Abstract
A new alkylated chalcone (1), a new 1,16-hexadecanediol diester (2), and eight known compounds, were isolated from a dichloromethane-soluble repository extract of the leaves and twigs of Cryptocarya rubra collected in Hawaii. The structures of the new compounds were determined by interpretation of their spectroscopic data, and the absolute configurations of the two known cryptocaryanone-type flavonoid dimers, (+)-bicaryanone A (3) and (+)-chalcocaryanone C (4), were ascertained by analysis of their electronic circular dichroism and NOESY NMR spectra. All compounds isolated were evaluated against HT-29 human colon cancer cells, and, of these, (+)-cryptocaryone (5) was found to be potently cytotoxic toward this cancer cell line, with an IC50 value of 0.32 µM. This compound also exhibited glucose transport inhibitory activity when tested in a glucose uptake assay.
The genus Cryptocarya belongs to the pantropical family Lauraceae, of which several species grow in the tropical rainforests of Pacific Rim countries.1 Different types of natural products showing considerable structural diversity have been isolated from species of this genus, including flavonoids and pyrones.2–12 The first flavanones bearing a reduced A-ring and possessing a five-membered lactone moiety at the C-5 and C-6 positions, namely, cryptocaryanone-type flavonoids, were found to occur in diastereomeric form in Cryptocarya species,3 and several symmetrical dimers of this compound type3,11 and some complex dimeric molecules composed of flavonoid and pyrone units2,4,6,10 have been characterized. Cryptocaryanone-type flavonoids have been reported to exhibit cytotoxicity toward a panel of human cancer cell lines,3,6,7 with (+)-cryptocaryone (5) found to be potently cytotoxic toward human P388 leukemia cells (IC50, 0.04 µM).6 This compound was reported to induce human prostate PC-3 cancer cell apoptosis through activation of caspases 3 and 88 and to inhibit NF-κB activation5 and tyrosine kinase.6
Thus far, there has been no report on either the phytochemical or the biological attributes of Cryptocarya rubra C.R. Skeels. As an extension of a search for new natural product anticancer agents from diverse organisms,13 a small-scale sample of a dichloromethane partition of an ethanol-soluble extract of the leaves and twigs of C. rubra was investigated. This was obtained from a repository donated to the Natural Products Discovery Institute (NPDI), Institute for Hepatitis and Virus Research (IHVR) by