Abstract
PA1 is a large hairpin polyamide (dImPyPy-β-PyPyPy-γ-PyPy-β-PyPyPyPy-β-Ta; Py = pyrrole, Im = imidazole, β = beta alanine) that targets the sequence 5′-WWGWWWWWWW-3′ (W = A or T) and is effective in eliminating HPV16 in cell culture (Edwards, T. G., Koeller, K. J., Slomczynska, U., Fok, K., Helmus, M., Bashkin, J. K., Fisher, C., Antiviral Res. 91 (2011) 177-186). Described here are its DNA binding properties toward a natural DNA, a 523 bp portion of HPV16 (2150-2672) containing three predicted perfect match sites. Strategies for obtaining binding data on large fragments using capillary electrophoresis are also described. Using an FeEDTA conjugate of PA1, 19 affinity cleavage (AC) patterns were detected for this fragment. In many cases, there are multiple possible binding sequences (perfect, single and double mismatch sites) consistent with the AC data. Quantitative DNase I footprinting analysis indicates that perfect and most single mismatch sites bind PA1 with Kds between 0.7-4 nM, indicating excellent tolerance for the latter. Double mismatch sites exhibit Kds between 12 and 62 nM. A large fraction of the accessible sequence is susceptible to PA1 binding, much larger than predicted based on the literature of polyamide-DNA recognition rules.
Keywords: polyamide, footprinting, affinity cleavage, HPV, binding constant
1. INTRODUCTION
N-methylpyrrole/imidazole-based polyamides (PA) can be considered higher homologs of the natural product distamycin A, a tris(pyrrole) reagent with a cationic C-terminus that binds to the minor groove of AT-rich DNA sequences and which has antitumor and antiviral properties along with high toxicity [1, 2]. Polyamides, the binding capabilities of which extend to GC-containing sequences via the incorporation of imidazole building blocks, have found extensive application as sequence-selective DNA binding agents for the control of gene expression [3, 4]. Typical modern PAs in use for such experiments employ the hairpin structure in which two polyamide strands are joined by aminobutyric acid, which can be thought of as a hairpin turn that allows two linked antiparallel polyamide strands to align in the minor groove of DNA and recognize the two DNA strands independently. Other building blocks have been used to construct analogous turns, including (R)-2,4-diaminobutyric acid, its acetamide and more [5-7]. In certain cases the PA strands have been linked at both ends, resulting in a cyclic PA [8-10].
Hairpin polyamides have long been used by Dervan [3, 11, 12], Sugiyama [6, 13], Lee [14-16], Kodadek [17] and others for their ability to recognize DNA sequences via hydrogen bonding interactions in the minor groove [5, 9, 18-30]. The properties of polyamides have been valuable for controlling gene expression, including the blocking gene-specific transcription factor binding sites and alkylating specific DNA regions [31, 32]. Polyamides have also been used to enhance gene transcription [33], and effects have been shown against pathogens and metabolic diseases at the cell culture level. Recently, whole animal efficacy, imaging and pharmacology or toxicology data have been reported for a number of disease targets [4, 10, 34-39].
The accessibility of PA to nucleosomes has been studied, the use of PA for specific disease states has been fruitful, and the number of in vivo studies (for safety and/or efficacy) of PA has been increasing. Adding alkylating functionality to polyamides has allowed them to act directly as gene silencing agents. Uptake of polyamides by cells has been studied and found to vary depending on cell type and PA characteristics in some cases [7, 40-44], while in other cases uptake was constant over a wide range of polyamide MW (from 400 to 4000) [44]. As described in the cited reports, uptake has been enhanced by a variety of means, such as blocking multidrug resistance efflux pumps with Verapamil® or modifying polyamide building blocks at either internal or terminal positions.
Recently, dramatic antiviral effects for human papillomavirus (HPV) were discovered for certain long polyamides that bound 10 or more bp of DNA [45, 46]. The active polyamides were designed to bind the dsDNA genome of HPV, an approximately 8 kb circular DNA molecule known as an episome. With IC50 values of ≤ 30-100 nM and no measurable cellular toxicity, the active polyamides function by eliminating viral DNA from infected human keratinocytes. Furthermore, the active compounds were shown to restore the phenotype of uninfected cells in keratinocyte tissue culture. Thus, HPV infections manipulate DNA synthesis by host enzymes, forcing suprabasal DNA synthesis to occur for infected keratinocyte tissue cultures; this effect is reversed by addition of properly-designed polyamides. PA1 (Fig. 1) is one of the lead compounds reported in this anti-HPV study, and shows excellent activity against HPV16 and 31 (IC50 = 100 and 108 nM). More recently, a detailed report has appeared on the mechanism of action of this class of anti-HPV compounds [46].
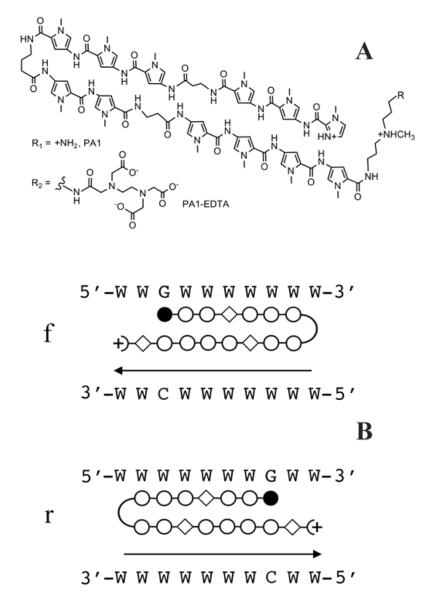
Fig. 1. Structure and Predicted Recognition Sequence for PA1.
A, Structure of PA1 and its EDTA conjugate. B, Target sequence for PA1 and alternative binding orientations. Closed circles = imidazole; open circles = pyrrole; open diamond = β alanine W=A/T. The arrowhead indicates the location of the polyamide tail and EDTA group. f refers to the forward binding orientation (amino terminus aligns with 5′ end of target sequence); r refers to the less favored reverse binding orientation (amino terminus aligns with the 3′ end of the target sequence.
The anti-HPV PAs are considerably larger than most reported hairpin polyamides and contain 14-26 rings. Broad-spectrum activity was a surprise because the analogous DNA sequences of the different HPV subtypes are not typically degenerate for polyamide binding. Therefore the origin of primary antiviral activity and broad spectrum activity are both of considerable interest from a range of perspectives, including cellular and biophysical perspectives as well as for the design of more-active second-generation anti-HPV compounds. In light of this broad-spectrum activity, the applicability of polyamide-DNA recognition rules developed mostly for smaller PA molecules (typically 8 rings) was of specific interest for the longer, antiviral polyamides [2, 3, 11, 12, 18, 19, 22, 26, 27, 29, 30, 47-49]. The use of a large natural sequence target is also important, as it better represents the biological setting than shorter sequences tailored to address specific questions about binding behavior [7]. In this genomic viral DNA setting, what level of detail can be achieved with commonly used assays and approaches? What is the relationship between predicted and observed binding sites? To begin to address this, the binding properties of the large, archetypal anti-HPV polyamide, PA1 (Figure 1), are therefore characterized here using an HPV16 DNA sequence as the target.
