each gene expression were restored to respective control levels by betaine supplementation.
Table 3.
Relative Gene Expressions of Dnmt1, Nos2, and Ppara.
| Diet | Dnmt1 | Nos2 | Pparα |
|---|---|---|---|
| Control | 0.126±0.77a (7) | -0.208±1.069a (10) | -0.609±0.469a (13) |
| Ethanol | 1.817±0.463b (8) | 7.610±2.616b (12) | -2.079±0.401c (12) |
| Betaine | -2.7±1.639ac (8) | -2. 694±2.445ac (14) | -0.095±0.756ac (14) |
Data are expressed as mean ± standard error of mean. In parentheses, the numbers of mice studied in each group. All data are represented by relative fold differences from control values within each genotype. Since there were no genotype effects on expressions of Nos2 and Ppara, values were calculated by pooling diet data from both genotypes, whereas Dnmt1 data refer to heterozygous mice only. Values with different letters are significantly different at p<0.05.
Immunohistochemistry of selected proteins
Immunohistochemical staining for protein levels of DNMT1 and iNOS in liver specimens from CβS heterozygous mice followed similar dietary patterns as found for their gene transcripts, with maximal and significant staining in the ethanol group that was normalized by betaine supplementation (Fig. 4). On the other hand, protein levels of Ppara did not follow the same trend as their gene expressions, since its mean level was increased by ethanol feeding and there was no difference after betaine treatment.
Figure 4. Immunohistochemical staining for iNOS, DNMT1, and PPARα in representative liver sections from heterozygous mice fed control (A), ethanol (B), and betaine-supplemented ethanol diets (C).
Both iNOS and DNMT1 are shown at 10X and PPARα is shown at 40 X magnification. Immunohistochemical intensity units are shown in the accompanying table. Values are expressed as mean ± standard deviation intensity units. Values with different letter superscripts are significantly different, p<0.05.
Pyrosequencing
Samples from CβS heterozygote mice were selected for pyrosequencing analyses of the same three genes with differential gene expressions according to diet, and DNA regions were chosen according to anticipated changes according to prior MethylC-seq analyses. Since there were no significant methylation differences among groups in promoter region CpG sites in any of the three genes, representative sites were selected according to CpG rich sites in gene bodies. As shown in Table 4, DNA methylation of CpG rich regions in Dnmt1 and Pparα were not significantly different in liver samples from the three diet groups, except for increased methylation of Dnmt1 in the betaine supplement group at a region over 1 kb upstream of the transcription start site in chromosome 9. On the other hand, the percent DNA methylation in a region spanning 7 CpG sites in the second intron of Nos2 was significantly reduced by ethanol feeding and was sustained at control levels by betaine supplementation. Among all three diet groups in this CβS heterozygote cohort, the percent DNA methylation of Nos2 in this region was correlated negatively with its relative expression (r=-0.5104; p<0.03) and positively with liver SAM (r=0.7538, p<0.0001) and the SAM:SAH ratio (r=0.79850, p<0.0001). Whereas a previous study found that the transcription of Nos2 in mouse mesangial cells was inhibited by hypermethylation of a specific promoter region in its DNA (Yu and Kone, 2004), we were unable to find methylation differences in the same Nos2 promoter region in the heterozygote livers of control, ethanol-fed, and betaine supplemented mice.
Table 4.
CpG methylation of Dnmt1, Nos2, and Ppara in heterozygous mice.
| Diet | Dnmt1 | Nos2 | Pparα |
|---|---|---|---|
| Control | 47.9±8.47a (5) | 43.81 ± 0.63c (6) | 39.5±7.89 (5) |
| Ethanol | 47.6±7.48a (5) | 41.17 ± 0.77d (7) | 36.4±4.53 (5) |
| Betaine | 60.5±5.5b (5) | 45.59 ± 1.73c (7) | 27.6±5.29 (5) |
Values are expressed as mean ± standard deviation. In parenthesis, the number of mice studied in each group. Dnmt1 chromosome 9, base pair position in gene body 20,766,199–20,766,217; Nos2: chromosome 11, second intron, base pair position 78,738,995 – 78,739,403, including 7 CpG sites; Pparα chromosome 15, base pair position in promoter region at 8,557,227–8,557,256. Values with different letter symbols are significantly different: a vs. b: p<0.04; c vs d p<0.01.
