Abstract
The human proteome has millions of protein variants due to alternative RNA splicing and post-translational modifications, and variants that are related to diseases are frequently present in minute concentrations. For DNA and RNA, low concentrations can be amplified using the polymerase chain reaction, but there is no such reaction for proteins. Therefore, the development of single molecule protein sequencing is a critical step in the search for protein biomarkers. Here we show that single amino acids can be identified by trapping the molecules between two electrodes that are coated with a layer of recognition molecules and measuring the electron tunneling current across the junction. A given molecule can bind in more than one way in the junction, and we therefore use a machine-learning algorithm to distinguish between the sets of electronic ‘fingerprints’ associated with each binding motif. With this recognition tunneling technique, we are able to identify D, L enantiomers, a methylated amino acid, isobaric isomers, and short peptides. The results suggest that direct electronic sequencing of single proteins could be possible by sequentially measuring the products of processive exopeptidase digestion, or by using a molecular motor to pull proteins through a tunnel junction integrated with a nanopore.
The proteome is likely a much better molecular indicator of the current health status of humans than the genome,2 but proteomic data are harder to acquire.3 Protein sequences deduced from cDNA lack information about alternative splicing and post-translational modifications. Low concentrations of DNA and RNA are readily amplified by PCR, but there is no similar technique available for proteins. Thus, there may be many rare protein variants yet to be discovered at concentrations that are well below the detection limits of current techniques.4 In view of this, a single-molecule technique for protein sequencing is critical in identifying biomarkers and enabling the real-time diagnostic possibilities that follow. We are currently developing recognition tunneling (RT) as an electronic single molecule sequencing method for DNA. Here, we show that the method also works to identify individual amino acids and peptides, and so may open the way to single-molecule protein sequencing.
Recognition Tunneling
In recognition tunneling (Figure 1a), two metal electrodes, separated by a gap of about two nanometers, are covered with a layer of recognition molecules that are strongly-bonded to the electrodes. The recognition molecules form weaker, non-covalent, contacts with target analyte molecules. Single molecule signals dominate when a sharp electrode is used because the signal from the shortest path is by far the largest. When a small bias (<1 V) is applied across the electrode gap, molecules captured by these non-covalent contacts produce a stochastic train of current spikes (pA - nA) at kHz rates,1,5–7 the trapped molecule remaining bound for about a second (as determined by dynamic force spectroscopy measurements1). Such weakly-bonded complexes can remain intact for long times because of their confinement.8 Thermal vibrations of the molecule generate current spikes (Figs. 1c, d), the distribution of which is characteristic of the bonding in the tunnel junction (Figs. 1e, f). Much as in classical spectroscopy, the temporal, spectral, and amplitude information contained within a signal train (“signal features” in machine-learning terminology) can be used as an electronic “fingerprint” with which to identify each molecule that enters the RT junction. The electronic “fingerprints” are decoded9 with high accuracy by a machine-learning algorithm (the “Support Vector Machine”, SVM10).
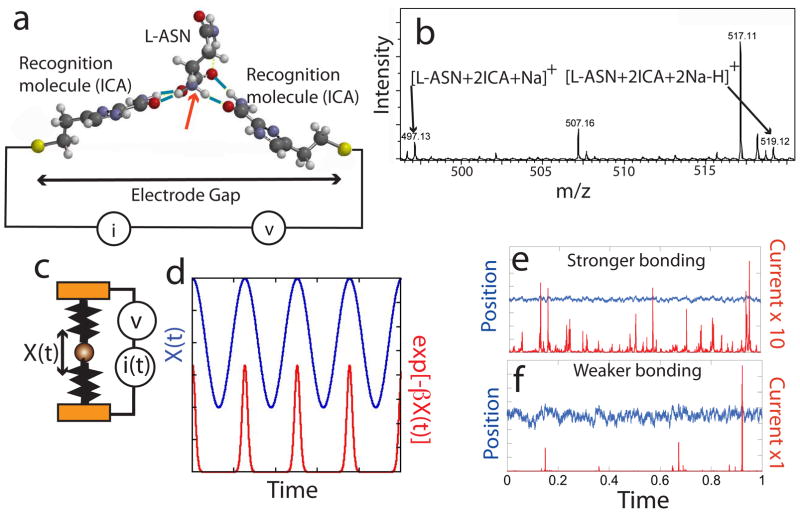
Figure 1. Recognition tunneling (RT).
