Abstract
In a recent study of Multiple Sclerosis (MS), we observed additive effects and epistatic interactions between variants of four genes that converge to induce T cell hyper-activity by altering Asn-(N) linked protein glycosylation: namely, the Golgi enzyme MGAT1, cytotoxic T-lymphocyte antigen 4 (CTLA-4), interleukin-2 receptor-α (IL2RA) and interleukin-7 receptor-α (IL7RA). As the CTLA-4, IL2RA and IL7RA variants are associated with Type 1 Diabetes (T1D), we examined for joint effects in T1D. Employing a novel conditional logistic regression for family-based datasets, epistatic and additive effects were observed using 1,423 multiplex families from the Type 1 Diabetes Genetic Consortium dataset. The IL2RA and IL7RA variants had univariate association in MS and T1D, while the MGAT1 and CTLA-4 variants associated with only MS or T1D, respectively. However, similar to MS, the MGAT1 variant haplotype interacted with CTLA4 (p=0.03), and a combination of IL2RA and IL7RA (p=0.01). The joint effects of MGAT1, CTLA4, IL2RA, IL7RA and the two interactions using a multiple conditional logistic regression were statistically highly significant (p<5×10−10). The MGAT1 - CTLA-4 interaction was replicated (p=0.01) in 179 trio families from the Genetics of Kidneys in Diabetes study. These data are consistent with defective N-glycosylation of T cells contributing to T1D pathogenesis.
Introduction
With the advancement of high-throughput genotyping technologies, hundreds of common genetic variants have been identified for human complex traits, such as Type 1 Diabetes [T1D, MIM 222100]. However, it has been reported that these genetic variants explain only a small proportion of heritability1. Gene-gene interactions are likely a major factor in explaining the mystery of missing heritability1 and thus, characterizing gene-gene interactions is of fundamental importance in unraveling the etiology of complex human diseases. However, successfully detecting gene-gene interactions faces many challenges. For example, a major constraint is the issue of multiple hypothesis testing. In a genome-wide search for gene-gene interactions, correcting for the very large number of tests greatly diminishes the power to detect interactions with moderate effects.
Single-gene disorders displaying Mendelian inheritance disrupt molecular pathways at a single step. However, a similar degree of pathway disruption may be obtained through small defects in multiple genes/environmental inputs that combine to disrupt a single pathway. These interactions may be epistatic or additive and may promote disease only when combined, and therefore poorly detected by GWAS. A functional approach that groups candidate variants based on a shared ability to alter a common molecular pathway provides an alternative method to identify interactions. Indeed, we recently reported that multiple environmental factors (vitamin D3 deficiency and metabolism) and multiple genetic variants (IL-7RA, IL-2RA, MGAT1, MGAT5 and CTLA-4) converge to dysregulate Golgi N-glycosylation and T cell function in Multiple Sclerosis (MS) 2-4. Causality of defective N-glycosylation in MS is supported by animal data, where genetic and metabolic induced alterations control T cell growth, TH1/TH17 differentiation and autoimmunity, including development of a spontaneous MS-like disease in Mgat5 deficient PL/J mice5-9. In MS, epistatic interactions and additive effects were observed between the four variants and environmental factors resulting in dysregulated N-glycosylation. For example, a haplotype of the Golgi N-glycosylation enzyme MGAT1 promotes MS, alters N-glycosylation, T cell activation thresholds, and surface expression of anti-autoimmune cytotoxic T-lymphocyte antigen 4 (CTLA-4) in a manner that is sensitive to metabolic conditions, Vitamin D3 signaling, the number of N-glycans attached to CTLA-4 (CTLA-4, rs231775) and interleukin-7/interleukin-2 signaling modulation by the IL-7R (rs6897932) and IL-2RA (rs2104286) variants. The interaction between the MGAT1 and CTLA-4 variants was epistatic, as CTLA-4 (rs231775) lacks univariate association with MS. In contrast, a non-additive interaction was observed between the MGAT1 risk variant and a combination of the IL-7R and IL-2RA risk variants, a result consistent with their opposing effects on mRNA levels of the MGAT1 enzyme. These data suggest that studies only examining univariate association, such as GWAS, are unlikely to adequately define heritability.