Merck Research Laboratories, Rahway, NJ, USA. This extract was found to be cytotoxic toward the HT-29 human colon cancer cell line and was selected for further investigation. Using activity-guided fractionation by column chromatography over silica gel, two new and eight known compounds were identified. The structures of all compounds were characterized by analysis of their spectroscopic data, and the cytotoxicity toward the HT-29 human colon cancer cell line was evaluated by a cytotoxicity assay, with the active compound 5 also being tested for its glucose transport inhibitory effects in a glucose uptake assay.
RESULTS AND DISCUSSION
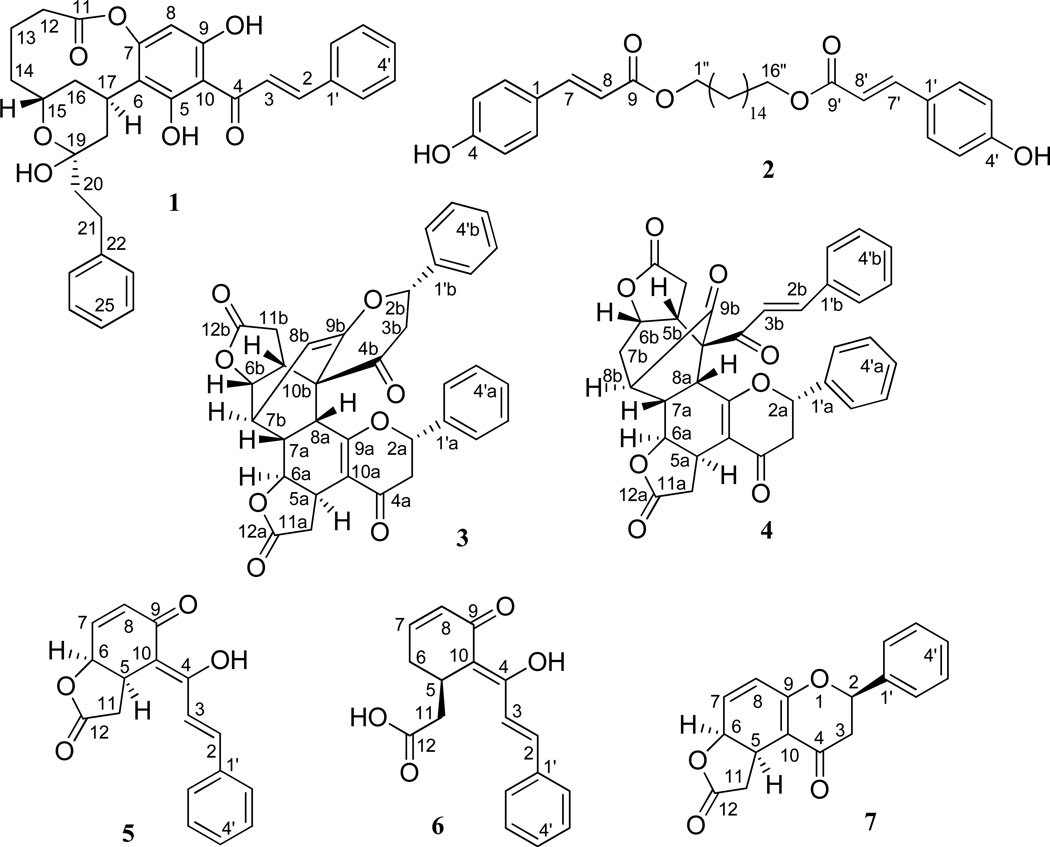
Silica gel chromatographic separation of a small-scale sample from a repository extract of C. rubra yielded two new compounds, (−)-rubrichalcolactone (1) and 1,16-hexadecanediol-di-p-coumaroate (2), and eight known compounds, (+)-bicaryanone A (3),3 (+)-chalcocaryanone C (4),3 (+)-cryptocaryone (5),3 (+)-desmethylinfectocaryone (6),6 (+)-cryptocaryanone A (7),3 (±)-5-hydroxy-7-methoxyflavanone,14 (±)-5,7-dihydroxyflavanone,15 and cinnamic acid.16,17
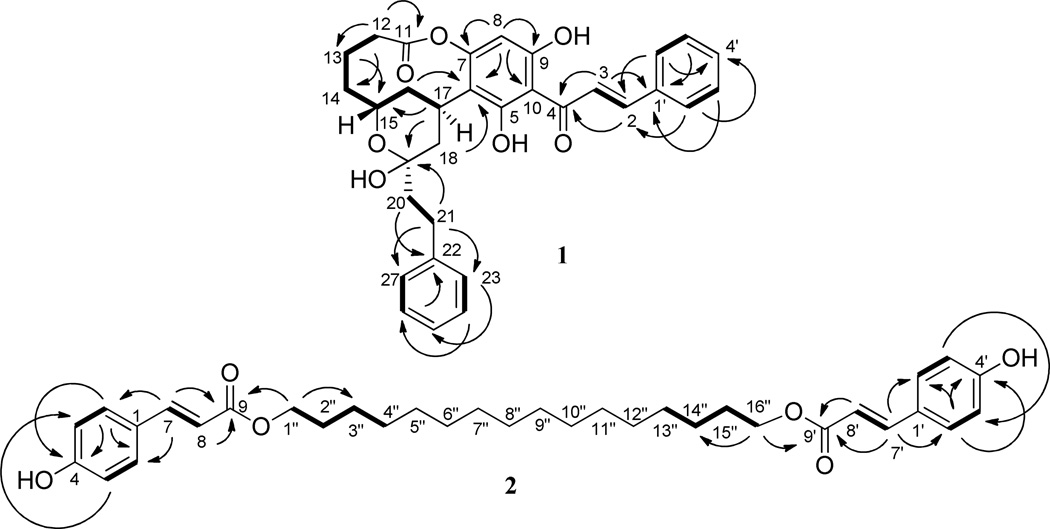
Compound 1 was isolated as an amorphous yellow powder that exhibited a sodiated molecular ion peak at m/z 551.2026, corresponding to a molecular formula of C32H32O7 (calcd for C32H32O7Na, 551.2046), with 17 indices of hydrogen deficiency. The IR spectrum showed absorption bands at 3297, 1708, 1625, and 1553 cm−1, indicating the presence of hydroxy, conjugated carbonyl vinylic and aromatic ring functionalities, respectively.2 The UV absorption maximum at 344 nm was consistent with 1 being a chalcone derivative.2 This was supported by the 1H NMR signals that appeared in the range δH 7.00–7.55 for 10 protons for two monosubstituted aromatic rings, and resonances at δH 7.79 (d, J = 16.0 Hz) and δH 8.01 (d, J = 15.6 Hz), assignable to protons of a trans double bond conjugated to a carbonyl group, and the signals for a chalcone unit displayed in the 13C NMR spectrum (Table 1).2 These preliminary assignments were supported by HMBC correlations of H-2/C-4, H-3/C-1′, H-3′/C-1′, and H-6′/C-4′ (Figure 1). Three signals at δC 158.8 (C-9), 159.3 (C-5), and 165.5 (C-7) observed in the 13C NMR spectrum indicated that the chalcone unit of 1 contains a trioxygenated ring A. A low-field singlet at δH 13.81 was consistent with the presence of a hydroxy group at C-5 hydrogen-bonded with a C-4 carbonyl group.
Table 1.