(a) Recognition molecules (1H-imidazole-2-carboxamide, ICA) are strongly attached to a pair of closely spaced electrodes, displacing contamination and forming a chemically well -defined surface. An analyte (here shown as L-ASN) is captured by non covalent interactions (blue bars show H-bonds) with the recognition molecules. The bonding pattern is specific to the analyte. The red arrow shows the orientation of the molecular dipole for L-ASN. This orientation is different when D-ASN is captured (Fig. S1). (b) ESIMS shows that stoichiometric adducts form between reader molecules here illustrated for 2:1 complexes of ICA and L-ASN. Data for other analytes are given in Tables S6 and S7. How RT signals are generated: (c) Picturing the analyte as amass (sphere) trapped by a pair of springs that represent the non-covalent bonds, the extent of analyte motion, X(t), depends on the strength of the springs. (d) A simple sinusoidal motion of the analyte (blue trace) produces a series of sharp current spikes (red trace) because of the exponential dependence of tunnel current on position. (e) and (f) are simulations for random thermal excitation of a strongly (e) and more weakly (f) bonded analyte, showing how the current fluctuations are much bigger when the bonding is weaker (red traces). The simulations are carried out as described in Huang et al.1
Tunneling measurements
We used a scanning tunneling microscope, operated in buffered aqueous solution, to create a tunnel gap set to a reproducible distance by controlling the gap conductance, collecting useful signals out to 25 kHz in frequency. Palladium (Pd) probes, partially insulated with polyethylene, and Pd substrates,11 functionalized with the recognition molecule, 4(5)-(2-mercaptoethyl)-1H-imidazole-2-carboxamide, ICA, were used as electrodes.12 We found that a tunnel current of 4 pA at a bias of 0.5V produced RT signals from all but two of the twenty naturally-occurring amino acids (Figs. S2 and S3) while controls using buffer alone were almost free of signals (Figure S2a). This result is surprising, because ICA molecules were designed to interact with DNA bases.13 Nonetheless, electrospray ionization mass spectrometry (ESIMS)14,15 clearly shows the presence of 2:1 adducts of ICA molecules with all of the seven amino acids analyzed in the present paper (Figures 1b, S11, S12 and Tables S6 and S7).
Identifying amino acids
We demonstrate the power of RT with three applications: distinguishing a modified amino acid, sarcosine (or N-methylglycine, mGLY, a potential cancer marker16) from glycine (GLY), two enantiomers (L- and D-asparagine, ASN) and two isobaric amino acids, leucine (LEU) and isoleucine (ILE). In addition, we examined a pool of data from all seven analytes to evaluate how well any one amino acid can be identified. This is an essential first step in developing a sequence-reading technique.
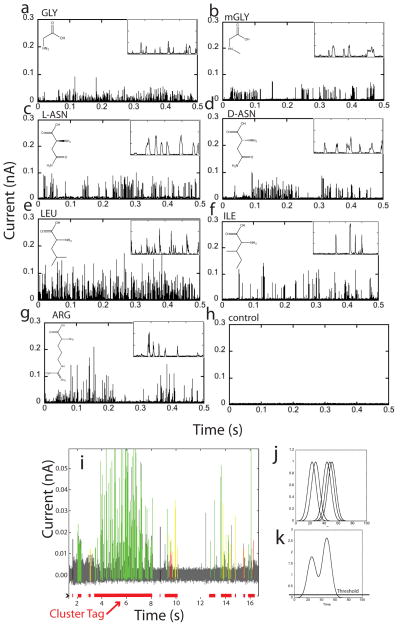
In a typical RT experiment, a solution of amino acid was added into the STM liquid cell after the tunnel junction had stabilized in buffer solution for ~ 2 hours. For each analyte, a minimum of 3 (usually 4) separate experiments were run with freshly-made probes, substrates and samples. Figure 2 illustrates representative signal trains. The spike shape carries significant information, and the insets show this in expanded traces. Signals occur in clusters and a computer algorithm (Fig. 2i) was developed to identify clustered data automatically. Clusters appear to correspond to single molecule binding events for the following reasons: (a) The duration of each cluster is on the order of 0.2 s (Figure S10b) comparable to the time for which hydrogen-bonded complexes remain bound in a nanogap.1,8,17 (b) Signals within clusters are much more strongly correlated than signals from different clusters (Figure S15). (c) In signals obtained from mixed samples (Fig. 5), each cluster gave signals from only one analyte or the other.
Figure 2.
Examples of RT signals from amino acids. GLY (a) and its N-methylated modification, sarcosine (mGLY) (b). Enantiomers L-ASN (c) and D-ASN (d). Isobaric isomers LEU (e) and ILE (f). (g) shows data for the charged amino acid, ARG. (h) is control data from buffer solution alone. The insets are expanded traces (current scale 150 pA, time scale 20 ms) displaying the complex peak shapes that are important features in the analysis of these data. (i) Signal trace for ARG, color-coded according to the peak assignments made by a machine learning algorithm (green = correct, red = wrong call, black = “water peak”, yellow = common to all amino acids). The red bars at the bottom mark signal clusters generated by a particular single-molecule binding event. Automatic cluster-identification was done by placing Gaussians of unit height and full-width of 4096 data points (1 data point = 20 us) at the location of each spike (j), summing them (k), and assigning a cluster to regions where this sum exceeds 0.05. This choice picks out obvious single molecule events well (cf. Fig. 5).