Studies have shown that genetic risk factors and pathways are frequently shared across different autoimmune diseases, albeit not always in the same direction10-14. For example, the IL2RA gene is significantly associated with both MS and T1D10; 11; however, the direction of the effect may be the same or opposite depending upon the specific variant examined11; 15. Similarly, HLA-DR15 is a risk marker for MS but is protective in T1D. These considerations, along with a common molecular target (ie N-glycosylation), motivated us to hypothesize that the four MS variants we detected2 may also interact in T1D to determine disease susceptibility. By borrowing the interaction information learned from MS, the burden of multiple testing present in a random genome-wide search is significantly reduced. The most common test for genetic association is the case-control design; however, this can be biased by population stratification. In contrast, a family-based design, such as the Type 1 Diabetes Genetics Consortium (T1DGC), provides inference of association that is robust against population stratification. A common way to analyze family data is with conditional logistic regression (CLR)16; 17. Cordell et al. 18 proposed the use of CLR to test genetic interaction between two variants by constructing 15 pseudo controls for each affected child. This approach is difficult to be generalized to examine multiple variants as the number of pseudo controls for each affected child grows exponentially with the number of variants. In addition, analyzing linked variants requires knowledge of recombination rates between variants. One way to avoid these complications is to match each affected child to the pseudo control whose genotype is formed by all of the other non-transmitted alleles by parents. Kotti et al. 19 used this matching strategy to test gene-gene interactions. We have recently shown that Kotti's matching strategy is suboptimal for testing gene-gene interactions20. Therefore, to test both additive and non-additive genetic effects using the multiplex family data collected by the T1DGC, we utilized an easy-to-implement yet efficient method of constructing pseudo controls. Using this method, we identified additive and non-additive effects of MGAT1, CTLA4, IL2RA, and IL7RA on T1D risk, with an overall p-value less than 5×10−10.
Results
A novel matching strategy
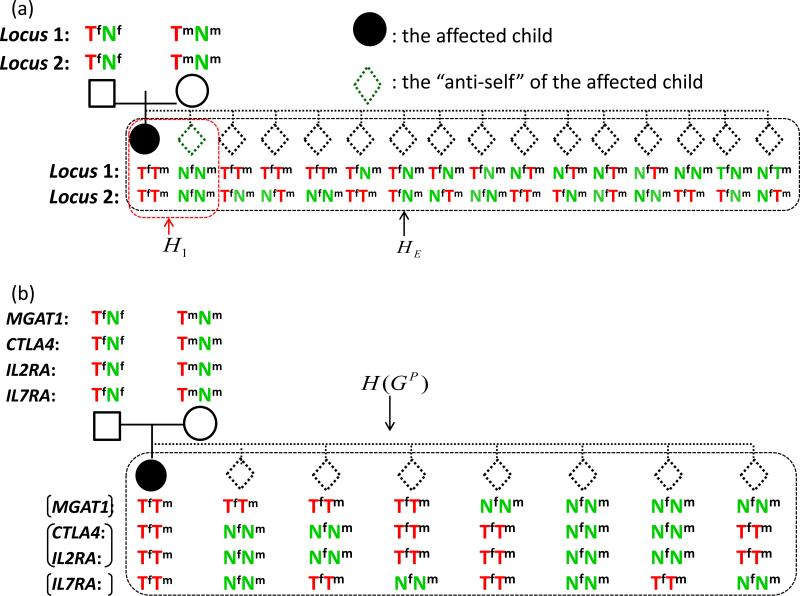
In recent theoretical work20 we examined and compared conditional logistic regressions under two matching strategies: 1:1 matching and exhaustive matching. Suppose that we are interested in testing L loci. In 1:1 matching, each affected child is matched to its “anti-self”, a pseudo control whose genotype is formed by the non-transmitted alleles. In exhaustive matching, each affected child is matched to 4L-1 pseudo controls. The two matching strategies at two loci for a case-parent trio are illustrated in Figure 1(a). Compared with exhaustive matching, the 1:1 matching strategy is simpler, more straightforward to implement, and computationally easier. Furthermore, 1:1 matching does not require knowledge of recombination rates between markers, whereas exhaustive matching does. Intuitively, 1:1 matching utilizes less information from the data. However, we found that 1:1 matching is as efficient as exhaustive matching when the true underlying genetic effects are additive, which requires that there are no intra- or inter-locus interactions20. Thus, when the focus is on additive genetic effects, we can safely use 1:1 matching; on the other hand, when the focus is non-additive effects, we should consider exhaustive matching.