2. MATERIALS AND METHODS
2.1 Synthesis
dIm-PyPy-β-PyPyPy-γ-PyPy-β-PyPyPyPy-β-Ta (PA1) was prepared and characterized as previously described [45]. Im = imidazole, dIm = desamino-imidazole, Py = pyrrole, β = beta alanine, γ = γ-aminobutyric acid, and Ta = amino tail at C-terminus formed from NMe(CH2CH2CH2NH2)2. The EDTA conjugate was prepared as follows: By analogy to the literature [50], to a vigorously-stirred mixture of EDTA dianhydride (23 mg, 0.088 mmol, 20 eq) in N,N-diisopropylethylamine (DIEA) (0.5 mL), DMF (0.25 mL), and DMSO (0.25 mL) at 55 oC was added a mixture of PA1 (10 mg, 0.0044 mmol, 1 eq) in DIEA (0.5 mL) and DMF (0.5 mL) dropwise over a period of 10 min. The reaction mixture was stirred at 55 oC for 20 min. Following addition of 0.1 N NaOH (1.8 mL), the mixture was stirred at 55 oC for an additional 15 min. The bottom layer of the biphasic mixture was removed from the reaction vessel and neutralized by addition of 0.35 M aq. TFA (1 mL). The mixture was diluted with 0.5 mL DMSO and divided into two equal portions, each of which was filtered through 20 μm polyethylene filters. Each portion was further diluted with 0.5 mL DMSO and purified by reversed-phase HPLC [H2O (0.1% TFA), MeOH (0.1% TFA)] using a Phenomenex Jupiter 250 × 21.2 mm, 4 μm, 90 Å, C12 column. Concentration of pooled fractions, followed by lyophilization afforded the PA1-EDTA conjugate (3.8 mg, 33% yield) as a white, fluffy solid: average molecular weight = 2169.27, experimental (ESI) [M+H]+ = 2169.8.
2.2. Footprinting
To generate the 523 bp DNA fragment comprised of the 2150-2672 region of the HPV16 genome (Accession # AF125673), two primers oligomers were ordered from Integrated DNA Technologies, Inc. (Coralville, Iowa) (top strand): 5′-FAM-AT GTG ATA GGG TAG ATG ATG GAG GTG; (bottom strand) 5′-HEX CT CAT ACA CTG GAT TTC CGT TTT CGT CAT ATG G-3′. To generate the smaller duplexes, the following primers were used: top strand FAM-TGG TGC AGC TAA CAC AGG TAA starting at 2291; top strand FAM-CTA CAG TGC CCT GTT GGA AC starting at 2434; bottom strand HEXCAA CAG GGC ACT GTA GCA TC starting at 2449; bottom strand HEX-CT CAT ACA CTG GAT TTC CGT TTT CGT CAT ATG G starting at 2672.
DNase I footprinting experiments were performed as previously described [49] in TKMC buffer with 10 mM CHAPS. DNA concentrations were adjusted between 200 pM and 10 nM as needed. A small range of DNase I units (0.01-0.02 U in a 250 μL reaction) and a 5 min reaction time was typically used, although the reactivities of batches of DNase I can vary. Single hit conditions were confirmed by CE. An illustration of this control reaction for the same sequence of DNA has been published recently [49]. Samples were analyzed via capillary electrophoresis (CE) at the DNA Core Facility at University of Missouri. Data were processed using Genemarker V1.97 software (Softgenetics LLC, State College, PA). DNase I cleavage products were mapped using Sanger sequencing (USB, Affymetrix, Santa Clara, CA). Peaks in the footprint were normalized to a peak not sensitive to PA concentration and plotted as fraction bound vs. PA concentration and fit to a binding isotherm as previously described [49].
2.3. Affinity Cleavage
The polyamide-EDTA-Fe conjugate was prepared by adding 0.8 eq of Fe(NH4)2(SO4)2 to a solution of the conjugate, incubating 5 min, adding the DNA fragment to 1 nM and incubating overnight at room temperature in 10 mM Tris, 10 mM CHAPS, pH 7.5. The PA-EDTA conjugate concentration was varied from 5 nM to 250 nM and reported at 50 nM. The cleavage reaction was initiated by adding 5 μL fresh made 100 mM DTT to a 100 μL reaction and incubating 30 min - ~ 4 hr. The reaction was quenched and products purified using a Qiagen PCR purification kit. Fragment analysis was conducted by CE. Affinity cleavage results were mapped using Maxam-Gilbert reactions on 5′ FAM endlabeled DNA [51].
3. RESULTS
3.1. PA1 Sites on a Natural DNA Target
PA1 was originally designed to bind to HPV16. Therefore for the first studies of PA1 on a natural DNA target, we chose a portion of the HPV16 genome at 2150-2672. This region is located within the E1 gene. It has three predicted perfect PA1 sites based on the literature rules, all with the sequence of either 5′-WGWWWWWWW-3′ or 5′-WWGWWWWWWW-3′ where W=A or T. These perfect binding sites are well spaced from one another (50 bp or more). There are also a number of predicted single mismatch and double mismatch sites. PA1-DNA interactions were characterized via two experiments: affinity cleavage (AC) via a PA1-EDTA conjugate and DNase I footprinting.
To achieve quantitative footprinting and affinity cleavage via capillary electrophoresis (CE), high signal-to-noise ratios are necessary in the electropherograms. Due to the loss of CE signal as fragment sizes increase past 250 bp and given the size of the duplex of interest (523 bp), it was necessary to work with 5′ dyes on both strands and smaller duplexes composed of the internal regions of the genome fragment. Fragments in the first half of the 523 bp duplex (5′ end of the top strand) were detected with 5′-FAM on the top strand at 2150; fragments in the last half (3′ end) of the 524 bp duplex were detected with 5′ labeling with HEX on the bottom strand (dye at 2672). Using both dyes on the same duplex DNA afforded information from both strands in one experiment, since the dyes are detected in different channels on the sequencer. However, since the signal becomes weaker as the fragment size increases, this led to low signal-to-noise in the middle of the duplex. This was addressed by preparing shorter duplexes spanning 2150-2449, 2434-2672, and 2291-2449 (Fig. 2). The last duplex was needed for affinity cleavage. Since it provides information about the most sites, affinity cleavage data will first be considered in isolation. Kd determinations and issues surrounding those measurements follow.
Fig. 2. Map of Duplexes Used in the Analysis.
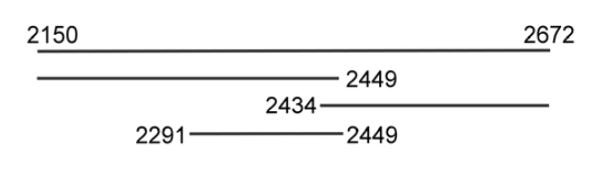
2150-2672 indicates the entire 523 bp region of the HPV16 genome (# AF125673) under study. The 2150-2249 and 2434-2673 fragments were used in the footprinting analysis. Another duplex at 2291-2449 was also used in affinity cleavage experiments. For clarity, duplexes are represented by one strand.