Discussion
Epigenetic regulation of genes relevant to liver injury in ASH may be closely related to underlying ethanol-induced changes in methionine metabolism. Our initial studies in micropigs fed ethanol-containing diets that were deficient in the folate methyl donor demonstrated accelerated onset of the histopathology of ASH with significant decreases in hepatic SAM and the SAM:SAH methylation ratio compared to those fed folate replete diet (Halsted et al., 2002) together with increased transcript levels of genes involved in endoplasmic reticulum stress and lipogenesis (Esfandiari et al., 2005). Subsequent intervention studies in the same micropig model showed that the histopathology of ASH, abnormal SAM:SAH ratio, and the activation of the same liver injury genes could be prevented by concurrent supplementation of ethanol diets with the methyl donor SAM (Esfandiari et al., 2007; Villanueva et al., 2007). More recently, we demonstrated that 4 wk intragastric feeding of ethanol to CβS-heterozygous mice reduced the SAM:SAH ratio while elevating the expressions of selected endoplasmic reticulum genes, each of which were associated with decreased H3K9me3 histone methylation (Esfandiari et al., 2010).
The main findings of the present study are, first, that 4- wk of ethanol feeding resulted in increased steatosis and total hepatic histopathology scores typical of early ASH in both genotypes, together with significant reductions in SAM and the SAM:SAH methylation ratio, all of which were prevented by supplementing the ethanol diet with the methyl donor betaine (Fig. 2 and Table 1). Contrasting genotype effects, plasma homocysteine levels were significantly increased in the ethanol fed heterozygotes compared to those in the wild type mice (Table 1), as has been shown by others who developed this model (Watanabe et al., 1995). These data showing greater perturbation of methionine metabolism among heterozygotes provide a rationale for the use of this genotype for several of the subsequent analyses, including MethylC-seq genomic and pyrosequencing methylation analyses. Unexpectedly, betaine supplementation did not significantly reduce ALT levels in either genotype despite the improvement of histological scores of inflammation and steatosis in both genotypes. A prior study on the effects of betaine supplementation in a similar intragastric mouse model with 6 weeks of ethanol feeding showed decreased serum homocysteine and prevention of the steatosis response to ethanol but non-significant reduction in ALT levels (Ji and Kaplowitz, 2003). We may speculate that longer dietary exposure in our mouse model could have been associated with greater histopathology in the ethanol groups and potential reductions in ALT in the betaine supplemented groups.
Second, there were no significant changes in global DNA methylation according to dot-blot analyses of all samples from both genotypes and all three diets. These findings are consistent with those from our prior study (Esfandiari et al., 2010) which used the liquid chromatography-tandem mass spectrometry (LC-TMS) method to calculate percentage of methyl-C among total cytosine residues (Quinlivan and Gregory, 2008). Both methylation dot blot and MethylC-seq specifically quantitate CpG site methylation, while LC-TMS measures all cytosine methylation. According to prior studies in animal models of other liver diseases, global DNA methylation may be regulated in part by underlying hepatic inflammation (Gonda et al., 2012; Medici et al., 2013; Stenvinkel et al., 2007; Wierda et al., 2010), which was minimal in our studies.
Third, our genome analysis of heterozygote samples by MethylC-seq showed that DNA hypomethylation was induced by ethanol feeding in each of 19 autosomes and was prevented in each autosome by betaine supplementation (Table 2). Furthermore, the results of this analysis showed that hypomethylation occurred exclusively in gene bodies, whereas no diet induced methylation effects were found in gene promoter regions (Fig. 3). These novel findings highlight the need to examine gene body methylation patterns when examining DNA methylation changes in other models of ASH. Our data are consistent with other genome-wide studies of DNA methylation patterns, in which CpG island promoters are predominantly un-methylated, in contrast to the high levels of methylation within gene bodies (Aran et al., 2011; Ball et al., 2009; Lister et al., 2009; Zemach et al., 2010). Recent studies indicate that gene body methylation levels are likely to be determined by the greater accessibility of their DNA to methyl donors and methyltransferases (Jjingo et al., 2012).