Figure 1.
(a) The 1:1 and the exhaustive matching strategies for a case-parents trio at two loci. Tf: the allele transmitted to the child by the father. Nf: the allele not transmitted to the child by the father. Tm: the allele transmitted to the child by the mother. Nm: the allele not transmitted to the child by the mother. H1: all possible offspring genotypes given the couple's genotypes under the 1:1 matching. HE: all possible offspring genotypes given the couple's genotypes under the exhaustive matching. Under the null hypothesis of no association, the two genotypes in H1 are equally likely; under the null hypothesis of no association and linkage equilibrium between the two SNPs, the 16 genotypes in HE are equally likely. (b) The matching strategy used to analyze the four SNPs in this study.
Based upon our prior understanding of MGAT1 and other genetic variants altering N-glycosylation in MS2, we expected both additive effects and gene-gene interactions between variants of the following four genes: MGAT1 (rs7726005 and rs2070924), CTLA4 (rs231775), IL2RA (rs2104286) and IL7RA (rs6897932). At the individual gene level, our studies in MS indicate that the MGAT1 IVA/VT-T haplotype (rs7726005 and rs2070924) has a dominant effect while SNPs rs231775 (CTLA4), rs2104286 (IL2RA), and rs6897932 (IL7RA) demonstrate additive effects. Between genes, we found that the MGAT1 IVA/VT-T haplotype interacts with two sets of SNPs, rs231775 (CTLA4), and a combination of rs2104286 (IL2RA) and rs6897932 (IL7RA). In the following, we show how this prior information can be used to facilitate our construction of pseudo controls for each affected child in the T1DGC study.
The rs2070924 SNP in MGAT1 is almost in complete linkage disequilibrium with rs7726005. The haplotype formed by these two rare SNPs shows a dominant effect, indicating that exhaustive matching would be more efficient than 1:1 matching; however, the frequency of the haplotype is rare and in this case a dominant model is close to an additive model. To reduce the complexity of matching, we used 1:1 matching at MGAT1. Both rs2104286 (IL2RA) and rs6897932 (IL7RA) show additive individual effects in our previous MS study2; therefore, we used 1:1 matching at each of the two genes. As there was no evidence of genetic interaction between rs231775 (CTLA4) and rs2104286 (IL2RA), we assumed that the alleles at rs231775 (CTLA4) and rs2104286 (IL2RA) are co-transmitted from parents to offspring. In addition, there was no evidence of genetic interaction between rs231775 (CTLA4) and rs6897932 (IL7RA). Thus, we could also let rs231775 (CTLA4) co-transmit with rs6897932 (IL7RA) when constructing pseudo controls. Indeed, the two methods give identical results. Finally, because we wanted to test gene-gene interactions, we considered exhaustive matching among the three groups (MGAT1), (IL2RA, CTLA4), and (IL7RA), leading to 1:7 matching. Note that the genes are in linkage equilibrium. Thus, under the null hypothesis, the 8 possible offspring genotypes, including that of an affected child and his/her 7 matched pseudo controls, are equally likely. We used H(GP) to denote the 8 genotypes given the parental genotype GP. The final matching strategy to identify both additive and non-additive multi-locus genetic effects of these genes is summarized in Figure 1(b).
Conditional logistic regressions (CLR)
Matched case-control data are often analyzed by conditional logistic regressions. Let be the genotype of the ith child among n total affected children and be the genotype of the parents of the ith affected child. Using the matching strategy we described above, the likelihood function of association parameters is
The form of and depends upon our model. For example, when testing the association of the dominant MGAT1 IVA/VT-T haplotype in T1D, we assigned to 1 if the offspring has at least one copy of the haplotype and 0 otherwise. Correspondingly, is the log of the genotype relative risk (GRR) for carriers of the haplotype to those non-carriers. As another example, when testing interaction between the MGAT1 IVA/VT-T haplotype and rs231775 (CTLA4), is a vector of numerical values with three elements indicating the presence of the MGAT1 IV/V haplotype, the number of copies of the G allele (i.e. the risk allele) of rs231775 (CTLA4) and the product of the first two numbers. Correspondingly, is a vector of coefficients corresponding to the main effect of the MGAT1 IV/V haplotype, the main effect of rs231775 (CTLA4), and the interaction of the two variants, respectively.