3.2. Affinity Cleavage
In the AC technique, the EDTA moiety is bound to iron to execute hydroxyl radical cleavage of DNA at the binding site [30]. While the PA-EDTA conjugate concentration was varied from 5 to 250 nM, all relevant sites appeared by 50 nM. This concentration is well above the Kds for most PA-DNA interactions (see below), which means that both high and low affinity sites are detected. Since the EDTA is positioned on the PA tail (open end of the hairpin; see Fig. 1), cleavage patterns indicate the location of the tail on the DNA sequence. AC also provides information about PA binding orientation, i.e., tail facing right or left. An orientation is termed “forward” if amino terminal end of the polyamide is aligned with the 5′ end of the target sequence [50, 52]. With a PA in the hairpin conformation, the tail of the PA will also be near the 5′ end of the binding sequence (see Fig. 1). In a “reverse” orientation, the amino terminal (and for hairpin, tail end) of the PA aligns with the 3′ end of the target sequence. Based on work with small PAs (6-8 rings) this is generally considered less favorable [52].
In addition, it is necessary to map the cleavage patterns using chemistry that best matches the Fenton chemistry of Fe EDTA. Therefore indexing of the electropherograms was achieved with Maxam Gilbert sequencing [53]. A sample electropherogram illustrating affinity cleavage for a perfect site (site 3) appears in Fig. 3A. Similar sample data for a single mismatch site (5) and double mismatch site (10) appear in Fig. S1.
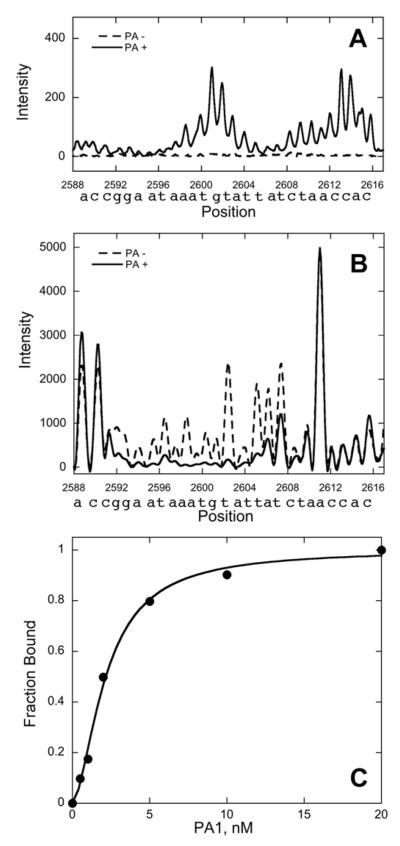
Fig. 3. Sample Binding Data.
A, Electropherogram detail illustrating affinity cleavage for Site 3. B, Electropherogram detail illustrating DNase I footprinting for Site 3. C, Isotherm obtained from electropherograms obtained as a function of PA1 concentration for Site 3.
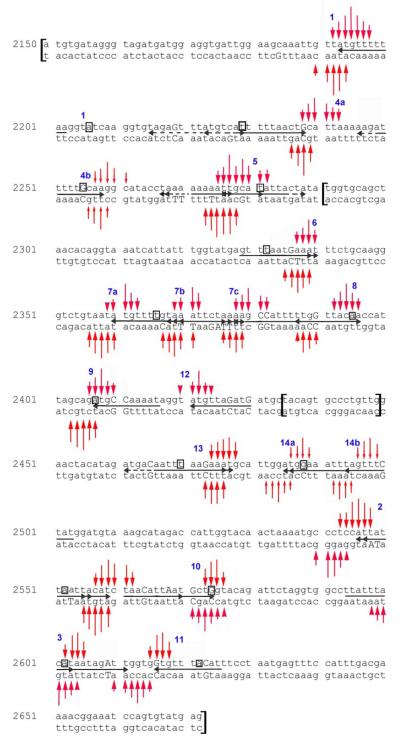
A sequence map of the AC patterns appears in Fig. 4. The 40-50 bp on the ends of the duplex are not accessible via CE because they are not well resolved under the experimental conditions employed for long DNA molecules. Ignoring the first and last 40-50 bp of the duplex, it is clear that there are many PA1 sites accessed at 50 nM PA1 conjugate. In Fig. 4, 19 AC patterns in the fragment are observed, and for all but site 12, bottom strand affinity cleavage information was obtainable with the duplexes used. We observed the expected 3′ shift between the top and bottom strand cleavage patterns. When detected using 5′ labeled DNA, this is characteristic of minor groove ligands [54]. Using these patterns and the established recognition rules, horizontal arrows were drawn to indicate binding sites consistent with the AC patterns (horizontal arrow with arrowhead at PA tail/EDTA and the 5′-WWG end of the recognition sequence). Of these, only two were perfect, discrete and isolated binding sites (1 and 3). The others represent ambiguous and/or unusual binding that is indicated with overlapping arrows. Since predicted double mismatch sequences cover most of the sequence, forward double mismatch sequences were only considered if the AC pattern could not be rationalized with a perfect or single forward mismatch site. While also fairly common in the sequence, reverse binding sites were neglected because all AC patterns could be explained with a forward binding mode, usually a single mismatch sequence. The AC sites are discussed in roughly numerical order and/or with respect to their position in the sequence, as is appropriate. Table 1 features predicted sequences of interest, and flanking sequences are included as context. A few for which there is no supporting experimental evidence for binding are included as appropriate (see below).
Fig. 4. Sequence Map of Affinity Cleavage (AC) and Footprinting.
Affinity cleavage was observed at 50 nM PA1-FeEDTA conjugate. Since signal-to-noise ratios vary with fragment length, vertical arrows indicate intensities relative to other arrows in a site, not to other sites. Small arrowheads reflect particularly weak affinity cleavage. Solid horizontal arrows indicate PA1 sites consistent with AC data. Mismatch nucleotides are indicated with a capital letter. For overlapping sites in which the mismatch occurs for only one site, the nt on only the bottom strand is capitalized. Boxed nt: DNase I integration site. Dashed horizontal arrows: predicted sites that are not supported by affinity cleavage. Brackets indicate where the various overlapping duplexes begin and end. See text for details. Possible reverse binding sites are omitted for clarity, as are forward double mismatches in regions where they overlap significantly with perfect or single mismatch sites.
Table 1.