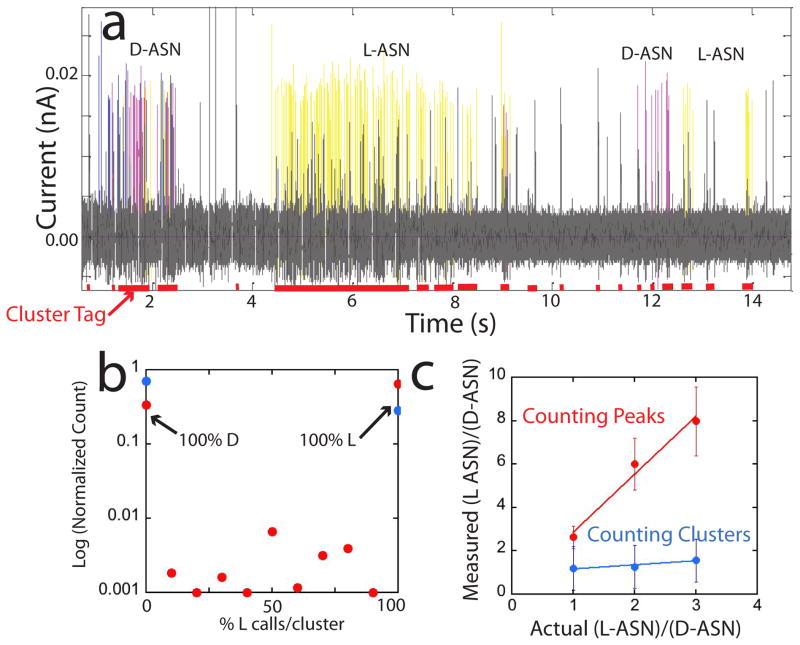
Figure 5.
A mixture produces alternating cluster signals as different molecules diffuse into and out of the gap. (a) Signal trace obtained with a 1:1 mixture of L- and D- asparagine. The Support Vector Machine assignments are coded purple (D-ASN) and yellow (L-ASN) (black spikes are unassigned). Each cluster (red tags) contains only one type of signal, as shown statistically in (b). The red points are for 556 raw data clusters and the blue points are for 400 clusters that remain after filtering for common signals. After filtering (blue points), no mixed clusters survive, with all of the clusters being 100% L- or D-ASN signals. Quantification of the L/D ratio using SVM trained on pure samples is shown in (c). The measured ratio increases with actual ratio in the samples but the calibration depends on whether the number of signal spikes (red) or clusters (blue) is used, probably reflecting differential binding. Error bars are from repeated runs and repeated samplings.
We illustrate how signal features can distinguish pairs of analytes using data obtained from mGLY and LEU (Figure 3). Tunnel-current amplitude distributions5,18,19 are largely overlapped (Figure 3a). Features associated with pulse shapes (Figures 3b,c) suffer less overlap, though the overlap is still limits the accuracy of calling single molecule events to around 70% (50% represents random calls). However, when these two signal features are used together to generate a 2D map of probability densities (Fig. 3d), only a small fraction of the data are overlapped (yellow, near the origin) leading to a 95% calling accuracy if signals in the red area are assigned to mGly and in the green area to Leu. This is an illustration of Cover’s theorem, which states that separability in pattern recognition increases in higher dimensions.20 We used the SVM,9,21 a machine-learning algorithm, to discover these relationships by training on a subset of the data. The SVM assigns data between pairs of classes by partitioning the feature space into two regions. For the data shown in Fig. 3d, this partition is the curve that best separates green from red regions. In this case SVM analysis yields an accuracy of less than 95% because a partition of the space cannot include all the “data islands”. However, SVM analysis can be extremely accurate when a large number of features are used.
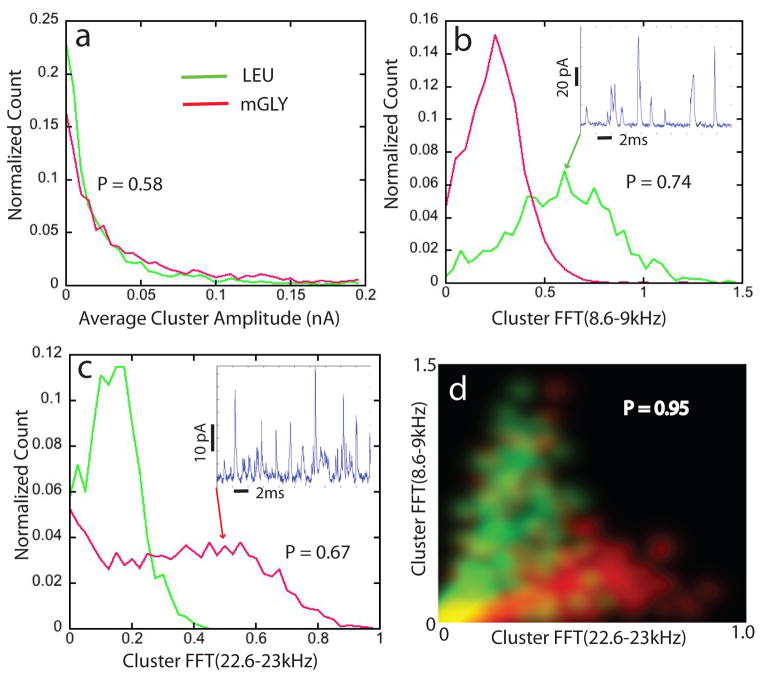
Figure 3.