We characterized the significance of additive and non-additive effects utilizing p-values. P-values of individual terms in a multiple CLR were calculated using the Wald test. When examining the joint effect of multiple terms, we use the likelihood ratio test. Conditional logistic regressions were fitted using the “clogit” function in the Survival package in R (http://cran.r-project.org/package=survival).
Gene-gene interactions and joint effects in the T1DGC study
We first examined the individual effects of the four variants. We used the risk alleles (column 3 of Table 1) as the test alleles and the protective alleles as the reference alleles. Many groups have reported association between T1D and CTLA4, IL2RA, and IL7RA21; 22. All variants are significantly associated with T1D except the MGAT1 IVA/VT-T haplotype (Table 1). This differs from MS, where all variants were associated except CTLA-4.
Table 1.
Individual genetic effects estimated from the T1DGC data.
| alleles | freq | GRR (95% CI) | p-value | |
|---|---|---|---|---|
| MGAT1 IVA/VT-T (rs7726005, rs2070924) | - | 0.039 | 1.06 (0.87-1.29) | 0.58 |
| rs231775 (CTLA-4) | A, G | G: 0.414 | 1.17 (1.08-1.26) | 5.3 ×10−5 |
| rs2104286 (IL2RA) | A, G | A: 0.775 | 1.25 (1.14-1.36) | 6.9 ×10−7 |
| rs6897932 (IL7RA) | C, T | C: 0.751 | 1.12 (1.02-1.22) | 1.3×10−2 |
Motivated by the interactions between MGAT1 and CTLA4, IL2RA, and IL7RA for MS susceptibility, we tested their genetic interactions for T1D susceptibility. Although the MGAT1 IVA/VT-T haplotype does not show univariate association with T1D, it is a protective, neutral, and risk allele for AA, AG, and GG CTLA-4 genotypes, respectively (Table 2). For subjects with the AG genotype at rs231775 (CTLA-4), the MGAT1 IVA/VT-T haplotype shows no association with T1D. For subjects with the AA genotype, i.e., the low risk group based on CTLA-4, the MGAT1 IVA/VT-T haplotype leads to increased risk of T1D with a GRR 1.53 (p-value=0.014). For subjects with the GG genotype, i.e., the high risk group based on CTLA-4, the MGAT1 VA/VT-T haplotype has a protective role for T1D with a GRR 0.58 (p-value=0.028). The different effects of the MGAT1 VA/VT-T haplotype under the three CTLA-4 genotypes suggest a gene-gene interaction between the two variants. The p-value for interaction is 0.029. Stratified point estimates of GRRs, 95% confidence intervals, and p-values can be found in Table 2.
Table 2.
Stratified analysis of MGAT1 IVA/VT-T using the T1DGC data. Stratification was based on CTLA4 genotypes.
| CTLA-4 genotypes | GRR (95% CI) | p-value |
|---|---|---|
| AA | 1.53 (1.09-2.15) | 1.4×10−2 |
| AG | 1.00 (0.75-1.33) | 1.0 |
| GG | 0.58 (0.36-0.95) | 2.8×10−2 |
The p-value for interaction is 2.9×10−2.
In MS, the MGAT1 IVA/VT-T haplotype also interacts with a combination of the IL2RA and IL7RA risk alleles2. Our conditional logistic regression indicates that the MGAT1 IVA/VT-T haplotype also shows differential effects on T1D susceptibility between subjects with four risk alleles of IL2RA and IL7RA versus other subjects (Table 3). The MGAT1 IVA/VT-T haplotype increases T1D risk for subjects at high risk based on IL2RA and IL7RA. It has a protective risk in the rest of the population, as can be seen from the point estimates of the GRRs in Table 3. Testing the interaction between them leads to a p-value of 0.013.
Table 3.
Stratified analysis of MGAT1 IVA/VT-T using the T1DGC data. Stratification was based on the number of risk alleles in IL2RA and IL7RA.
| IL * | GRR (95% CI) | p-value |
|---|---|---|
| 4 | 1.30 (0.95-1.77) | 0.10 |
| <4 | 0.90 (0.69-1.16) | 0.40 |
The p-value for interaction is 1.3×10−2.