1H- and 13C NMR Spectroscopic Data for (<)-Rubrichalcolactone (1) in CDCl3
| positiona | δH (mult., J, Hz)b | δC (mult.)c | positiona | δH (mult., J, Hz)b | δC (mult.)c |
|---|---|---|---|---|---|
| 2 | 7.79 d (16.0) | 142.4 CH | 18 | 1.83 m, 2.04 m | 32.1 CH2 |
| 3 | 8.01 d (15.6) | 127.9 CH | 19 | 100.7 C | |
| 4 | 193.0 C | 20 | 2.23 m 2.60 m | 42.6 CH2 | |
| 5 | 13.81br s (OH) | 159.3 C | 21 | 2.83 t (9.6) | 29.5 CH2 |
| 6 | 104.7 C | 22 | 141.5 C | ||
| 7 | 165.5 C | 23 | 7.00 d (6.8) | 128.3 CH | |
| 8 | 6.03 br s | 96.2 CH | 24 | 7.15 overlapped | 128.5 CH |
| 9 | 158.8 C | 25 | 7.15 overlapped | 125.9 CH | |
| 10 | 105.4 C | 26 | 7.15 overlapped | 128.5 CH | |
| 11 | 173.5 C | 27 | 7.00 d (6.8) | 128.3 CH | |
| 12 | 2.35 m | 33.7 CH2 | 1′ | 135.5 C | |
| 13 | 1.66 m, 1.72 m | 21.0 CH2 | 2′ | 7.55 d (7.6) | 128.5 CH |
| 14 | 1.49 m, 1.53 m | 34.9 CH2 | 3′ | 7.22 overlapped | 129.0 CH |
| 15 | 3.74 br s | 69.8 CH | 4′ | 7.30 overlapped | 130.2 CH |
| 16 | 1.66 m, 1.72 m | 35.6 CH2 | 5′ | 7.22 overlapped | 129.0 CH |
| 17 | 3.49 br s | 23.3 CH | 6′ | 7.55 d (7.6) | 128.5 CH |
Assigned by analysis of the 1H, 13C, DEPT 90, DEPT 135, COSY, HSQC, and HMBC NMR spectra.
Recorded at 400.1 MHz and referenced to the solvent residual peak at δ 7.26.24 J values are presented in Hz and omitted if the signals overlapped as multiplets. The overlapped signals assigned from the 1H-1H COSY, HSQC, and HMBC spectra are presented without designating multiplicity.
Recorded at 150.9 MHz and referenced to the solvent residual peak at δ 77.16.24 CH3, CH2, CH, and C multiplicities were determined by DEPT 90 and DEPT 135 and HSQC experiments.
Figure 1.
COSY (─, 1H →1H) and key HMBC (↷, 1H → 13C) correlations of compounds 1 and 2.
To meet 17 indices of hydrogen deficiency, a bicyclic system composed of a 10-membered ring and a tetrahydropyran ring connected by a phenylethyl group was proposed for 1 (10 indices for the chalcone residue, four for the phenylethyl group, one for an ester carbonyl group, and two for a bicyclic system). The 10-membered ring containing C-6, C-7, oxygen, and C-11–C-17 was proposed to be linked to the chalcone unit at the C-6 and C-7 positions, as indicated by 1H-1H COSY sequences between H2-12/H2-13/H2-14/H-15/H2-16/H-17 and HMBC correlations of H-16/C-6, H-18/C-6, H-12/C-11, and H-12/C-14 (Figure 1). The tetrahydropyran ring between C-15 and C-19 was found to be fused with the 10-membered ring through C-15 and C-17 and to be connected with a phenylethyl group at C-19, as implied by the 1H-1H COSY sequence of H-15/H2-16/H-17/ H2-18 and HMBC correlations of H-17/C-15, H-17/C-19, H-20/C-22, H-21/C-19, H-21/C-23, H-21/C-27, H-23/C-25, and H-26/C-22 (Figure 1). Thus, compound 1 was identified as an analogue of kurzichalcolactone, a compound known to occur in Cryptocarya kurzii,2 C. obovata,4 and C. konishii.6 Comparison of the 1H- and 13C NMR spectra of 1 with those of kurzichalcolactone2 indicated that both compounds contain a comparable chalcone unit and a closely related side chain. In the 1H- and 13C NMR spectra of 1, the signals due to a phenylethenyl group of kurzichalcolactone2 were replaced by the signals assignable to a phenylethyl group.
The electronic circular dichroism (ECD) spectrum of 1 showing a negative Cotton effect (CE) at 230 nm and positive CEs at 247 and 344 nm was indicative of exciton coupling arising from the α,β-unsaturated carbonyl conjugated with a benzene ring chromophore at C-17, similar to that of chartaceone A1,18 indicating a 17R configuration for this compound. From a Dreiding model (Figure S5, Supporting Information), it was evident that both H-15 and OH-19 of 1 should be assigned axial orientations to avoid steric hindrance (no NOESY correlation between H-15 and H-17 observed in the NOESY 2D NMR spectrum of 1), which indicated 15R and 19S absolute configurations, respectively. Such a determination was supported by the closely comparable NMR data and specific rotation value of 1 with those of kurziflavolactones A–D and kurzichalcolactone.2 Accordingly, the structure of (−)-rubrichalcolactone (1) was assigned as shown.