Signal features identify analytes: (a) Peak amplitudes are exponentially distributed so provide little discrimination. Assigning the larger spikes to mGLY (red curve) yields an accuracy (p=0.58) only slightly better than random (0.5). Particular Fourier components (Table S1) of the clusters (b and c) show more separation, producing 74% (b) and 67% (c) accuracies if called solely on the more probable value of the feature. The way in which these Fourier components reflect peak shapes in a cluster is illustrated by the signal traces inset in (b) and (c), each trace having the feature value pointed to. The high amplitude of high-frequency components of the mGLY signals (inset in c) is evident in the sharper spikes. Accuracy improves when multiple features are used together. (d) Shows a 2D plot of probability density as a function of the two FFT feature values. The color scale shows mGLY data points as red and LEU points as green. Calling all the spikes with pairs of feature values that fall in the green regions as LEU and all the spikes with pairs of features that fall in the red regions as mGLY produces a correct call 95% of the time. Only the yellow regions yield ambiguous calls.
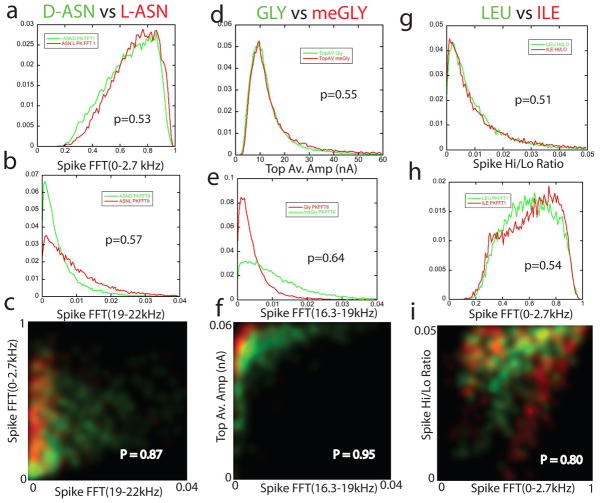
The effect of combining data from two or more parameters is even more dramatic in the case of chemically similar pairs of analytes (Figure 4). The six feature distributions shown in Figs. 4a,d,g,b,e and h are quite overlapped for each pair of analytes (the probability of a correct identification among each pair is marked on each plot with “p=”, and values are typically only a little over 50%). Using the probability densities plotted as a function of the values of pairs of features (Figs 4 c, f and i) increase the identification accuracy to 80% or greater in all three cases. (The separation of stereoisomers is presumably a consequence of the local-chiral adsorption geometry on surfaces.22)
Figure 4.
Closely related pairs of analytes can be significantly separated (>80%) using just two signal features together. All data are for pure solutions of one analyte. Chiral enantiomers D-ASN and L-ASN (a,b,c), GLY and mGLY (d,e,f), and the isobaric isomers LEU and ILE (g,hi) are quite well separated in a 2D probability density maps (c, f and i) even when the distributions of any one signal feature are almost completely overlapped in 1D (a,d,g,b,e and h see Methods and Table S1 for a description of these features). The 2D maps plot probability densities for the analyte pairs (color coded as listed at the top) as a function of both the features that, by themselves, produce separations only a little above random (0.51 to 0.64). Probabilities of making a correct call based on the probability densities are marked on c, f and I, and calculated as described in the caption for Figure 3.
SVM analysis with a large set of signal features is carried out as follows: Each signal spike, represented by N feature values, is plotted in an N dimensional space. A subset of known data is used to find the support vectors (of N-1 dimension) that best partition the known data and thus train the SVM. Data from subsequent analyses are then identified according to which side of the partition they reside on. Thus far we have described the SVM as a binary classifier, separating data into one of two classes, but multiclass SVMs are readily constructed. In the version of the SVM used here, training for multiple analytes works by determining the support vector set that best separates signals from each analyte from a pool of signal feature values taken from the remainder of the analytes. This process was repeated seven times to cover each of the analytes studied, generating seven different support vector Once trained, signals from an unknown sample are fed to all seven SVM’s sequentially, and a confidence level returned for assignment to each amino acid (as opposed to the remainder). Each signal spike is assigned to the amino acid corresponding to the highest confidence level.
Reproducibility of the SVM analysis
Key questions are: How reproducible are the tunneling data? How transferrable is the SVM training? To address these questions, we have analyzed multiple sets of data for each analyte, selecting signal features and settings for the SVM parameters that give robust results across multiple data sets. Each spike, and the cluster that contains it, were characterized by values of 161 signal features (Table S1). This large number of features includes parameters that describe amplitudes and amplitude fluctuations in both individual spikes and clusters as well spike and cluster shapes as described by Fourier and cepstrum23 components. (Fourier and cepstrum components were corrected for the frequency response of the instrument - Methods and Fig. S8). 30,000 data spikes (corresponding to about 3,000 clusters) were collected for each of the seven analytes. A correlation analysis (Fig S4) was used to identify groups of signal features that are linearly dependent, with each group represented one of the strongly correlated signal features. This reduced the total feature set by 40 (Table S2) to 121. A second correlation analysis identified features that vary from experimental run to run on the same analyte, and those that do not vary from one analyte to another. 15 such unreliable features were found (Table S3), and removing them reduces the sensitivity to experimental artifacts, and reduces the feature set to 106. Noise spikes (1 to 15% of the total data, varying from run to run) were eliminated by training the SVM to find signals common to all seven analytes. This last stage of noise filtering adjusted by varying the soft margin (broadening of the partition boundaries) of the SVM parameters. Increasing the soft margin improved accuracy at the cost of rejecting more signals (Fig. S5). The SVM was then trained on a small subset (~10%) of the data and then tested on the remainder. This process was repeated using randomly chosen training data to ensure that fluctuations in the outcomes were small. Finally, the analysis was repeated with smaller numbers of signal features to see how the final accuracy depended on the number of features used. Table 1 shows how a single signal spike can be assigned to any one of the seven analytes with 95% accuracy compared to 14% probability of a correct random call. Spikes within a cluster are highly correlated (Figure S15) so sampling multiple peaks within a cluster cannot be used to improve accuracy. However, spikes from different clusters constitute independent reads, and can be used to improve accuracy when reads are known to come from molecules of the same type as in the output from a chromatography column (or some other sequential separation). Cluster correlations were removed by randomizing the order of the spikes, and analytes assigned based on a majority vote of successive spikes (Table 1) to yield accuracies that approach 100%.