NV1028 PA1 Sites on 2150-2673 Fragment of HPV16a
| AC | Sequence | Position | Site Type | Integration nt | Kd DNase I | Comment |
|---|---|---|---|---|---|---|
| 1 | TGTT ATGTTTTTAA GGTA | 2193-2202 | Perfect | 2205 | 2.2 ± 1 | |
| 3 | TATT ATGTAAATAA GGCC | 2603-2594 | Perfect | 2602 | 0.7 ± 0.2 | |
| 2 | TTAG ATGTAATTAA TAAT | 2559-2550 | Perfect | 2552 | 1.7 ± 0.7 | |
| 2 | CTCC ATTATTAATT ACAT | 2546-2555 | single | |||
| 2 | TCCA TTATTAATTA CATC | 2547-2556 | single | |||
| 2 | GATG TAATTAATAA TGGA | 2555-2546 | single | |||
| 2 | ATGT AATTAATAAT GGAG | 2556-2547 | single | |||
| 4a | AATG CAGTTAAAAA TGAC | 2238-2229 | single | 2229 | 3.0 ± 1.8 | |
| 4 | GGTG TAGAGTTTAT GTCA | 2215-2224 | single | No ACb | ||
| 4 | GTTT ATGTCATTTT TAAC | 2223-2232 | single | No ACb | ||
| 4 | AAAA ATGACATAAA CTCT | 2229-2220 | single | No ACb | ||
| 4 | TAAA AAGATTTTTG CAAG | 2246-2255 | single | |||
| 4b | TGCC TTGCAAAAAT CTTT | 2258-2249 | single | 2255 | 3.8 ± 1 | |
| 5 | TACC TAAAAAAAAT TGCA | 2267-2276 | single | No ACb | ||
| 5 | ACCT AAAAAAAATT GCAT | 2268-2277 | single | No ACb | ||
| 5 | TGCA ATTTTTTTTA GGTA | 2276-2267 | single | |||
| 5 | AAAA TTGCATATTA CTAT | 2276-2285 | single | 2281 | 2.5 ± 1 | |
| 5 | ATGC AATTTTTTTT AGGT | 2277-2267 | single | |||
| 5 | TAAT ATGCAATTTT TTTT | 2281-2272 | single | |||
| 5 | CATA TAGTAATATG CAAT | 2288-2279 | single | No AC or FPb | ||
| 6 | AGAA ATTTCATTAA ACTC | 2240-2331 | double | 2.7 ± 0.2 | ||
| 6 | GAAA TTTCATTAAA CTCA | 2339-2330 | double | |||
| 7a | TAAT ATGTTTTGTA AATT | 2360-2369 | single | 2366 | 3.8 ± 1 | |
| 7b | TTTG TAAATTCTAA AAGC | 2368-2377 | double | |||
| 7b | TTGT AAATTCTAAA AGCC | 2369-2378 | double | |||
| 7b | TGTA AATTCTAAAA GCCA | 2370-2379 | double | |||
| 7c | CTTT TAGAATTTAC AAAC | 2376-2366 | single | |||
| 7c | GCTT TTAGAATTTA CAAA | 2377-2368 | double | |||
| 7c | CTAA AAGCCATTTT TGGT | 2378-2387 | double | |||
| 7c | GGCT TTTAGAATTT ACAA | 2378-2369 | double | |||
| 7c | TGGC TTTTAGAATT TACA | 2379-2370 | double | |||
| 8 | ATTT TTGGTTACAA CCAT | 2387-2396 | double | |||
| 8 | ATGG TTGTAACCAA AAAT | 2396-2387 | double | 2395 | 62 ± 3 | |
| 9 | GCAG ATGCCAAAAT AGGT | 2407-2416 | double | 2407 | 32 ± 9 | |
| 10 | CAGC ATTAATGTTA GATG | 2570-2561 | double | |||
| 10 | GTAC CAGCATTAAT GTTA | 2574-2565 | double | 2574 | 41 ± 24 | |
| 11 | CACC AATCTATTAT GTAA | 2611-2602 | double | |||
| 11 | GGTG GTGTTTACAT TTCC | 2616-2625 | double | 2622 | 12 ± 2 | |
| 12 | AGGT ATGTTAGATG ATGC | 2421-2430 | double | c | ||
| 13 | ATAG ATGACAATTT AAGA | 2461-2470 | single | No ACb | ||
| 13 | TGCA TTTCTTAAAT TGTC | 2476-2468 | double | |||
| 13 | ATGC ATTTCTTAAA TTGT | 2477-2469 | double | 2470 | 12 ± 3 | |
| 14a | TTGG ATGGAAATTT AGTT | 2485-2494 | single | 2488 | 25 ± 1 | |
| 14a | TGGA TGGAAATTTA GTTT | 2486-2495 | single | |||
| 14b | AATT TAGTTTCTAT GGAT | 2494-2503 | single | Weak AC |
Strong sites assayed at 200 pM DNA; weak Kds assayed at 2 and 10 nM DNA.
Position corresponds to PA arrowhead/where the sequence begins.
No AC, no affinity cleavage pattern observed to support binding to this sequence. FP, footprint.
Not determined. Site 12 is too close to the end of a duplex.
Sites 1 and 3 are straightforward, that is, isolated and perfect match sites. The sequence most consistent with the AC pattern at site 4a is a single mismatch site at 2238-2229 (the convention is the leading nt position is at the arrowhead). There are a number of other single mismatch sites nearby (indicated with dashed arrows), but no reliable affinity cleavage patterns above the noise were discerned to support binding in those regions. Although the AC signal is relatively weak, Site 4b is a straightforward single mismatch site.
For site 5, the affinity cleavage on the top strand has two maxima (at 2277 and 2279); this is less obvious in the corresponding pattern on the bottom strand. This is consistent with multiple PA binding events in this region. Indeed, there are 4 predicted forward single mismatch PA sites that could conceivably contribute to the pattern (solid arrows with arrowhead at the cleavage pattern), with those beginning at 2276-77 being most attractive. The other sites (dashed arrows) are not supported by discernable affinity cleavage patterns. Fig S2 features an expanded view of the possible binding sites around site 5.
The AC pattern at site 6 is best rationalized with forward double mismatch sequences. Due to the especially high number of AT base pairs, these sequences overlap significantly and cannot be distinguished.
Site 7 is composed of three AC patterns that are very close together (2359-2382). While cleavage at sites 7a and 7c can be rationalized with forward single mismatch sequences, double mismatch sites are needed to explain the AC pattern at 7b. Three such sequences are indicated in Fig. 4. Since these overlap significantly, they cannot be distinguished. Fig S2 features an expanded view of the possible binding sites around site 7.
AC 8 is unusual because top and bottom strand affinity cleavage patterns are farther apart from one another than is typically observed [30, 50, 55], and indexing was verified. The AC pattern at site 8 can be rationalized with a double mismatch PA binding site (2396- 2387 bp).
The AC pattern at site 9 can be most easily rationalized with a double mismatch forward binding site (2407-2416). This part of the genome is relatively GC rich, ruling out many alternative and overlapping binding sequences. The pattern at site 12 is similar: One double mismatch forward site (2421-2430 bp) is consistent with this pattern.
The AC pattern at site 13 can be rationalized with a forward double mismatch sequence, and two of these overlap extensively. Although there is a forward single mismatch site beginning at 2461, there is no discernable affinity cleavage pattern to support it (dashed arrow).
There are overlapping single mismatch sites at 2485-2494, 2486-2495, and 2494-2503 (site 14), and we observe weak affinity cleavage signals to support them.
The AC patterns at site 2 suggest a specific region of the DNA for PA1 binding, but alternative binding modes are possible. The AC pattern centered at 2546 suggests a single mismatch site and there are two highly overlapping possibilities. The AC pattern at the other (3′) end of the same sequence aligns well to a perfect site (2559-2550). There are two nearby overlapping single mismatch all A/T sites nearby which cannot be ruled out (2555-2546, 2556-2547). Clearly PA1 binds in both the orientations (i.e., with the tail facing different directions).
There are multiple double mismatch sites consistent with the AC pattern at site 10, although the one at 2574-2565 is closest to the AC pattern and most probable. For site 11, there are multiple double mismatch sites consistent with the AC pattern.