Compound 2 was isolated as an amorphous colorless powder with a molecular formula of C34H46O6, as determined by HRESIMS (m/z 573.3183 [M + Na]+, calcd for C34H46O6Na, 573.3192). It showed UV (λmax 228 and 311 nm) and IR [νmax 3351 (hydroxy), 1683 and 1632 (α,β-unsaturated ester), and 1605, 1586, and 1444 (aromatic ring)] absorption bands, which indicated the presence of an α,β-unsaturated enoyl group.19 The 1H NMR spectrum displayed signals due to a trans double bond conjugated with a ketone group at δH 6.32 (d, J = 16.0 Hz) and 7.64 (d, J = 15.6 Hz) and two pairs of para-coupled aromatic protons at δH 6.85 (d, J = 8.4 Hz) and 7.44 (d, J = 8.8 Hz),20,21 together with overlapped signals for a long-chain alkyl unit at δH 1.26 (Table 2).22–24 In turn, in the 13C NMR spectrum, seven signals for a p-coumaroyl group were observed in the range δC 167.7–115.9,20,21 as well as a signal for an oxymethylene group at δC 64.8, and several signals for methylene groups of a long-chain system (Table 2). An oxygen bridge between the p-coumaroyl group and the long-chain alkyl system was deduced from the HMBC correlations from H-1″ to C-9 and C-3″ and H-16″ to C-9′ and C-14″ (Figure 1). Inspection of the MS and NMR spectra of 2 indicated that this compound is composed by two p-coumaroyl groups connected to two primary hydroxy groups of a 1,16-hexadecanediol residue, which was supported by COSY and key HMBC correlations shown in Figure 1. Based on these observations, the structure of 2 was determined as 1,16-hexadecanediol-di-p-coumaroate.
Table 2.
1H- and 13C NMR Spectroscopic Data for 1,16-Hexadecanediol-di-p-coumaroate (2) in CDCl3
| positiona | δH (mult., J, Hz)b | δC (mult.)c |
|---|---|---|
| 1/1′ | 127.5 C | |
| 2/2′ | 7.44 d (8.8) | 130.1 CH |
| 3/3′ | 6.85 d (8.4) | 116.0 CH |
| 4/4′ | 157.8 C | |
| 5/5′ | 6.85 d (8.4) | 116.0 CH |
| 6/6′ | 7.44 d (8.8) | 130.1 CH |
| 7/7′ | 7.64 d (15.6) | 144.4 CH |
| 8/8′ | 6.32 d (16.0) | 115.9 CH |
| 9/9′ | 167.7 C | |
| 1″/16″ | 4.21 t (6.8) | 64.8 CH2 |
| 2″/15″ | 1.56 m | 28.9 CH2 |
| 3″/14″ | 1.41 m | 26.1 CH2 |
| 4″–13″ | 1.26 overlapped | 29.3–29.9 CH2 |
Assigned by analysis of the 1H, 13C, DEPT 90, DEPT 135, COSY, HSQC, and HMBC NMR spectra.
Recorded at 400.1 MHz and referenced to the solvent residual peak at δ 7.26.24 J values are presented in Hz.
Recorded at 100.6 MHz and referenced to the solvent residual peak at δ 77.16.24 CH3, CH2, CH, and C multiplicities were determined by DEPT 90 and DEPT 135 and HSQC experiments.
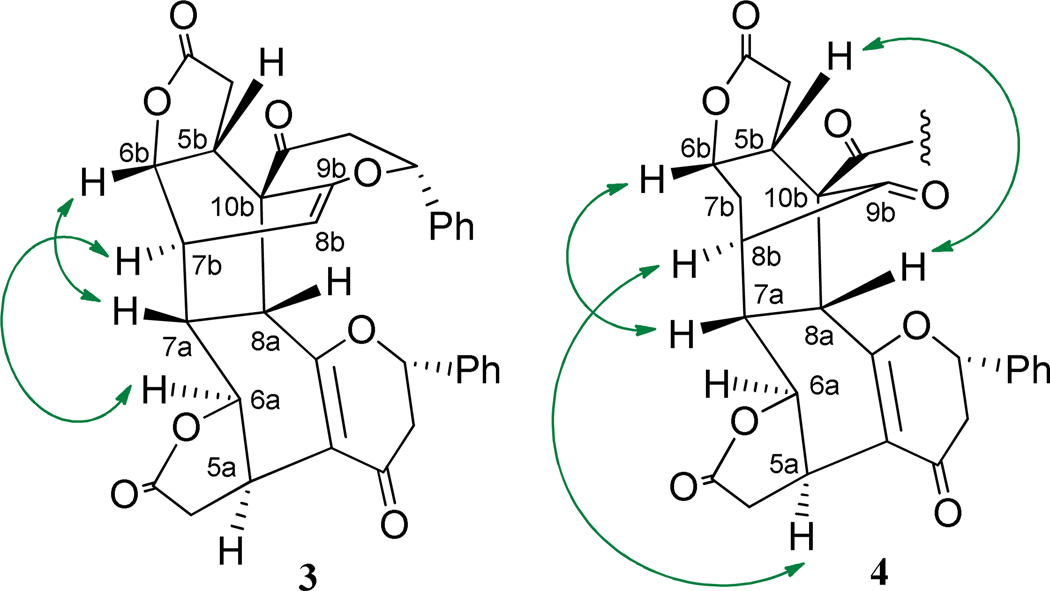
(+)-Bicaryanone A (3) and (+)-chalcocaryanone C (4) were characterized by comparison of their spectroscopic data (Figure 3, Table S1 and Figure S6, Supporting Information) with their literature values.3 These two cryptocaryanone-type flavonoid dimers were isolated and identified originally from Cryptocarya infectoria3 and then from C. chingii,11 with a Diels-Alder (DA) cycloaddition occurring between two identical cryptocaryanone A molecules being proposed for the biogenesis of 3.3 The structure of (+)-bicaryanone D, a C-2a and C-2b epimer of (+)-bicaryanone A (3), has been established by analysis of its single-crystal X-ray data.3 However, in the literature,3,11 the structures of (+)-bicaryanone A (3), (+)-bicaryanone D, and (+)-chalcocaryanone C (4) do not match the crystal structure of bicaryanone D,3 since the stereocenters at C-7a, -7b/-8b, -8a, and -10b of these compounds were not indicated correctly, and the absolute configurations at these positions of 3 and 4 were not demonstrated unequivocally. In the present study, a structure showing the conformation of the central ring system of bicaryanone D has been generated by a Mercury program (Mercury Cambridge Structural Database (CSD) version 3.1.1)25 based on the reported crystal structure of bicaryanone D,3 and the structures of 3 and 4 were modified accordingly. The absolute configurations of both 3 and 4 have been determined by measurement of their NOESY 2D NMR and ECD spectra and comparison of these data with those of bicaryanone D,3 together with a consideration of the Diels-Alder cis cycloaddition reaction.26,27
Figure 3.