Table 1.
Accuracy with which any one of seven pure analytes is identified from the total pool of data taken from all seven pure samples using 52 signal features together. Results in the first row are based on a single spike. The subsequent rows are based on a majority vote using 3 and 5 spikes taken from different signal clusters. These results were obtained with the noise-filter soft margin set to reject ~ 70% of the data spikes.
| Number Spikes | ARG_L | ASN_D | ASN_L | GLY | ILE | LEU | mGLY |
|---|---|---|---|---|---|---|---|
| 1 | 95.14 | 94.99 | 96.99 | 97.24 | 96.87 | 94.36 | 96.45 |
| 3 | 98.77 | 99.62 | 99.99 | 99.62 | 99.99 | 99.55 | 99.99 |
| 5 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 |
Accuracy is reduced in the more challenging (and more realistic) case where the SVM is trained on data from one run and tested on data from other runs, because data filtering is not as stringent. In this case, accuracies of 90% or greater could be attained for pooled data from all 7 analytes, based on a single read. Once again, accuracies increased rapidly when repeated reads were made of spikes from different clusters.
Analyzing mixtures of analytes
Thus far we have confined our analysis to pure samples of one type of analyte, distinguishing one pure sample from six other pure samples. Can the SVM, trained on a pure analyte, recognize it in a mixture? To address this question, we made mixtures of L- and D-ASN with stoichiometric ratios of 1:1, 2:1 and 3:1, repeating measurements at each concentration twice. We then used the support vectors developed for pure L- and D-ASN to assign the spikes in the signal train obtained from mixtures. Figure 5a shows a stream of raw data that has been color-coded according to this assignment (yellow = L-ASN, purple = D-ASN, black = common). The red bars at the bottom of the trace mark the identified clusters, and it is clear that each cluster corresponds to just one analyte or the other. This is summarized statistically for 556 clusters in Figure 5b. Essentially all of the clusters consist of all L- or all D-ASN spikes with less than a percent in total containing more than one type of spike. When the common noise filter is applied only pure clusters remain (blue points). This further supports the view that clusters reflect single molecule binding events.
The measured stoichiometric ratio is quite sensitive to signal filtering and the method used to count molecules. Figure 5c shows the measured L/D ratio based on the total number of spikes of each type (red data points) and the total number of clusters of each type (blue data points). Spikes overcount the L-ASN content (the slope of the linear fit in Fig. 5a is 2.7) while clusters undercount it (the slope is 0.2). In the case where spikes are counted, the excess assigned to L-ASN is a consequence of longer clusters, probably reflecting stronger binding of this analyte to the recognition molecules. In the case where clusters are counted, the undercounting of L-ASN molecules may reflect a local reduction in concentration owing to preferential binding of L-ASN on the surfaces of the electrodes. Nonetheless, the relationship between measured and actual stoichiometry is monotonic and reproducible to better than about 20%.
RT signals from peptides
The obvious hydrogen bonding sites for amino acids are the zwitterionic centers (Fig. 1a). In a peptide, N and C termini are more spatially separated, so it is not at all clear that amino acids, as parts of peptides, will produce RT signals. We found that 100uM solutions of the short peptides GGGG and GGLL readily produced RT signals (Fig S6). Interestingly, the SVM trained on pure amino acids did not recognize either of these peptides as their constituent amino acids (Table S5a). However, each peptide produced distinctive signals, allowing one to be separated from the other, and also from any of the amino acids (Table S5a). Thus, the binding motifs of the amino acid residues in a peptide are clearly different from those of the same amino acids free in solution. We also obtained signals from the trimer, GGG. An SVM analysis of all three peptides (GGG, GGGG and GGLL) together (Table S5b) yielded > 90% accuracy (with 65% of the signals rejected as common, as might be expected given the sequence homology). Thus multiple peptides may be separated from each other, even when the difference is just one residue in four. This suggests that amino acid variants of proteins can be detected. In addition single molecule sequencing of proteins may be possible, particularly if residues can be presented to a tunnel gap sequentially.