3.4. Quantitative Footprinting Studies
All PA1 sites on this fragment were also examined using DNase I footprinting chemistry on the 2150-2672 region of the HPV 16 genome. Integration peaks were chosen based first on their location in the footprint, and then secondly for optimum signal-to-noise so that integration was as accurate as possible. Given that DNase I is much larger than PA1 (29 kD), in some cases the integration peak was located outside the recognition sequence for PA1. In assessing integration site locations, it is also important to note that DNase I is not entirely sequence neutral and has preferences for DNA shapes and sequences (e.g. highly AT-rich sequences are disfavored for cutting) [56, 57]. Indeed, we found regions in the sequence that were not sensitive to DNase I, that is, we could not detect cleavage by the enzyme even in the PA-free controls. These are noted where appropriate.
A sample footprint electropherogram and isotherm for a perfect site (site 3) appears in Fig. 3B and C. Similar sample data for a single mismatch site (5) and double mismatch site (10) appear in Fig. S1. The integration sites for DNase I appear as solid boxes in Fig. 4. Using the above dye-labeled duplexes (Fig. 2) and the sites indicated in the figure for integration, 14 nucleotide positions were integrated to obtain Kd’s as detected by DNase I (Table 1). In most cases, a single Kd represents each quantitated AC site, even if there are multiple possible PA binding locations and/or orientations for an AC pattern. Although there are three distinct affinity cleavage patterns for site 7, the degree of coverage by PA1 made only two DNase I integration sites possible. Site 4 was evaluated using multiple integration sites (see below). Site 12 was not amenable to quantitation, even with the smaller internal duplexes generated for this purpose (Fig. 2).
In reconciling footprinting data with affinity cleavage DNA, we first found it helpful to obtain a Kd for the PA1-EDTA conjugate as measured by footprinting and compare it to that obtained for the non-modified polyamide. Using DNase I as the footprinting agent, we find that the Kd for the 2193-2202 site (site 1) is 1.5 ± 0.6 nM for PA1-EDTA, quite similar to that obtained for PA1. Thus under these conditions, there is no appreciable difference in affinity between PA and PA-EDTA, despite the fact that the conjugated EDTA moiety removes one positive charge and adds (three) negative charges to the polyamide when not bound by Fe(II).
Kd values obtained by DNase I are summarized in Table 1. The Kds for the perfect match sites range from 0.7 to 2.2 nM. Values for sites 1 and 3 (2.2 and 0.7 nM, respectively) are very comparable to that obtained using a TAMRA-labeled DNA hairpin in the presence of DNA carrier [53].
Most integration sites associated with single mismatch sequences (AC sites 4ab, 5, 7a and 7c) exhibit Kds around 3 nM, indicating good tolerance for this configuration. Both integrated peak positions at site 4 (4a and 4b) give similar Kds. PA1 binding at the sequence starting at 2238 can be legitimately quantitated at 2229 since the PA extends toward the integration site.
Kds for most of the double mismatch sites (sites 6 and greater) were obtained by increasing the DNA concentration as needed. While this is only a small sampling of possible double mismatch sequences, these are generally weaker Kds than for perfect and single mismatch sites by at least two-fold, with greater variability among them (12-62 nM). An exception is site 6, assigned to a forward double mismatch sequence with an unexpectedly low Kd of 2.7 nM. All other possible binding sequences in this region have more mismatches and also represent reverse binding modes; thus they are significantly disfavored by one or both of these factors.
In contrast, site 8 has the weakest Kd among the double mismatch sites (62 nM). This is mapped to a forward double mismatch sequence (2396-2387), and there is another double mismatch sequence (or more precisely, orientation) at 2387-2396 that could explain the affinity cleavage centered around 2390. There are other potential binding sites in this area near the AC pattern, but they are reverse sites with at least two mismatches. With the experimental information available, it is not possible to discriminate among these recognition sequences.
Lastly, we were able to integrate at 2488 at site 14, for which we did observe two weak AC patterns and could obtain a Kd of 25 nM. All possible binding sequences are single mismatch sequences in this region. The weak value of a Kd for a single mismatch site is notable in this context; we report many other single-mismatch sites with much tighter Kd values.
4. DISCUSSION
4.1. Studying Ligand Binding on Long Natural Sequences
In working with a long natural and A/T rich sequence, we encountered a number of challenges. The most obvious is that 523 bp is actually a bit long for easily evaluating all binding sites with a single, doubly endlabeled duplex with capillary electrophoresis. Using dyes on the 5′ end of each strand left little useful signal-to-noise ratios in the center of the sequence. This contrasts with the ability of CE to sequence DNA molecules of 1000 bp or more: much higher S/N is tolerated for sequencing than for quantitative footprinting (fragment analysis). In this study, the sequence length issue was remedied by making smaller internal duplexes; however, working with a smaller duplex of about 350 bp initially would have been more efficient.
In spite of these challenges, we were able to quantitatively characterize a large number of PA1 binding sites in this region of the genome via affinity cleavage and DNase I footprinting. In many regards, the results were as expected: perfect match sites bind with low nM Kd’s. Single mismatch sites were also well tolerated, exhibiting Kd’s in the same range. While there is a range of tolerances for single mismatch sites in the literature (see below), we rationalize that PA1 tolerates these sites well due to its unusually large size relative to most literature PAs. Put more simply, there are more opportunities for hydrogen bonds that can compensate for a few local, less-than-optimal contacts. One immediate conclusion is that the concept of affinity for PA-DNA binding is actually undefined in general terms: the PA-DNA binding is dependent on the arrangement of purportedly degenerate A and T nucleotides and on the sequence context in which the target DNA is located. This conclusion is further explained in the paragraphs below.
Of added interest are the more unexpected behaviors. Given how dense the binding sequences are and how many were detected, we were surprised to find some regions where there was no evidence of binding by any technique, even though reasonable binding sequences were present. Regions near the ends must be excluded, as the fragments are too short to be analyzed by CE. Given the density of documented PA sites, there are two relatively large and noteworthy “dead spots” at 2301-2328 and 2501-2540. These contain a number of potential double mismatch sequences, but no data to support actual binding. In the former region, DNase I control samples indicate poor cutting by the enzyme, and thus no conclusions can be drawn about footprints or the lack thereof. However, it should be noted that affinity cleavage also failed to show binding in region 2301-2328, and is not subject to the sequence preference of a nuclease. In the latter region, DNase I controls were normal, but no footprints were observed. The sequence is unusually GC rich in this region compared to most other parts of the sequence. In spite of that, a number of double mismatch recognition sites can be rationalized.
Another consequence of the high density of possible PA1 sites is that it was not always possible to assign a distinct 10 bp binding sequence and binding mode for PA1 binding. This is particularly dramatic at site 5 (near 2276) and site 7 (near 2369, 2376), where there are three or more possible overlapping sequences that are consistent with the experimental observations. See also Fig. S2.
Complicating assignments further, any perfect reverse binding site could actually be a forward single mismatch site where C replaces G. Data do not permit us to distinguish between these possibilities. In addition, an all-A/T sequence could be bound in either forward or reverse orientations, or both, for the ensemble of molecules studied. Two such regions are present: one at site 5 (2267-77) and another at site 2 (2546-2556). Since it was always possible to assign a reasonable forward binding sequence to the data, reverse modes were neglected in the figures to streamline the presentation. Of course, this does not mean that PA1 fails to (also) bind in this reverse fashion.