Fourth, among the 21 selected genes related to liver injury and methionine metabolism, only Nos2, Pparα, and Dnmt1 presented patterns of changes in transcript levels that reflected opposing responses to ethanol and betaine supplementation. In particular, ethanol feeding increased both the transcript and protein levels of Dnmt1 in heterozygous mice and of Nos2 in pooled genotype samples, each of which were reduced by provision of the methyl donor betaine, whereas the transcript levels of Pparα were reduced by ethanol and maintained at control levels by betaine (Table 3 and Fig. 4).
Fifth, the findings of significant methylation changes in the gene body of Nos2 (Table 4) are supported by the novel observation from methylome analysis that the effects of ethanol on mouse chromosomes are confined to gene bodies and are not found in promoter regions (Fig. 3).
The significant correlations of these findings with SAM:SAH ratios from all three diet groups supports a potential link between the effects of ethanol and betaine with hepatic methionine metabolism and gene specific methylation. However, since gene expression is regulated by many factors, we cannot infer a specific causative relationship between the methylation pattern of Nos2 (Table 4) at this gene body site and its expression (Table 3), whereas the clinical relevance of this finding may only be established by additional studies in liver biopsies from ASH patients.
iNOS is required for the sustained generation of nitric oxide, which, in turn, causes a broad spectrum of injury, including processes of lipid peroxidation, mitochondrial disruption, and DNA damage with pro-apoptotic effects (Davis et al., 2001). Others correlated increased liver Nos2 expression and activity in rats fed high ethanol diets with the severity of steatosis and necroinflammation in association with activation of nuclear factor kappa beta and induction of tumor necrosis factor alpha (Yuan et al., 2006). Furthermore, mitochondrial dysfunction and the histopathology of ASH were significantly attenuated in ethanol-fed Nos2 knock-out mice (Venkatraman et al., 2004) and in mice treated with a selective iNOS inhibitor (McKim et al., 2003). Consistent with our present findings, others found more than 40-fold elevation in hepatic Nos2 transcript levels in mice fed an intragastric ethanol and high fat diet, together with more severe histopathology than shown in the present experiment (Xu et al., 2011), whereas betaine supplementation reversed liver histopathology, elevated iNOS levels, and nitric oxide accumulation in an ethanol fed rat model of ASH (Kharbanda et al., 2012).
PPARα functions as a transcription factor to induce the expressions of a series of genes involved in fatty acid transport, mitochondrial fatty acid oxidation, catabolism, and inflammatory responses. Our present data showed a discrepancy between the transcript and protein expressions of Pparα in response to ethanol feeding (Table 3 and Fig. 4) and no changes among diet groups in its DNA methylation according to pyrosequencing (Table 4). Another study showed that Pparα gene expression was both increased and associated with increased DNA methylation in ethanol fed rats (Khachatoorian et al., 2013), while others showed that down-regulation of Pparα gene expression is involved in the development of steatosis associated with ASH (Aoyama et al., 1998). The regulation of Pparα expression is complex and is influenced by fatty acid accumulation (Kersten et al., 2000), whereas its protein level is unstable and is rapidly degraded through the ubiquitin-proteasome pathway (Blanquart et al., 2002). The observation that the ubiquitin-proteasome pathway is inhibited by ethanol (French, 2000) could account for our findings of increased PPARα protein levels according to immunohistochemistry after ethanol diet (Fig. 4).