IL is the number of risk alleles in IL2RA and IL7RA.
Since the variants we considered here are in linkage equilibrium, the interactions we observed cannot be explained by each other. To confirm this and evaluate the overall impact of the variants, we fit a multiple CLR with the four variants and the two interactions. Table 4 indicates that the point estimates of the GRRs and p-values for the four variants in the multiple CLR are similar to those from individual CLRs.
Table 4.
Multivariate analysis of the four genes, including main effects and two gene-gene interactions, using the T1DGC data.
| Variable | GRR (95% CI) | P-value |
|---|---|---|
| MGAT1 IVA/VT-T | 1.08 (0.80-1.46) | 0.63 |
| CTLA-4 | 1.19 (1.10-1.28) | 2.5×10−5 |
| IL2RA | 1.21 (1.11-1.33) | 2.1 ×10−5 |
| IL7RA | 1.10 (1.00-1.20) | 4.0×10−2 |
| MGAT1 IVA/VT-T × CTLA-4 | 1.58 (1.14-2.18) | 6.0×10−3 |
| MGAT1 VA/VT-T × (IL=4) | 0.76 (0.60-0.97) | 2.7×10−3 |
The p-value (LRT) for the two interaction terms is 2.0×10−3. The p-value (LRT) for the joint effect and additive and non-additive effects is 3.9×10−10.
The two interaction terms are significant in the multiple CLR, indicating they are still important after accounting for the main effects, consistent with what we observed in Tables 2 and 3. Therefore, we used a likelihood ratio test of two degrees of freedom to examine the joint effects of the two interaction terms. The p-value based on the likelihood ratio test is 0.002. Finally, we used a likelihood ratio test of seven degrees of freedom to test the joint effect of both main and interaction effects. The p-value is less than 5×10−10.
Replication analysis in the GoKinD study
The MGAT1 IVA/VT-T haplotype is not available in public GWAS datasets. Therefore, to attempt replicate the results above, we genotyped all 4 variants in 379 trios form the Genetics of Kidneys in Diabetes study (GoKinD). The results show that the only significant individual term is IL7RA (p-value 0.016). This is not surprising. First, the sample size is much smaller than that of the T1DGC study. Second, the T1D offspring in the GoKinD study have either clear-cut kidney disease or normal renal status despite long-term diabetes23. Thus, the combination of the two cohorts does not fully represent the T1D population. It has been estimated that about 30% of T1D patients have kidney disease23. We therefore hypothesized that those T1D patients without kidney disease are more similar to the general T1D population and conducted a separate analysis of the two cohorts. Due to the small sample size and the low frequency of the MGAT1 IVA/VT-T haplotype, we grouped the genotypes AG and GG at rs231775 (CTLA-4) when analyzing the interaction between the MGAT1 IVA/VT-T haplotype and CTLA-4.
Our results reveal that the MGAT1 IVA/VT-T haplotype has a significant interaction with CTLA-4 (p-value 0.012) in the cohort free from kidney disease, with the direction of interaction consistent with the one we found in the T1DGC study (Table 5). Specifically, in both the T1DGC study and the GoKinD trios free from kidney disease, the MGAT1 IVA/VT-T haplotype increases the risk of T1D in patients with the AA genotype at CTLA-4 and plays a protective role in patients with the AG or GG genotype. The interaction is not significant in the GoKinD trios with kidney disease and it is estimated that the MGAT1 IVA/VT-T haplotype has a similar role in the AA and AG/GG groups (Table 5).
Table 5.
Stratified analysis of MGAT1 IVA/VT-T using the GoKinD data. Stratification was based on CTLA4 genotypes.
| GRR of MGAT1 IVA/VT-T | ||||
|---|---|---|---|---|
| CTLA-4 genotypes | kidney patients* | kidney controls | ||
| GRR (95% CI) | p-value | GRR (95% CI) | p-value | |
| AA | 1.75 (0.51-5.98) | 0.37 | 0.83 (0.25-2.73 | 0.76 |
| AG/GG | 0.40 (0.08-2.06) | 0.27 | 0.78 (0.29-2.09) | 0.62 |
The p-value for interaction is 1.2×10−2.