↶↷, 1H →1H) correlations of compounds 3 and 4. Data were recorded at a DRX-800 MHz NMR spectrometer for 3 and at a DRX-600 MHz NMR spectrometer for 4.
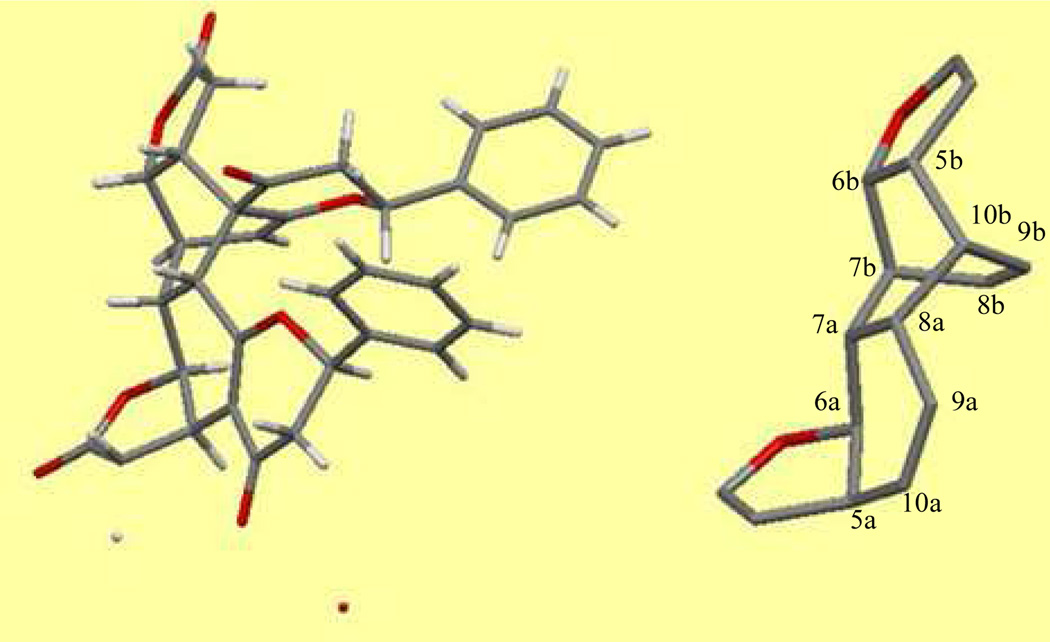
As shown in Figure 2, the structure of (+)-bicaryanone D is composed of two cryptocaryanone-type flavonoid units connected by a C-10b-spiro multiple-ring system adopting a boat-boat conformation, with C-5a through to C-10a comprising the first boat conformation with a near-planar cis-fused lactone ring at the C-5a and C-6a positions, and C-7a, C-8a, C-10b, and C-5b through to C-7b representing the second boat conformation, composed of a near-planar cis-fused lactone ring at the C-5b and C-6b positions. The olefinic C-8b-C-9b bond bridges the C-7b and C-10b positions. Also, H-2a, H-2b, H-5a, H-6a, and H-7b, and the bridge of C-8b-C9b are α-oriented, while H-5b, H-6b, H-7a, and H-8a, and C-10b-C-4b bond are β-oriented. The (2aR, 2bR) absolute configuration of (+)-bicaryanone D was established by analysis of its ECD spectrum, especially the negative CE value at 337 nm.3 Thus, the (2aR, 5aR, 6aS, 7aR, 8aR, 2bR, 5bR, 6bS, 7bR, 10bR) absolute configuration was defined by the collective analysis of its reported single-crystal structure and the preferred conformation shown in Figure 2.
Figure 2.