Dnmt1 encodes the principal mammalian DNMT and functions in maintenance of methylation status. Our study showed that ethanol feeding of heterozygous mice increased the transcript and protein levels of Dnmt1 (Table 3, Fig. 4), which were each reduced by provision of the methyl donor betaine, whereas pyrosequencing demonstrated that methylation targeted an upstream non-promoter region of the gene body after betaine treatment (Table 4). Limited data are available on the role of Dnmt1 in the development of ASH. Ethanol fed Dnmt1 N/+ hypomorphic mice that express reduced levels of Dnmt1 presented less severe hepatic steatosis than wild type mice with normal expression of Dnmt1, together with concomitant dysregulation of key enzymes involved in lipid metabolism and oxidative stress (Kutay et al., 2012). Consistent with our findings, the methylation of the promoter regions of the genes for ethanol and lipid metabolism in that study were not different among genotypes and dietary treatments, suggesting that factors other than DNA methylation regulated gene transcript levels in absence of Dnmt1 with or without ethanol feeding.
In summary, ethanol feeding of the CβS-deficient mouse model reduced the hepatic SAM:SAH ratio of methylation capacity in association with reduced gene body methylation in all autosomes and in a specific gene body site in Nos2, each of which were prevented by betaine supplementation. Even though this is a new finding, its clinical significance remains uncertain and other factors linking methylation to gene expression changes in ASH include histone modifications (Esfandiari et al., 2010; Mandrekar, 2011; Shukla et al., 2008) and/or potential effects of ethanol induced liver injury and inflammation on epigenetic marks as described by others (Gonda et al., 2012; Medici et al., 2013; Stenvinkel et al., 2007; Wierda et al., 2010).
Supplementary Material
Acknowledgments
Financial support: This research was supported by National Institutes of Health grant numbers K08DK084111 (to V.M.), R03AA020577-01 (to C.H.H.), P50AA11991 Southern California Research Center for ALPD and Cirrhosis (to HT), P50AA11991 Morphology Core (to S.W.F.), R01ES021707 (to J.M.L. and D.S.), and by a Biomedical Laboratory Research and Development National Merit Review grant 1 101 BX001155 from the Department of Veterans Affairs, Office of Research and Development (to K.K.K.). V.M. is a member of the University of California San Francisco Liver Center (Liver Center grant number P30 DK026743) and received funds from the Division of Gastroenterology and Hepatology at UC Davis.
Footnotes
The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Aoyama T, Peters JM, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez FJ. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha) J Biol Chem. 1998;273(10):5678–84. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20(4):670–80. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84(2):113–21. doi: 10.1016/j.yexmp.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart C, Barbier O, Fruchart JC, Staels B, Glineur C. Peroxisome proliferator-activated receptor alpha (PPARalpha) turnover by the ubiquitin-proteasome system controls the ligand-induced expression level of its target genes. J Biol Chem. 2002;277(40):37254–9. doi: 10.1074/jbc.M110598200. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S, Banfield K. S-adenosylmethionine-dependent methyltransferases. In: Carmel R, Jacobsen D, editors. Homocysteine in health and disease. Cambridge University Press; Cambridge: 2001. pp. 63–78. [Google Scholar]