Also unexpected was the low Kd for site 6 of 2.7 nM assigned to a forward double mismatch sequence. All other double mismatch binding sites were significantly weaker. Clearly there is a large number of possible double mismatch sequences and only a few of these are sampled here. It is possible that other factors contribute to unusual affinities, including flanking sequence effects (e.g., minor groove width and shape) and/or subtle DNA conformational behavior [58]. Additional studies would be required to clarify this issue.
Finally, there are a number of what appear to be reasonable sites for PA1 binding for which there is no discernable support from affinity cleavage, and any nearby integration could reflect another adjacent site. These unbound sites are found beginning at 2215 and 2223 near site 4, at 2267 and 2288 near site 5, and at 2461 near site 13 (all indicated with dashed arrows in Fig. 4).
It would be insightful to understand why these anomalies are observed and whether any are functions of the techniques (reagent reactivities, issues surrounding the location of the sequence in the duplex relative to the labeled ends (i.e. sensitivity), etc.) or something more significant like unusual local DNA structures. The former issues were addressed as much as possible through experimental design and controls. As for the latter, we are hopeful that additional data we are collecting and analyzing on other natural sequences will shed some light on these issues.
PA Mismatch Behavior in Other PA Systems
In the literature, there are extensive studies about polyamide specificity and the discrimination against mismatch sites. For example, these include a report on the effects of AT/TA recognition degeneracy on PADNA recognition [30]. In this report, only a 12-fold difference in binding was found when an 6-ring PA designed against 5′-TG(A,T)3-3′ was examined against eight possible DNA targets. This library was assembled by changing the parenthetic portion of the sequence from TTT to AAA. TTA, TAA, TAT. ATT, ATA and AAT comprised the parenthetic portion of the sequence for the other targets examined. The PA orientation was constant as determined by MPE•Fe(II)•EDTA affinity cleavage.
Another early report used aliphatic/aromatic amino acid pairings in polyamides for recognition of the minor groove of DNA [26]. In this particular study, a 14-ring polyamide was unable to distinguish between T (a mismatch), and G (a perfect match), in an eight bp sequence. However, upon replacing two specific pyrrole heterocycles with β-alanines (β), the binding specificity for the perfect match versus the mismatch increased very significantly. Thus, a study [26] of the base-pair specificity for paired aliphatic/aromatic polyamide building blocks reported that 12-ring PA dIm-β-ImPyPyPy-γ-ImPyPyPy-β-Py-β-Dp exhibited a greater than 48-fold specificity for 5′-TGTGAACA-3′ vs. a single bp mismatch, 5′-TGTTAACA-3′. This was compared to the parent, allheterocyclic 12-ring PA dImPyImPyPyPy-γ-ImPyPyPyPyPy-β-Dp, which could not distinguish the same perfect match and single-bp-mismatch sequences at all [26]. This work seemed to provide an elegant roadmap for the use of polyamides to recognize sequences of DNA with significant GC content. Such sequences that are obviously far removed from the AT rich DNA that distamycin and its higher homologs bind most readily. However, examination of the reported data shows at least two binding sites that were not discussed for several PAs: multiple such sites are found in Fig. 6a of Ref. [26]. Additional recognition complexity is presented without discussion in the supplementary material: For example, Fig. S1a of this paper shows four “extra,” unannotated binding sites for DNA recognition by dImPyPy-β-PyPy-γ-ImPy-β-PyPyPy-β-Dp.
In related but more recent reports, libraries of polyamides were examined for their DNA binding preferences using traditional and microarray-based methodologies [59]. For the initial library study, single bp mismatches were discriminated against relative to perfect match sites by factors of 4-100 [60].
For the most part, except for recent work such as that by Hsu et al. [60], literature studies of polyamide specificity have focused on relatively narrow ranges of DNA sequences and sequence contexts, and have both asked and answered eloquent, specific questions about the molecular recognition of DNA by these minor groove binding agents. For the purposes of such studies, DNA sequences of about 50 to 250 bp were employed, and the number of possible binding sites was generally limited to several. We found ourselves faced with a different type of problem than had typically been addressed in the literature: we wished to determine where a polyamide binds to a completely natural genomic sequence (HPV16), and our interest was driven by the anti-HPV16 and anti-HPV31 activity of PA1 as discovered in the collaboration between the JKB group and NanoVir, LLC [45]. Surprisingly, we have found prolific binding of PA1 to nearly half of the available base pairs in a 524-mer. Given that the first 40 and last 50 base pairs were not accessible to us by capillary electrophoresis (CE), it is more accurate to say that PA1 binds approximately half (196) of the visible 434 base pairs. The differences in dissociation constant for perfect match, single mismatch and double mismatch sites are often not dramatic. For example, the number of mismatches does not correlate particularly well with the binding strength: single mismatch site 7a has a Kd of ca. 3.8 nM, while double mismatch site 8 has a Kd of 62 nM, and site 11, a double mismatch site, has an intermediate Kd of 12 nM.
We have begun to wonder about the binding specificity of relatively long antiviral polyamides to HPV because of the broad spectrum anti-HPV activity discovered with NanoVir. At this point, a considerable number of NanoVir polyamides show excellent antiviral efficacy in both cell and tissue culture against HPV 16, 18 and 31. While there is certainly sequence homology for these three forms of HPV, it is not a type of homology that would be recognized by the same polyamide because the position of G is not generally conserved. We expect that the results presented here will help shed light on possible mechanisms of action, especially when combined with recent biological studies done at NanoVir [61]. Of note is that addition of an active PA to HPV-infected cells was shown to cause physical changes to the HPV genome (episome) [61], and the ability of active polyamides to bind far more sites than predicted is consistent with such major structural effects on the entire viral episome. These new viral DNA structures may be caused by massive binding of particular polyamides to the viral DNA, and these binding events may be precisely what induces the DNA Damage Response (DDR) to destroy that viral DNA [61]. It is notable that the DDR was neither induced by an inactive polyamide in HPV-positive cells nor by an active PA in HPV negative cells. The large size of the PA under investigation here confers a special tolerance for mismatch sites. This may limit accessibility of the PA to human DNA, which is generally very tightly wound in chromatin structures leading to the observed, non-cytotoxic antiviral effect [45, 46, 61].
Supplementary Material
Perfect match sites for a 14 ring antiviral PA on the HPV 16 genome bind with low nM affinity.
Single mismatch sites are bound with affinities similar to those observed with perfect sites.
Affinities for double mismatch PA sites on the HPV 16 genome vary from low to mid nM.
Most of the accessible sequence from the HPV sequence under study is susceptible to PA binding.
ACKNOWLEDGMENTS
We thank NIH for AI083803 awarded to JKB and NanoVir, LLC for financial support. JKB is co-founder part owner of NanoVir, LLC. We thank NanoVir, LLC for the loan of equipment and the use of active antiviral compounds. We thank the Danforth Plant Science Center for HRMS [National Science Foundation (NSF) Grant DBI 0922879].
Footnotes
SUPPLEMENTARY INFORMATION AVAILABLE: Fig. S1: Affinity cleavage, footprinting, and binding isotherms for a sample single mismatch site (5) and double mismatch site (10). Fig S2: detailed binding maps for PA1 around site 5 and site 7.