The structure of bicaryanone D and partial structure showing the conformation of the central ring system as generated by Mercury CSD Version 3.1.1,25 and based on the reported crystal structure of bicaryanone D.3
A (2aS, 5aR, 6aS, 2bS, 5bR, 6bS) absolute configuration for 3 was established by comparison of its NMR and ECD data with those of cryptocaryanones A and B and (+)-bicaryanone D,3 and a (2aS, 5aR, 6aS, 5bR, 6bS) absolute configuration could be determined for 4 by comparing its NMR and ECD spectra with those of 3 (Figures 3 and 4) and (+)-bicaryanone D.3 The correlations between H-6a/H-7b and H-7a/H-6b (Figure 3) in the NOESY NMR spectrum of 3 indicated a (7aR, 7bR) absolute configuration, while the correlations between H-6b/H-7a, H-5a/H-8b, and H-5b/H-8a (Figure 3) in the NOESY NMR spectrum of 4 indicated a (7aS, 8aR, 8bR) absolute configuration. Both 3 and 4 are proposed to be generated from an intermolecular [π4s + π2s] cycloaddition between two cryptocaryanone-type flavonoid units, with the preferred cis ring juncture being formed,26,27 as supported by the crystal structure of bicaryanone D (Figure 2).3 Therefore, compounds 3 and 4 should contain a central ring system showing the same conformation and configuration as those of bicaryanone D (Figure 3), with a (8aR, 10bR) absolute configuration for 3 and a 10bS absolute configuration for 4. This proposal was supported by the similar NMR data, consistent ECD curves (Figure 4), and NOESY correlations (Figure 3) of 3 and 4 with those of (+)-bicaryanone D.3 Therefore, the (2aS, 5aR, 6aS, 7aR, 8aR, 2bS, 5bR, 6bS, 7bR, 10bR) absolute configuration for 3 and the (2aS, 5aR, 6aS, 7aS, 8aR, 5bR, 6bS, 8bR, 10bS) absolute configuration for 4 were determined.
Figure 4.
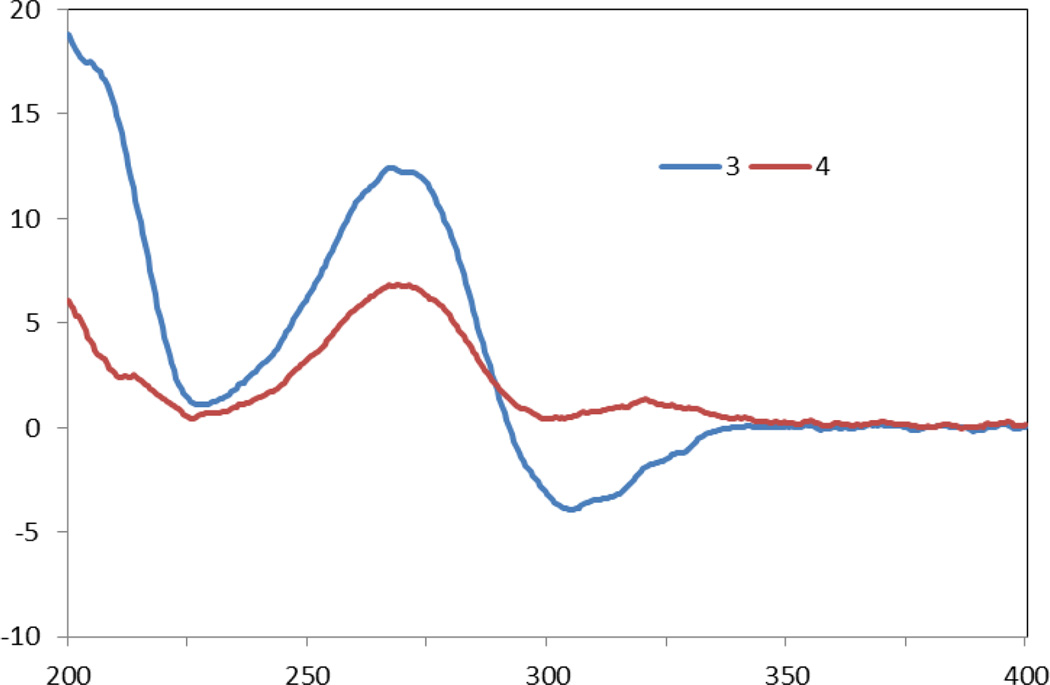
ECD spectra of 3 and 4. The data were obtained in MeOH corrected by subtracting a spectrum of the appropriate solution in the absence of the samples recorded under identical conditions.
All compounds isolated in the present study were tested for their cytotoxicity against the HT-29 human colon cancer cell line following a previous method,19 using paclitaxel as the positive control. Consistent with a previous report,6 (+)-cryptocaryone (5) showed potent cytotoxicity toward this cell line, with an IC50 value of 0.32 µM, but its close analogue, (+)-desmethylinfectocaryone (6), was inactive (IC50 >10 µg/mL), indicating that the five-membered lactone ring connected to the reduced A ring of a flavanone unit is required for the cytotoxicity of this type of compound. (+)-Cryptocaryanone A (7) exhibited borderline cytotoxicity toward this cell line, with an IC50 value of 10.9 µM (3.1 µg/mL), which is much lower than that of 5, implying that a conjugated system in this type of compound can improve the cytotoxicity. All other compounds were found to be non-cytotoxic toward this cell line (IC50 >10 µM), including the two dimeric cryptocaryanone-type flavonoids, 3 and 4.