- Davis KL, Martin E, Turko IV, Murad F. Novel effects of nitric oxide. Annu Rev Pharmacol Toxicol. 2001;41:203–36. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- Dayal S, Wilson KM, Leo L, Arning E, Bottiglieri T, Lentz SR. Enhanced susceptibility to arterial thrombosis in a murine model of hyperhomocysteinemia. Blood. 2006;108(7):2237–43. doi: 10.1182/blood-2006-02-005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari F, Medici V, Wong DH, Jose S, Dolatshahi M, Quinlivan E, Dayal S, Lentz SR, Tsukamoto H, Zhang YH, French SW, Halsted CH. Epigenetic regulation of hepatic endoplasmic reticulum stress pathways in the ethanol-fed cystathionine beta synthase-deficient mouse. Hepatology. 2010;51(3):932–41. doi: 10.1002/hep.23382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari F, Villanueva JA, Wong DH, French SW, Halsted CH. Chronic ethanol feeding and folate deficiency activate hepatic endoplasmic reticulum stress pathway in micropigs. Am J Physiol Gastrointest Liver Physiol. 2005;289(1):G54–63. doi: 10.1152/ajpgi.00542.2004. [DOI] [PubMed] [Google Scholar]
- Esfandiari F, You M, Villanueva JA, Wong DH, French SW, Halsted CH. S-adenosylmethionine attenuates hepatic lipid synthesis in micropigs fed ethanol with a folate-deficient diet. Alcohol Clin Exp Res. 2007;31(7):1231–9. doi: 10.1111/j.1530-0277.2007.00407.x. [DOI] [PubMed] [Google Scholar]
- French SW. Mechanisms of alcoholic liver injury. Can J Gastroenterol. 2000;14(4):327–32. doi: 10.1155/2000/801735. [DOI] [PubMed] [Google Scholar]
- French SW, Nash J, Shitabata P, Kachi K, Hara C, Chedid A, Mendenhall CL. Pathology of alcoholic liver disease. VA Cooperative Study Group 119. Semin Liver Dis. 1993;13(2):154–69. doi: 10.1055/s-2007-1007346. [DOI] [PubMed] [Google Scholar]
- Gonda TA, Kim YI, Salas MC, Gamble MV, Shibata W, Muthupalani S, Sohn KJ, Abrams JA, Fox JG, Wang TC, Tycko B. Folic acid increases global DNA methylation and reduces inflammation to prevent Helicobacter-associated gastric cancer in mice. Gastroenterology. 2012;142(4):824–833. e7. doi: 10.1053/j.gastro.2011.12.058. [DOI] [PubMed] [Google Scholar]
- Halsted CH, Medici V. Vitamin-dependent methionine metabolism and alcoholic liver disease. Adv Nutr. 2011;2(5):421–7. doi: 10.3945/an.111.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsted CH, Villanueva JA, Devlin AM, Niemela O, Parkkila S, Garrow TA, Wallock LM, Shigenaga MK, Melnyk S, James SJ. Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci U S A. 2002;99(15):10072–7. doi: 10.1073/pnas.112336399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124(5):1488–99. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3(4):462–74. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405(6785):421–4. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- Khachatoorian R, Dawson D, Maloney EM, Wang J, French BA, French SW. SAMe treatment prevents the ethanol-induced epigenetic alterations of genes in the Toll-like receptor pathway. Exp Mol Pathol. 2013;94(1):243–6. doi: 10.1016/j.yexmp.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda KK. Alcoholic liver disease and methionine metabolism. Semin Liver Dis. 2009;29:155–165. doi: 10.1055/s-0029-1214371. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, Rogers DD, 2nd, Mailliard ME, Siford GL, Barak AJ, Beckenhauer HC, Sorrell MF, Tuma DJ. Role of elevated S-adenosylhomocysteine in rat hepatocyte apoptosis: protection by betaine. Biochem Pharmacol. 2005;70(12):1883–90. doi: 10.1016/j.bcp.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, Todero SL, King AL, Osna NA, McVicker BL, Tuma DJ, Wisecarver JL, Bailey SM. Betaine treatment attenuates chronic ethanol-induced hepatic steatosis and alterations to the mitochondrial respiratory chain proteome. Int J Hepatol. 2012;2012:962183. doi: 10.1155/2012/962183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay H, Klepper C, Wang B, Hsu SH, Datta J, Yu L, Zhang X, Majumder S, Motiwala T, Khan N, Belury M, McClain C, Jacob S, Ghoshal K. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PLoS One. 2012;7(8):e41949. doi: 10.1371/journal.pone.0041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279(1):G178–85. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol. 2011;17(20):2456–64. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–12. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- McKim SE, Gabele E, Isayama F, Lambert JC, Tucker LM, Wheeler MD, Connor HD, Mason RP, Doll MA, Hein DW, Arteel GE. Inducible nitric oxide synthase is required in alcohol-induced liver injury: studies with knockout mice. Gastroenterology. 2003;125(6):1834–44. doi: 10.1053/j.gastro.2003.08.030. [DOI] [PubMed] [Google Scholar]