In this pilot study, high concentrations (~100 μM) were used to ensure rapid diffusion of analytes into the tunnel gap (though concentrations down to 1 μM worked). Sample concentrations can easily be reduced by micro- or nano-fluidic injection of samples into the tunnel gap. Of significant interest is incorporation of tunnel junctions into nanopores, where capture assisted by electrophoresis24 or electroosmosis25, could reduce this concentration to the pM range.
Bonding in the RT Junctions
All seven analytes form stoichiometric adducts with one or two ICA molecules, as demonstrated by ESIMS (Figures S11 and S12 and Tables S6 and S7). ICA was designed to bond DNA bases, but the DFT calculated structures in Figures 1a and S1 show that amino acids can be captured by hydrogen bonding to ammonium and carboxylate groups of their zwitterionic centers. However, the observation that peptides generate RT signals that are different from those generated by amino acids (Table S5a), suggests that other types of binding motif are possible. How many such motifs might occur? To address this question for the case of the amino acids, we used an algorithm that identifies clusters of data26 to locate such clusters in the 24 dimensional-space occupied by the most significant signal features from single spikes (Figure S16). Three distinct clusters were found for six of the amino acids (Table S9) with only two for N-methylglycine, methylation presumably blocking a bonding site.
We examined bonding further using force spectroscopy. A dipeptide (Cys-Gly) was attached to an AFM tip through a PEG linker via the thiol of the cysteine residue and single-molecule rupture forces recorded (a hexane terminated PEG was used as a control). When the peptide retracted from an ICA coated gold surface, bond-rupture events were observed. The distribution of rupture forces (Figure S14) is consistent with two hydrogen bonds.17
Conclusions
In conclusion, RT generates complex signals, the basis of a new form of molecular spectroscopy for identifying a potentially vast range of chemicals at the single molecule level. It discriminates between members of molecular classes, like enantiomers and isobaric isomers, analytes which present challenges to other analytical techniques. RT has the potential for numerous significant advantages over a variety of current types of instrumentation and analytical methods that require chemical labeling or complex and expensive instrumentation such as mass spectrometers. Instrumentation, for single molecule analysis and protein sequencing that integrate RT and nanopore technologies on a solid-state device platform, would likely be substantially smaller, less expensive, have lower operating costs and be more robust. Individual amino acids can be identified with high accuracy. In the near-term, a microreactor containing an exo-peptidase should be able to identify the terminal sequence of proteins by feeding the digest to the tunnel gap and analyzing the time dependent signal (Fig. S7). We have also shown that peptide chains generate distinctive and reproducible signals. With RT tunnel junctions integrated into nanopores, it may very well prove possible to carry out continuous strand sequencing of proteins (a molecular motor being used to feed entire proteins into nanopores27). The real power of RT as a chemical spectroscopy lies in the possibility of massively parallel detection using large-scale integration of solid-state devices. Such devices are under development in our lab.
Methods
Preparation of analytical solutions
Amino acids were obtained from Sigma Aldrich (>98% purity) and dissolved in 1mM phosphate buffer (pH 7.4) made using water from a Milli-Q system with specific resistance of ~18 MΩ-cm and total organic carbon contamination below 5 ppb. Peptides were obtained from CPC Scientific and solutions prepared as for the amino acids.
Preparation of probes and substrates
Pd substrates were deposited on a 750μm silicon wafer using e-beam evaporation of 100 nm of Pd onto a 10 nm Ti adhesion layer. Probes were etched11 from 0.25 mm Pd wire (California Fine Wires). They were insulated with polyethylene to leave the metal end open with a linear dimension a few tens of nm. Probes were tested to ensure that leakage current was < 1pA in the standard buffer solution at 0.5V bias. This is because ionic leakage current cannot be simply subtracted from the signal because of its distance dependence,28 so leaky probes result in errors in the set point current. For functionalization, insulated probes and the Pd substrates were first cleaned by rinsing them with ethanol and H2O, dried with nitrogen, and then immersed in a solution of ICA13 (0.5 mM) in ethanol. After ~16 h, the probe and substrate were removed, rinsed with ethanol, gently dried with nitrogen, and used immediately.
Tunneling measurements
We used two different PicoSPM’s (Agilent Technologies, Chandler) equipped with custom LabView interfaces for data acquisition. Tunnel current was sampled at 50 kHz. The -3dB bandwidth of the current to voltage converter was 7 kHz, but useful signals were obtained out to the Nyquist limit of 25 kHz after correction for the instrumental response (Figure S8). The liquid cells were cleaned in Piranha (note solutions are potentially explosive and must be handed with extreme care) and rinsed with Milli-Q water and ethanol. The current set point was set to 4 pA with 0.5V bias applied (probe positive, as this results in less leakage), and the probe approached with integral and proportional gains set to 1.0. The surface was scanned to ensure that the grain structure of the Pd was clearly visible (Fig. S9). The microscope was left to stabilize for at least two hours before signals were recorded and then the integral and proportional gains reduced to 0.1.The control (1.0 mM phosphate buffer at pH 7.4) was run before an amino acid solution was measured. Recordings are distorted by movement of the Z PZT during runs in which a series of high amplitude spikes are recorded, but this artifact is common to all analytes and incorporated into the training of the SVM. We used different batches of substrates and probes for each run, usually recording four runs for each analyte. We also alternated measurements between different instruments. In this way, the influence of small changes in experimental conditions could be removed from the final analysis.