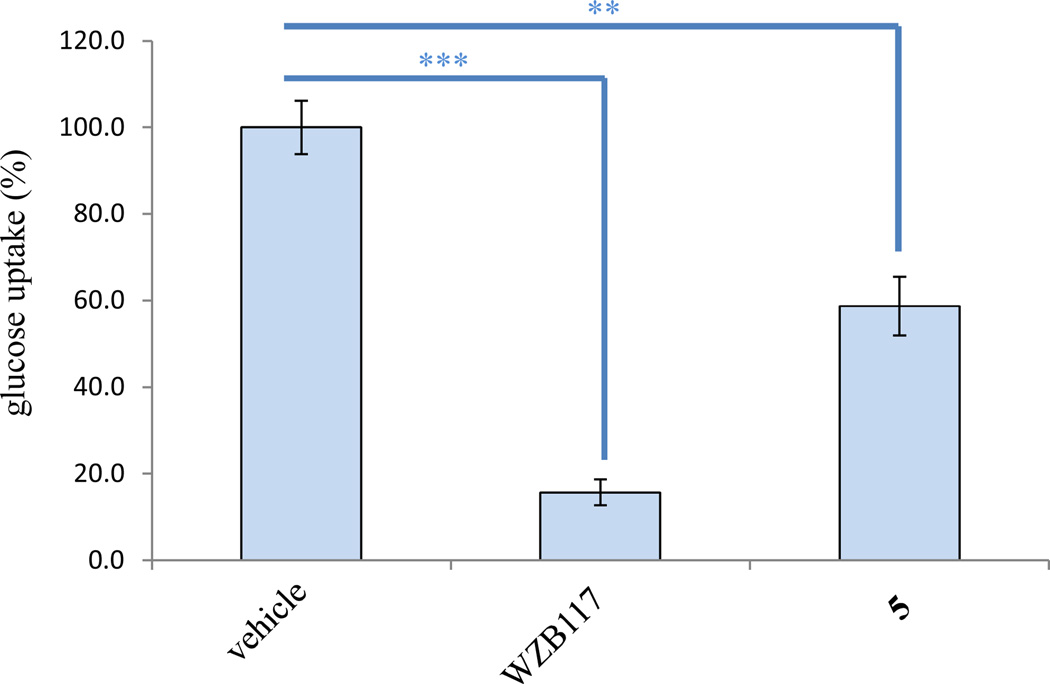
Cancer cells use glycolysis rather than oxidative phosphorylation to meet their energy and biomass synthesis needs for fast proliferation rates, hence glucose-uptake inhibition has been regarded as a potential target for discovery of new anticancer agents.28–32 A new strategy targeted to GLUT-1 inhibition to treat cancer is emerging, and down-regulation of GLUT-1 was proposed as the potential anticancer mechanism for the flavone, apigenin.31,32 The major cytotoxic principle of C. rubra, (+)-cryptocaryone (5), isolated in the present study, was tested in a glucose uptake assay, using the vehicle solvent as the negative control and WZB117, a potent glucose transport inhibitor showing antitumor efficacy in vivo identified in our previous study,30 as the positive control. The results showed that the human cancer cell transported 100, 58.7, and 15.7% of glucose (Figure 5), respectively, when treated by vehicle, 5 (30 µM), and WZB117 (30 µM). The glucose uptake value of the cells treated by 5 was significantly different from that of the cells treated by the vehicle control (p ≤ 0.01). Our previous studies have shown that some small molecules exhibit both cytotoxicity toward a panel of human cancer cells and glucose-uptake inhibition,28–30 of which WZB117 suppressed tumor growth in a nude mouse model by a glucose deprivation-like mechanism targeting GLUT-1-mediated glucose transport.30 The present study showed that (+)-cryptocaryone (5) exhibited a glucose transport inhibitory effect, implying that cryptocaryone-type compounds might have potential for further investigation as potential anticancer agents targeted to glucose deprivation.
Figure 5.
Inhibition of glucose transport by compound 5. H1299 human lung cancer cells were treated by vehicle, WZB117 (30 µM, positive control), or 5 (30 µM) for 15 min. After glucose uptake was initiated by 2-deoxy-d-[3H] glucose, cells were lysed and the radioactivity of the cell lysates was measured. The result showed that compound 5 inhibited glucose transport (columns, means, n = 3; bars, SE; ** p ≤ 0.01 and *** p ≤ 0.001).
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotation values were obtained on a Perkin-Elmer model 343 polarimeter. UV spectra were recorded on a Hitachi U2910 UV spectrophotometer. ECD measurements were performed using a JASCO J-810 spectropolarimeter. IR spectra were recorded on a Nicolet 6700 FT-IR spectrometer. 1H- and 13C-, DEPT, HSQC, HMBC, NOESY, and COSY NMR spectra were recorded at room temperature on a Bruker Avance DRX-400, DRX-600, or DRX-800 MHz NMR spectrometer. ESIMS and HRESIMS were measured on a Q-TOF mass spectrometer in the positive-ion mode. Column chromatography was conducted using silica gel (65 × 250 or 230 × 400 mesh, Sorbent Technologies). Analytical thin-layer chromatography (TLC) systems were performed on precoated silica gel 60 F254 plates (Sorbent Technologies, Atlanta, GA). Sephadex LH-20 was purchased from Amersham Biosciences, Uppsala, Sweden. For visualization of TLC plates, sulfuric acid reagent was used. All procedures were carried out with solvents purchased from commercial sources that were used without further purification.
Plant Material
The leaves and twigs of C. rubra were collected and identified by Dr. Hans Beck in Manoa Valley, Oahu County, Hawaii in August 1992. A voucher herbarium specimen representing this collection has been deposited at the New York Botanical Garden, Bronx, NY, under accession number HB01243.