- Medici V, Shibata NM, Kharbanda KK, LaSalle JM, Woods R, Liu S, Engelberg JA, Devaraj S, Torok NJ, Jiang JX, Havel PJ, Lonnerdal B, Kim K, Halsted CH. Wilson’s disease: changes in methionine metabolism and inflammation affect global DNA methylation in early liver disease. Hepatology. 2013;57(2):555–65. doi: 10.1002/hep.26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Olineka T, Hodzic E, Golino D, Rowhani A. Comparative procedures for sample processing and quantitative PCR detection of grapevine viruses. J Virol Methods. 2012;179(2):303–10. doi: 10.1016/j.jviromet.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Quinlivan EP, Gregory JF., 3rd DNA methylation determination by liquid chromatography-tandem mass spectrometry using novel biosynthetic [U-15N]deoxycytidine and [U-15N]methyldeoxycytidine internal standards. Nucleic Acids Res. 2008;36(18):e119. doi: 10.1093/nar/gkn534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, LaSalle JM. The human placenta methylome. Proc Natl Acad Sci U S A. 2013;110(15):6037–42. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Lott P, Korf I, LaSalle JM. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res. 2011;21(10):1583–91. doi: 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32(9):1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimburger O, Barany P, Alvestrand A, Nordfors L, Qureshi AR, Ekstrom TJ, Schalling M. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261(5):488–99. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Mkrtchyan H, Dynnyk A. Intragastric ethanol infusion model in rodents. Methods Mol Biol. 2008;447:33–48. doi: 10.1007/978-1-59745-242-7_3. [DOI] [PubMed] [Google Scholar]
- Venkatraman A, Shiva S, Wigley A, Ulasova E, Chhieng D, Bailey SM, Darley-Usmar VM. The role of iNOS in alcohol-dependent hepatotoxicity and mitochondrial dysfunction in mice. Hepatology. 2004;40(3):565–73. doi: 10.1002/hep.20326. [DOI] [PubMed] [Google Scholar]
- Villanueva JA, Esfandiari F, White ME, Devaraj S, French SW, Halsted CH. S-Adenosylmethionine Attenuates Oxidative Liver Injury in Micropigs Fed Ethanol With a Folate-Deficient Diet. Alcohol Clin Exp Res. 2007;31:1934–1943. doi: 10.1111/j.1530-0277.2007.00511.x. [DOI] [PubMed] [Google Scholar]
- Villanueva JA, Halsted CH. Hepatic transmethylation reactions in micropigs with alcoholic liver disease. Hepatology. 2004;39(5):1303–10. doi: 10.1002/hep.20168. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Osada J, Aratani Y, Kluckman K, Reddick R, Malinow MR, Maeda N. Mice deficient in cystathionine beta-synthase: animal models for mild and severe homocyst(e)inemia. Proc Natl Acad Sci U S A. 1995;92(5):1585–9. doi: 10.1073/pnas.92.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. 2010;14(6A):1225–40. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Vallero RO, Golub MS, Suarez JK, Ta TA, Yasui DH, Chi LH, Kostyniak PJ, Pessah IN, Berman RF, Lasalle JM. Long-lived epigenetic interactions between perinatal PBDE exposure and Mecp2308 mutation. Hum Mol Genet. 2012;21(11):2399–411. doi: 10.1093/hmg/dds046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lai KK, Verlinsky A, Lugea A, French SW, Cooper MP, Ji C, Tsukamoto H. Synergistic steatohepatitis by moderate obesity and alcohol in mice despite increased adiponectin and p-AMPK. J Hepatol. 2011;55(3):673–82. doi: 10.1016/j.jhep.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Kone BC. Hypermethylation of the inducible nitric-oxide synthase gene promoter inhibits its transcription. J Biol Chem. 2004;279(45):46954–61. doi: 10.1074/jbc.M407192200. [DOI] [PubMed] [Google Scholar]
- Yuan GJ, Zhou XR, Gong ZJ, Zhang P, Sun XM, Zheng SH. Expression and activity of inducible nitric oxide synthase and endothelial nitric oxide synthase correlate with ethanol-induced liver injury. World J Gastroenterol. 2006;12(15):2375–81. doi: 10.3748/wjg.v12.i15.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.