We also tested interaction between the MGAT1 IVA/VT-T haplotype and the IL2RA and IL7RA polymorphisms. No interaction was identified for either the separate or combined analysis of the GoKinD families. This is not surprising given the lower effect size of the interaction and reduced power relative to the T1DGC cohort.
Discussion
In this article, we presented two potential gene-gene interactions involved in regulating N-glycosylation and possibly T1D susceptibility. Our analysis is knowledge-driven and is motivated by the fact that gene-gene interactions were observed in MS. It is known that both MS and T1D are autoimmune diseases and they share many common pathways. Different from a genome-wide search for gene-gene interactions, our analysis avoids multiple testing thus potentially improves power. We have observed gene-gene interaction between the MGAT1 IVA/VT-T haplotype and CTLA-4 in both the T1DGC study and the GoKinD study. This provides encouraging evidence that the observed interaction is likely to be true. The interaction between the MGAT1 IVA/VT-T haplotype and IL2RA and IL7RA is observed in the T1DGC families but not in the GoKinD families. As the T1DGC study is larger than the GoKinD and identifying gene-gene interaction requires large sample sizes, it is likely that the interaction is true but the GoKinD study is not powerful enough to detect it. Independent studies are required to further examine how MGAT1, IL2RA, and IL7RA jointly affect T1D susceptibility.
There were several important differences between the results in MS and T1D. Univariate association of the MGAT1 IVA/VT-T haplotype was observed in MS but not T1D while the G allele of CTLA-4 (rs231775) associated with T1D but not MS. Yet, in both diseases epistatic interaction was observed between the two variants. The MGAT1 IVA/VT-T haplotype enhanced risk of MS in combination with the GG and AG genotypes of CTLA-4 (rs231775), whereas a protective interaction with the GG genotype was present in T1D. These differences are not surprising as many confirmed loci of several autoimmune diseases often show opposite directions11; 13. Such differences may provide important clues and new tools for understanding the pathogenic process of complex traits. In this case, the observed differences may arise from variances in metabolism between T1D and MS, coupled with the molecular mechanisms by which the MGAT1 IVA/VT-T haplotype and CTLA-4 (rs231775) alter N-glycosylation and cell surface expression of the CTLA-4 protein in T cells. The MGAT1 IVA/VT-T haplotype is a gain of function that increases mRNA and protein levels of the Golgi enzyme Mgat1. When metabolism limits substrate availability (i.e. UDP-GlcNAc derived from glucose) to the Golgi, the MGAT1 IVA/VT-T gain of function haplotype paradoxically lowers N-glycan branching by limiting UDPGlcNAc availability to downstream Golgi enzymes, resulting in reduced cell surface expression of the anti-autoimmune CTLA-4 protein. The G allele of CTLA-4 (rs231775) decreases the number of N-glycans attached to CTLA-4 by 50%, thereby also reducing surface expression of the CTLA-4 protein. Thus, when metabolism limits Golgi substrate (UDP-GlcNAc) availability, the MGAT1 IVA/VT-T haplotype and the G allele of CTLA-4 (rs231775) combine to both lower CTLA-4 cell surface expression2, consistent with the genetic interaction observed in MS. In contrast, when metabolism increases Golgi UDP-GlcNAc substrate supply, as occurs with high glucose levels7, the MGAT1 IVA/VT-T haplotype has the opposite effect on N-glycan branching and CTLA-4 surface expression. Thus, under high glucose levels often present in early stages of T1D, the MGAT1 IVA/VT-T haplotype is expected to counteract the G allele of CTLA-4 (rs231775) to promote CTLA-4 protein expression and inhibit T cell function, consistent with the protective genetic interaction observed in T1D.
Most existing multi-locus methods for family data only provide an overall significance of multiple loci or specific combinations of alleles. Examples of such methods include haplotype-based methods24-30, genotype-based methods30-34, family-based multiple dimension reduction35, and contrasting linkage disequilibrium 36. In comparison, the method we utilized here has two advantages. First, most of the existing multi-locus methods are based upon 1:1 matching, which is not efficient for testing non-additive effects. Second, these methods are mainly for hypothesis testing. In contrast, our method not only provides the significance level of effects, but also provides point estimates of GRRs.