SVM Analysis
We used the kernel-mode support vector machine10 available from https://github.com/vjethava/svm-theta. Each spike above 15pA in amplitude was characterized using the features listed in Table S2. The shape of each spike was characterized by constructing a FFT. The resulting Fourier amplitude distribution was then down sampled using linear interpolation into nine bins of equal frequency interval from zero to 25 kHz. FFT amplitudes (prior to downsampling) were averaged across three equally-spaced frequency intervals (0–2.7 kHz, 8.4–11.1 kHz and 22.3–25 kHz) and these averages were used as additional features.as was the ratio of the highest to lowest FFT bins useful (Peak Hi/Lo ratio, Fig. 4g).
Clusters contain additional information. They are identified with a Gaussian-broadening algorithm as described in Figure 2. 9 The peaks used to locate the clusters were subject to a 15 pA threshold, but once a cluster was identified, all of the data in it was used for the analysis, so amplitudes down to the baseline were included (c.f., Fig. 3a). Distributions of cluster lengths for various analytes are shown in Figs. S10a and b. We also developed a series of features to describe these clusters (Table S2). These included the spike frequency within a cluster, as well as the Fourier spectrum of the whole cluster (deconvolved for instrumental response by spectral division). Clusters contain many more data points than individual spikes, so the downsampling of the FFT was much finer, with a total of 61 bins used (each one corresponding to 25 kHz/61 or 410 Hz in width). The method of Noll23 was used to calculate the cepstrum amplitudes from the Fourier transform of the power spectrum, downsampling again to 61 frequency bins.
In order not to bias the analysis towards features with bigger numerical values and ranges, we rescaled all features as follows: The distribution of each signal feature was measured for one amino acid (in this case, arginine for the amino acid analysis, and glycine for the peptide analysis), The scale factor and additive constant required to move the mean of the distribution to zero and the standard deviation to 1.0 was calculated. Feature values for all of the other analytes were remapped using the same linear transformation.
Feature selection was performed in 3 stages. First, those features that showed too much linear correlation were removed. The normalized correlation between different pairs of features (x,y) was defined in the usual way, σxy = 〈(x − x̄)(y − ȳ)〉 where we normalized the components using σxx = 1. All of the data from the entire pool was used to generate a correlation matrix where correlations are shown by off-diagonal elements (Fig. S4). Trial and error resulted in rejecting all feature combinations for which σxy ≥ 0.7. We chose one feature from each overly correlated set to represent the set in the next stage of analysis.
Second, a comparison was performed for each feature for its variation over repeated experiments on the same analyte versus the variation between the different analytes. Histograms of all feature values (c.f. Figs 3 and 4) were compiled for each experimental run for a given analyte. The absolute values of the differences between the normalized histograms were accumulated to give an “in-group” fluctuation. The same procedure was carried out for all possible pairs of analytes to give an “out-group” measure of fluctuation. Parameters were then ranked by the magnitude of the ratio of out-group to in-group fluctuation and the bottom 15 parameters dropped (Table S3). Last, the usefulness of the remaining features was evaluated by determining the identification accuracy obtained with a randomly selected group of them. A tree-search was used to maximize the efficiency of this process. This led to the 52 features (Table S4) used in the analysis reported in Table 1.
Full details of the SVM (written in Matlab) can be found in a download of the data analysis code available from https://svmsignalanalysis.codeplex.com/.
ESIMS
Solutions of amino acids (100 μM), ICA (100 μM), and ICA (100 μM) mixed with each of the amino acids in 1:1 and 2:1 molar ratios (200 μM ICA) were prepared by dissolving them into water. Samples were injected into a Bruker MicrOTOF-Q electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass spectrometer, and tandem mass spectrometry was used to confirm the composition of mass peaks from adducts. We checked that the lack of buffer did not hinder the acquisition of RT signals (Figure S13). Full details of the methods and analysis are given in the supporting information.
Force Spectroscopy
A Cys-Gly dipeptide was ligated to a PEG tether (N=36) which was in turn attached to a SiN AFM probe (VeecoProbes) using click chemistry.29 Force curves (Figure S14) were collected in aqueous buffer over a gold-coated mica substrate covered with a monolayer of ICA. Full details of the sample preparation, data acquisition and analysis are given in the supporting information.
Supplementary Material
Acknowledgments
Shuai Chang assisted in the original survey of amino acids. We thank Pei Pang, Predrag Krstic, Carlos Hernandez-Suarez and William Offenberg for useful discussions. This work was supported in part by a DNA sequencing technology grant from the NHGRI, HG 006323.
Footnotes
Author Contributions
YZ and HL carried out tunneling measurements with assistance from SS, WS and JI. BA wrote the SVM code and analyzed data. BG contributed to the analysis. SM carried out force spectroscopy experiments. CB and SB carried out the electrospray MS. PZ and SL designed experiments and SL wrote the paper.
Competing Financial Interests
YZ, PZ and SL are named as inventors in patent applications. SL is cofounder of a company based on this technology.
Additional Information
Supplementary Information accompanies this paper.