DISCLOSURE OF CONFLICTS JKB is co-founder and part owner of NanoVir, LLC, holding a significant equity position.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA. 1985;82:1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Neidle S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- [3].White S, Baird EE, Dervan PB. On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem. Biol. 1997;4:569–578. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- [4].Nickols NG, Szablowski JO, Hargrove AE, Li BC, Raskatov JA, Dervan PB. Activity of a Py-Im polyamide targeted to the estrogen response element. Molecular Cancer Therapeutics. 2013 doi: 10.1158/1535-7163.MCT-12-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dose C, Farkas ME, Chenoweth DM, Dervan PB. Next generation hairpin polyamides with (R)-3,4-diaminobutyric Acid turn unit. J. Am. Chem. Soc. 2008;130:6859–6866. doi: 10.1021/ja800888d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vaijayanthi T, Bando T, Hashiya K, Pandian GN, Sugiyama H. Design of a new fluorescent probe: Pyrrole/imidazole hairpin polyamides with pyrene conjugation at their γ-turn. Bioorg. Med. Chem. 2013;21:852–855. doi: 10.1016/j.bmc.2012.12.018. [DOI] [PubMed] [Google Scholar]
- [7].Meier JL, Montgomery DC, Dervan PB. Enhancing the cellular uptake of Py-Im polyamides through next-generation aryl turns. Nucleic Acids Res. 2012;40:2345–2356. doi: 10.1093/nar/gkr970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chenoweth DM, Harki DA, Phillips JW, Dose C, Dervan PB. Cyclic pyrroleimidazole polyamides targeted to the androgen response element. J. Am. Chem. Soc. 2009;131:7182–7188. doi: 10.1021/ja901309z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Q, Dwyer TJ, Tsui V, Case DA, Cho J, Dervan PB, Wemmer DE. NMR structure of a cyclic polyamide-DNA complex. J. Am. Chem. Soc. 2004;126:7958–7966. doi: 10.1021/ja0373622. [DOI] [PubMed] [Google Scholar]
- [10].Raskatov JA, Hargrove AE, So AY, Dervan PB. Pharmacokinetics of Py-Im polyamides depend on architecture: Cyclic versus linear. J. Am. Chem. Soc. 2012;134:7995–7999. doi: 10.1021/ja302588v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Parks ME, Baird EE, Dervan PB. Recognition of 5′-(A,T)GG(A,T)2-3′ Sequences in the minor groove of DNA by hairpin polyamides. J. Am. Chem. Soc. 1996;118:6153–6159. [Google Scholar]
- [12].Turner JM, Baird EE, Dervan PB. Recognition of Seven Base Pair Sequences in the minor groove of DNA by ten-ring pyrrole-imidazole polyamide hairpins. J. Am. Chem. Soc. 1997;119:7636–7644. [Google Scholar]
- [13].Tao Z-F, Fujiwara T, Saito I, Sugiyama H. Rational design of sequence-specific DNA alkylating agents based on duocarmycin A and pyrrole-imidazole hairpin polyamides. J. Am. Chem. Soc. 1999;121:4961–4967. [Google Scholar]
- [14].Harris D, Stewart M, Sielaff A, Mulder K, Brown T, Mackay H, Lee M. Solution phase synthesis of imidazole- and pyrrole-containing hairpin polyamides. Heterocyclic Commun. 2007;13:17–24. [Google Scholar]
- [15].Henry JA, Le NM, Nguyen B, Howard CM, Bailey SL, Horick SM, Buchmueller KL, Kotecha M, Hochhauser D, Hartley JA, Wilson WD, Lee M. Targeting the inverted CCAAT box 2 in the topoisomerase IIα promoter by JH-37, an imidazole-pyrrole polyamide hairpin: Design, synthesis, molecular biology, and biophysical studies. Biochemistry. 2004;43:12249–12257. doi: 10.1021/bi048785z. [DOI] [PubMed] [Google Scholar]
- [16].Sielaff A, Cooper A, Mackay H, Brown T, O’Hare C, Kluza J, Kotecha M, Le M, Hochhauser D, Hartley JA, Lee M. Binding of f-PIP and JH-37 to the inverted CCAAT box-2 of the topoisomerase IIα promoter, Abstracts of Papers. 233rd ACS National Meeting; Chicago, IL, US. March 25-29, 2007; 2007. MEDI-134. [Google Scholar]
- [17].Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. A cell-permeable synthetic transcription factor mimics, Angewandte Chemie. International Edition Eng. 2007;46:2865–2868. doi: 10.1002/anie.200604485. S2865/2861-S2865/2810. [DOI] [PubMed] [Google Scholar]
- [18].Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- [19].Dervan PB, Burli RW. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- [20].Floreancig PE, Swalley SE, Trauger JW, Dervan PB. Recognition of the minor groove of DNA by hairpin polyamides containing α-substituted-β-amino Acids. J. Am. Chem. Soc. 2000;122:6342–6350. [Google Scholar]
- [21].Kielkopf CL, Bremer RE, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. Structural Effects of DNA Sequence on T.A Recognition by hydroxypyrrole/pyrrole pairs in the minor groove. J. Mol. Biol. 2000;295:557–567. doi: 10.1006/jmbi.1999.3364. [DOI] [PubMed] [Google Scholar]
- [22].Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- [23].Marques MA, Doss RM, Foister S, Dervan PB. Expanding the repertoire of heterocycle ring pairs for programmable minor groove DNA recognition. J. Am. Chem. Soc. 2004;126:10339–10349. doi: 10.1021/ja0486465. [DOI] [PubMed] [Google Scholar]
- [24].Marques MA, Doss RM, Urbach AR, Dervan PB. Toward an understanding of the chemical etiology for DNA minor-groove recognition by polyamides. Helvetica Chimica Acta. 2002;85:4485–4517. [Google Scholar]
- [25].Rucker VC, Melander C, Dervan PB. Influence of β-alanine on hairpin polyamide orientation in the DNA minor groove. Helvetica Chimica Acta. 2003;86:1839–1851. [Google Scholar]
- [26].Turner JM, Swalley SE, Baird EE, Dervan PB. Aliphatic/aromatic amino acid pairings for polyamide recognition in the minor groove of DNA. J. Am. Chem. Soc. 1998;120:6219–6226. [Google Scholar]
- [27].Urbach AR, Dervan PB. Toward rules for 1:1 polyamide:DNA recognition. Proc. Natl. Acad. Sci. USA. 2001;98:4343–4348. doi: 10.1073/pnas.081070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Urbach AR, Love JJ, Ross SA, Dervan PB. Structure of a beta-alanine-linked polyamide bound to a full helical turn of purine tract DNA in the 1:1 motif. J. Mol. Biol. 2002;320:55–71. doi: 10.1016/S0022-2836(02)00430-8. [DOI] [PubMed] [Google Scholar]
- [29].Warren CL, Kratochvil NCS, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr., Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Nat. Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].White S, Baird EE, Dervan PB. Effects of the A.T/T.A degeneracy of pyrrole--imidazole polyamide recognition in the minor groove of DNA. Biochemistry. 1996;35:12532–12537. doi: 10.1021/bi960744i. [DOI] [PubMed] [Google Scholar]
- [31].Saha A, Pandian GN, Sato S, Taniguchi J, Hashiya K, Bando T, Sugiyama H. Synthesis and biological evaluation of a targeted DNA-binding transcriptional activator with HDAC8 inhibitory activity. Bioorg. Med. Chem. 2013;21:4201–4209. doi: 10.1016/j.bmc.2013.05.002. [DOI] [PubMed] [Google Scholar]