Methods
Datasets
We analyzed Caucasian multiplex or trio families collected by the Type 1 Diabetes Genetics Consortium (T1DGC) and the Genetics of Kidneys in Diabetes (GoKinD)23. The SNPs were genotyped using a method previously described2. We excluded families with missing parents or genotyping errors at any of the four SNPs. For T1DGC, this lead to 2,858 affected offspring and their parents from 1,423 multiplex families. The original goal of the GoKinD study was to identify genetic factors associated with kidney disease within T1D patients. Here, we used these families to examine the four genes. The GoKinD analysis is based upon 379 trio families with both parents genotyped, divided into two cohorts: (1) 200 T1D patients with kidney disease and their parents and (2) 179 T1D patients without kidney disease and their parents. Other information, such as genotyping, can be found in a report from the consortium23. To test genetic effects, we used conditional logistic regression with a novel matching strategy, as described in the following section.
Statistics
To test genetic effects, conditional logistic regression with a novel matching strategy was used and is described in detail in the results section. The significance of additive and non-additive effects was characterized by p-values. P-values of individual terms in a multiple CLR are calculated using the Wald test. When examining the joint effect of multiple terms, we used the likelihood ratio test. Conditional logistic regressions were fitted using the “clogit” function in the Survival package in R (http:// cran.r-project.org/package=survival).
Acknowledgments
The authors thank the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) for access to the T1DGC and GoKinD DNA samples. Research was supported in part by grant R01HG004960 from the National Human Genome Research Institute to Z.Y. and grant R01AI082266 from the National Institute of Allergy and Infectious Diseases to M.D.
Footnotes
Conflict of Interest
None reported.
References
- 1.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mkhikian H, Grigorian A, Li CF, Chen HL, Newton B, Zhou RW, Beeton C, Torossian S, Tatarian GG, Lee SU, et al. Genetics and the environment converge to dysregulate N-glycosylation in multiple sclerosis. Nat Commun. 2011;2:334. doi: 10.1038/ncomms1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li CF, Zhou RW, Mkhikian H, Newton BL, Yu Z, Demetriou M. Hypomorphic MGAT5 polymorphisms promote multiple sclerosis cooperatively with MGAT1 and interleukin-2 and 7 receptor variants. Journal of neuroimmunology. 2013 doi: 10.1016/j.jneuroim.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigorian A, Mkhikian H, Li CF, Newton BL, Zhou RW, Demetriou M. Pathogenesis of multiple sclerosis via environmental and genetic dysregulation of N-glycosylation. Seminars in immunopathology. 2012;34:415–424. doi: 10.1007/s00281-012-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- 6.Lee SU, Grigorian A, Pawling J, Chen IJ, Gao G, Mozaffar T, McKerlie C, Demetriou M. N-glycan processing deficiency promotes spontaneous inflammatory demyelination and neurodegeneration. J Biol Chem. 2007;282:33725–33734. doi: 10.1074/jbc.M704839200. [DOI] [PubMed] [Google Scholar]
- 7.Grigorian A, Lee SU, Tian W, Chen IJ, Gao G, Mendelsohn R, Dennis JW, Demetriou M. Control of T Cell-mediated autoimmunity by metabolite flux to N-glycan biosynthesis. J Biol Chem. 2007;282:20027–20035. doi: 10.1074/jbc.M701890200. [DOI] [PubMed] [Google Scholar]
- 8.Grigorian A, Araujo L, Naidu NN, Place D, Choudhury B, Demetriou M. N-acetylglucosamine inhibits T-helper 1 (Th1) / T-helper 17 (Th17) responses and treats experimental autoimmune encephalomyelitis. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M111.277814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigorian A, Mkhikian H, Demetriou M. Interleukin-2, Interleukin-7, T cell-mediated autoimmunity, and N-glycosylation. Annals of the New York Academy of Sciences. 2012;1253:49–57. doi: 10.1111/j.1749-6632.2011.06391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 11.Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abecasis GR, Barrett JC, Behrens T, Cho J, et al. Pervasive Sharing of Genetic Effects in Autoimmune Disease. Plos Genet. 2011;7(8):e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JHM, Howson JMM, Stevens H, McManus R, Wijmenga C, et al. Shared and Distinct Genetic Variants in Type 1 Diabetes and Celiac Disease. New Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune Disease Classification by Inverse Association with SNP Alleles. Plos Genet. 2009;5(12):e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd JA. Etiology of Type 1 Diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, Clark PM, Healy B, Walker N, Aubin C, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Self SG, Longton G, Kopecky KJ, Liang KY. On estimating HLA/disease association with application to a study of aplastic anemia. Biometrics. 1991;47:53–61. [PubMed] [Google Scholar]