References
- 1.Huang S, et al. Identifying single bases in a DNA oligomer with electron tunneling. Nature Nanotechnol. 2010;5:868–873. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlen M, Ponten F. Antibody-based Proteomics for Human Tissue Profiling. Molecular & Cellular Proteomics. 2005;4:384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.National Research Council (US) Committee on Intellectual Property Rights in Genomic and Protein Research and Innovation. Reaping the Benefits of Genomic and Proteomic Research: Intellectual Property Rights, Innovation, and Public Health. National Academies Press (US); 2006. [PubMed] [Google Scholar]
- 4.Archakov AI, Ivanov YD, Lisitsa AV, Zgoda VG. AFM fishing nanotechnology is the way to reverse the Avogadro number in proteomics. Proteomics. 2007;7:4–9. doi: 10.1002/pmic.200600467. [DOI] [PubMed] [Google Scholar]
- 5.Chang S, et al. Electronic Signature of all four DNA Nucleosides in a Tunneling Gap. Nano Letts. 2010;10:1070–1075. doi: 10.1021/nl1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S, et al. Recognition tunneling measurement of the conductance of DNA bases embedded in self-assembled monolayers. J Phys Chem C. 2010;114:20443–22044. doi: 10.1021/jp104792s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay SM, et al. Recognition Tunneling. Nanotechnology. 2010;21:262001–262013. doi: 10.1088/0957-4484/21/26/262001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friddle RW, Noy A, De Yoreoa JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci (USA) 2012;109:13573–13578. doi: 10.1073/pnas.1202946109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang S, et al. Chemical Recognition and Binding Kinetics in a Functionalized Tunnel Junction. Nanotechnology. 2012;23:235101–235115. doi: 10.1088/0957-4484/23/23/235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CC, Lin CJ. LIBSVM : a library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2:27–52. [Google Scholar]
- 11.Chang S, et al. Palladium Electrodes for Molecular Tunnel Junctions. Nanotechnology. 2012;23:425202. doi: 10.1088/0957-4484/23/42/425202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuchband M, He J, Huang S, Lindsay SM. Insulated gold scanning tunneling microscopy probes for recognition tunneling in an aqueous environment. Rev Sci Instrum. 2012;83:015102. doi: 10.1063/1.3673640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang F, Li S, Lindsay SM, Zhang P. Synthesis, Physicochemical Properties and Hydrogen Bonding of 4(5)- Substituted-1H-imidazole-2-carboxamide A Potential Universal Reader for DNA Sequencing by Recognition Tunneling. Chemistry - a European Journal. 2012;18:5998–6007. doi: 10.1002/chem.201103306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel JRM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. International Journal of Mass Spectrometry. 2002;216:1–27. [Google Scholar]
- 15.Nesatyy VJ. Mass spectrometry evaluation of the solution and gas-phase binding properties of noncovalent protein complexes. International Journal of Mass Spectrometry. 2002;221:147–161. [Google Scholar]
- 16.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17.Fuhrmann A, et al. Long lifetime of hydrogen-bonded DNA basepairs by force spectroscopy. Biophys J. 2012;102:2381–2390. doi: 10.1016/j.bpj.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identification of Single Nucleotide via Tunnelling Current. Nature Nanotechnol. 2010;5:286–290. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 19.Zwolak M, Di Ventra M. Electronic Signature of DNA Nucleotides via Transverse Transport. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 20.Cover TM. Geometrical and Statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Transactions on Electronic Computers. 1965;EC-14:326–334. [Google Scholar]
- 21.Jethava V, Martinsson A, Bhattacharyya C, Dubhashi D. The Lovász θ function, SVMs and finding large dense subgraphs. Neural Information Processing Systems (online) 2012:1169–1177. [Google Scholar]
- 22.Kühnle A, Linderoth TR, Hammer B, Besenbacher F. Chiral recognition in dimerization of adsorbed cysteine observed by scanning tunnelling microscopy. Nature. 2002;415:891–893. doi: 10.1038/415891a. [DOI] [PubMed] [Google Scholar]
- 23.Noll AM. Short-Time Spectrum and Cepstrum Techniques for Vocal-Pitch Detection. J Acoustical Soc Am. 1964;36:296–302. [Google Scholar]
- 24.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keyser U. Controlling molecular transport through nanopores. J Roy Soc Interface. 2011;8:1369–1378. doi: 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu S. Fuzzy model identification based on cluster estimation. Journal of Intelligent & Fuzzy Systems. 1994;2:267–278. [Google Scholar]
- 27.Nivaia J, Marks DB, Akeson M. Unfoldase-mediated protein translocation though an alpha-hemolysin pore. Nature Biotechnol. 2013;31:247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bard AJ, Fan FRF, Mirkin MV. In: Electroanalytical Chemistry. Bard AJ, editor. Vol. 18. 1994. pp. 243–371. [Google Scholar]
- 29.Senapati S, Manna S, Lindsay SM, Zhang P. Application of catalyst-free click reactions in attaching affinity molecules to tips of atomic force microscopy for detection of protein biomarkers. Langmuir. 2013 doi: 10.1021/la4039667. dx.doi.org/10.1021/la4039667. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.