- [32].Ueno T, Fukuda N, Tsunemi A, Yao E-H, Matsuda H, Tahira K, Matsumoto T, Matsumoto K, Matsumoto Y, Nagase H, Sugiyama H, Sawamura T. A novel gene silencer, pyrrole-imidazole polyamide targeting human lectin-like oxidized low-density lipoprotein receptor-1 gene improves endothelial cell function. J. Hypertens. 2009;27:508–516. doi: 10.1097/hjh.0b013e3283207fe1. [DOI] [PubMed] [Google Scholar]
- [33].Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- [34].Fukasawa A, Nagashima T, Aoyama T, Fukuda N, Matsuda H, Ueno T, Sugiyama H, Nagase H, Matsumoto Y. Optimization and validation of a high-performance liquid chromatographic method with UV detection for the determination of pyrrole-imidazole polyamides in rat plasma. J. Chrom., B: Analytical Tech. Biomed. Life Sci. 2007;859:272–275. doi: 10.1016/j.jchromb.2007.09.032. [DOI] [PubMed] [Google Scholar]
- [35].Harki DA, Satyamurthy N, Stout DB, Phelps ME, Dervan PB. In vivo imaging of pyrrole-imidazole polyamides with positron emission tomography. Proc. Nat. Acad. Sci. USA. 2008;105:13039–13044. doi: 10.1073/pnas.0806308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pandian GN, Shinohara K.-i., Ohtsuki A, Nakano Y, Masafumi M, Bando T, Nagase H, Yamada Y, Watanabe A, Terada N, Sato S, Morinaga H, Sugiyama H. Synthetic small molecules for epigenetic activation of pluripotency genes in mouse embryonic fibroblasts. ChemBioChem. 2011;12:2822–2828. doi: 10.1002/cbic.201100597. [DOI] [PubMed] [Google Scholar]
- [37].Raskatov JA, Nickols NG, Hargrove AE, Marinov GK, Wold B, Dervan PB. Gene expression changes in a tumor xenograft by a pyrrole-imidazole polyamide. Proc. Nat. Acad. Sci. USA. 2012;109:16041–16045. doi: 10.1073/pnas.1214267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Synold T, Xi B, Wu J, Yen Y, Li B, Yang F, Phillips J, Nickols N, Dervan P. Single-dose pharmacokinetic and toxicity analysis of pyrrole–imidazole polyamides in mice. Cancer Chemother. Pharmacol. 2012;70:617–625. doi: 10.1007/s00280-012-1954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Nagase H, Watanabe T, Nobusue H, Suzuki T, Asami Y, Shinojima Y, Kawashima H, Takagi K, Mishra R, Igarashi J, Kimura M, Takayama T, Fukuda N, Sugiyama H. Inhibition of MMP-9 transcription and suppression of tumor metastasis by pyrrole-imidazole polyamide. Cancer Sci. 2010;101:759–766. doi: 10.1111/j.1349-7006.2009.01435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Crowley KS, Phillion DP, Woodard SS, Schweitzer BA, Singh M, Shabany H, Burnette B, Hippenmeyer P, Heitmeier M, Bashkin JK. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg. Med. Chem. Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- [41].Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci., USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nishijima S, Shinohara K.-i., Bando T, Minoshima M, Kashiwazaki G, Sugiyama H. Cell permeability of Py–Im-polyamide-fluorescein conjugates: Influence of molecular size and Py/Im content. Bioorg. Med. Chem. 2010;18:978–983. doi: 10.1016/j.bmc.2009.07.018. [DOI] [PubMed] [Google Scholar]
- [45].Edwards TG, Koeller KJ, Slomczynska U, Fok K, Helmus M, Bashkin JK, Fisher C. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 2011;91:177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Edwards TG, Helmus MJ, Koeller K, Bashkin JK, Fisher C. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J. Vir. 2013;87:3979–3989. doi: 10.1128/JVI.03473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang S, Nanjunda R, Aston K, Bashkin JK, Wilson WD. Correlation of local effects of DNA sequence and position of β-alanine inserts with polyamide-DNA complex binding affinities and kinetics. Biochemistry. 2012;51:9796–9806. doi: 10.1021/bi301327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dupureur CM, Bashkin JK, Aston K, Koeller KJ, Gaston KR, He G. Fluorescence assay of polyamide-DNA interactions. Anal. Biochem. 2012;423:178–183. doi: 10.1016/j.ab.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bashkin JK, Aston K, Ramos JP, Koeller KJ, Nanjunda R, He G, Dupureur CM, David Wilson W. Promoter scanning of the human COX-2 gene with 8-ring polyamides: Unexpected weakening of polyamide-DNA binding and selectivity by replacing an internal N-Me-pyrrole with beta-alanine. Biochimie. 2013;95:271–279. doi: 10.1016/j.biochi.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].White S, Baird EE, Dervan PB. Orientation Preferences of Pyrrole-Imidazole Polyamides in the Minor Groove of DNA. J. Am. Chem. Soc. 1997;119:8756–8765. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- [51].Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- [52].Herman DM, Baird EE, Dervan PB. Stereochemical control of the DNA binding affinity, sequence specificity, and orientation preference of chiral hairpin polyamides. J. Am. Chem. Soc. 1998;120:1382–1391. [Google Scholar]
- [53].He G, Vasilieva E, Bashkin JK, Dupureur CM. Mapping small DNA ligand hydroxyl radical footprinting and affinity cleavage products for capillary electrophoresis. Anal. Biochem. 2013;439:99–101. doi: 10.1016/j.ab.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schultz P, Taylor J, Dervan P. Design and synthesis of a sequence-specific DNA cleaving molecule. (Distamycin-EDTA)iron(II) J. Am. Chem. Soc. 1982;104:6861–6863. [Google Scholar]
- [55].Parks ME, Baird EE, Dervan PB. Optimization of the hairpin polyamide design for recognition of the minor groove of DNA. J. Am. Chem. Soc. 1996;118:6147–6152. [Google Scholar]
- [56].Koohy H, Down TA, Hubbard TJ. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS One. 2013;8:e69853. doi: 10.1371/journal.pone.0069853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Heddi B, Abi-Ghanem J, Lavigne M, Hartmann B. Sequence-Dependent DNA Flexibility Mediates DNase I Cleavage. J. Mol. Biol. 2010;395:123–133. doi: 10.1016/j.jmb.2009.10.023. [DOI] [PubMed] [Google Scholar]
- [58].Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Meier JL, Yu AS, Korf I, Segal DJ, Dervan PB. Guiding the design of synthetic DNA-binding molecules with massively parallel sequencing. J. Am. Chem. Soc. 2012;134:17814–17822. doi: 10.1021/ja308888c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CC, Dervan PB. Completion of a programmable DNA-binding small molecule library. Tetrahedron. 2007;63:6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Edwards TG, Vidmar TJ, Koeller K, Bashkin JK, Fisher C. DNA damage repair genes controlling human papillomavirus (HPV) episome levels under conditions of stability and extreme instability. PLoS ONE. 2013;8:e75406. doi: 10.1371/journal.pone.0075406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.