- 17.Schaid DJ, Sommer SS. Genotype relative risks: methods for design and analysis of candidate-gene association studies. Am J Hum Genet. 1993;53:1114–1126. [PMC free article] [PubMed] [Google Scholar]
- 18.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotti S, Bickeboller H, Clerget-Darpoux F. Strategy for detecting susceptibility genes with weak or no marginal effect. Hum Hered. 2007;63:85–92. doi: 10.1159/000099180. [DOI] [PubMed] [Google Scholar]
- 20.Yu ZX, Deng L. Pseudosibship methods in the case-parents design. Stat Med. 2011;30:3236–3251. doi: 10.1002/sim.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nierras CR, Todd JA, Rich SS, Nerup J. Genetics of Type 1 Diabetes: What's Next? Diabetes. 2010;59:1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santiago JL, Alizadeh BZ, Martinez A, Espino L, de la Calle H, Fernandez-Arquero M, Figueredo MA, de la Concha EG, Roep BO, Koeleman BPC, et al. Study of the association between the CAPSL-IL7R locus and type 1 diabetes. Diabetologia. 2008;51:1653–1658. doi: 10.1007/s00125-008-1070-4. [DOI] [PubMed] [Google Scholar]
- 23.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, et al. Genetics of Kidneys in Diabetes (GoKinD) study: A genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Zhang S, Merikangas KR, Trixler M, Wildenauer DB, Sun F, Kidd KK. Transmission/disequilibrium tests using multiple tightly linked markers. Am J Hum Genet. 2000;67:936–946. doi: 10.1086/303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen AS, Satten GA. Statistical models for haplotype sharing in case-parent trio data. Hum Hered. 2007;64:35–44. doi: 10.1159/000101421. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Sha Q, Chen HS, Dong J, Jiang R. Transmission/disequilibrium test based on haplotype sharing for tightly linked markers. Am J Hum Genet. 2003;73:566–579. doi: 10.1086/378205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Meulen MA, te Meerman GJ. Haplotype sharing analysis in affected individuals from nuclear families with at least one affected offspring. Genet Epidemiol. 1997;14:915–920. doi: 10.1002/(SICI)1098-2272(1997)14:6<915::AID-GEPI59>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 28.Dudbridge F, Koeleman BP, Todd JA, Clayton DG. Unbiased application of the transmission/disequilibrium test to multilocus haplotypes. Am J Hum Genet. 2000;66:2009–2012. doi: 10.1086/302915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knapp M, Becker T. Family-based association analysis with tightly linked markers. Hum Hered. 2003;56:2–9. doi: 10.1159/000073727. [DOI] [PubMed] [Google Scholar]
- 30.Fan R, Knapp M, Wjst M, Zhao C, Xiong M. High resolution T2 association tests of complex diseases based on family data. Ann Hum Genet. 2005;69:187–208. doi: 10.1046/j.1529-8817.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- 31.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: A class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Zhang S, Sha Q. A multi-marker test based on family data in genome-wide association study. Bmc Genet. 2007;8:65. doi: 10.1186/1471-2156-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi M, Umbach DM, Weinberg CR. Identification of risk-related haplotypes with the use of multiple SNPs from nuclear families. Am J Hum Genet. 2007;81:53–66. doi: 10.1086/518670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee WC. Testing for candidate gene linkage disequilibrium using a dense array of single nucleotide polymorphisms in case-parents studies. Epidemiology. 2002;13:545–551. doi: 10.1097/00001648-200209000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Cattaert T, Urrea V, Naj AC, De Lobel L, De Wit V, Fu M, Mahachie John JM, Shen H, Calle ML, Ritchie MD, et al. FAM-MDR: a flexible family-based multifactor dimensionality reduction technique to detect epistasis using related individuals. Plos One. 2010;5:e10304. doi: 10.1371/journal.pone.0010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Z, Wang S. Contrasting linkage disequilibrium as a multilocus family-based association test. Genet Epidemiol. 2011;35:487–498. doi: 10.1002/gepi.20598. [DOI] [PubMed] [Google Scholar]