Extraction and Isolation
The ground dried leaves and twigs of C. rubra were extracted exhaustively with EtOH at room temperature. The solvent was evaporated in vacuo. The resultant dried EtOH extract was suspended in 10% H2O in MeOH and partitioned with n-hexane three times. The spent aqueous layer was concentrated under a vacuum to approximately 40% aqueous MeOH and extracted three times with equal volumes of dichloromethane (DCM).
A small aliquot of a cytotoxic DCM extract (1.2 g; IC50, 13.3 µg/mL) was subjected to passage over a silica gel column, and eluted with gradient mixtures of n-hexane–acetone (100:1→1:1; 50 mL each). Fractions were pooled by TLC analysis to yield 11 combined fractions. Of these, fractions 4–10 were deemed cytotoxic toward the HT-29 cell line (IC50 < 10 µg/mL). Fractions 4 and 5 were combined and chromatographed over a silica gel column, and eluted with gradient mixtures of n-hexane–acetone (10:1→3:1), to yield three sub-fractions. The precipitate (2.0 mg) obtained from the first sub-fraction was chromatographed over silica gel using n-hexane-acetone (3:1) as solvent, and then purified by separation over a Sephadex LH-20 column, eluted with DCM-MeOH (1:1), to afford (±)-5-hydroxy-7-methoxyflavanone (1.0 mg). Fraction 6 was chromatographed over a silica gel column, and eluted with gradient mixtures of n-hexane–acetone (10:1→3:1), and then purified by separation over a Sephadex LH-20 column, eluted with DCM-MeOH (1:1), furnishing (+)-cryptocaryone (5, 15.0 mg). The combined fractions 7 and 8 were chromatographed over a silica gel column that was eluted with gradient mixtures of n-hexane–acetone (10:1→3:1). The precipitate obtained from sub-fractions 5–7 was purified by separation over a Sephadex LH-20 column, eluted with DCM-MeOH (1:1), to yield (±)-5,7-dihydroxyflavanone (4.0 mg). Fraction 9 was chromatographed over a silica gel column eluted with gradient mixtures of n-hexane–acetone (10:1→3:1), and then further separated using a Sephadex LH-20 column, eluted with DCM-MeOH (1:1), affording pure (+)-cryptocaryanone A (7, 0.8 mg). Fraction 10 was chromatographed over a silica gel column, eluted with gradient mixtures of n-hexane–acetone (10:1→1:1), to yield two combined sub-fractions. The first sub-fraction (5.0 mg) was chromatographed over silica gel using n-hexane–ethyl acetate (10:1→1:1) as solvent, and then finally purified by passage over a Sephadex LH-20 column, eluted with DCM-MeOH (1:1), providing (+)-bicaryanone A (3, 1.0 mg) and (+)-chalcocaryanone C (4, 1.0 mg). In the same manner, the second sub-fraction (10.0 mg) was chromatographed over silica gel and then purified by passage over a Sephadex LH-20 column, yielding (−)-rubrichalcolactone (1, 1.0 mg), 1,16-hexadecanediol-di-p-coumaroate (2, 1.0 mg), (+)-desmethylinfectocaryone (6, 0.8 mg), and cinnamic acid (1.0 mg).
(−)-Rubrichalcolactone (1)
Amorphous yellow powder; [α]20 d −126.6 (c 0.06, MeOH); UV (MeOH) λmax (log ε) 344 (4.11) nm; ECD (MeOH, nm) λmax (fε) 344.0 (+1.70), 289.5 (+0.79), 247.0 (+0.37), 230.0 (−0.74), 200.0 (−4.63); IR (dried film) νmax 3297, 2924, 2853, 1708, 1625, 1553, 1496, 1228, 730 cm−1; 1H- and 13C NMR data, see Table 1; positive-ion sodiated HRESIMS m/z 551.2026 (calcd for C32H32O7Na, 551.2046).
1,16-Hexadecanediol-di-p-coumaroate (2)
Amorphous colorless powder; UV (MeOH) λmax (log ε) 311 (4.27), 228 (4.02) nm; IR (dried film) νmax 3351, 2924, 2852, 1683, 1632, 1605, 1586, 1444, 983, 832 cm−1; 1H- and 13C NMR data, see Table 2; positive-ion sodiated HRESIMS m/z 573.3183 (calcd for C34H46O6Na, 573.3192).
Cytotoxicity Assay
Cytotoxicity of the samples was performed against HT-29 human colon cancer cells using a reported procedure.18 The IC50 values of the test samples in serial dilutions were calculated using nonlinear regression analysis (Table Curve2Dv4; AISN Software, Inc., Mapleton, OR). Measurements were performed in triplicate and are representative of two independent experiments in which the values generally agreed within 10%. Paclitaxel was used as a positive control, with an IC50 value of 1 nM.
Glucose Uptake Inhibitory Assay
This assay was conducted following a protocol used in previous reports.28–30 In brief, H1299 human lung cancer cells grown in 24-well plates were washed and incubated with 0.5 mL of serum-free DMEM in 10% CO2 at 37 °C for 2 h and then with 0.45 mL of KRP buffer at 37 °C for 30 min. WZB117 (used as the positive control)30 or the test compound was added to cells at a final concentration of 30 µM. The cells were incubated at 37 °C for 15 min, and then glucose uptake was initiated by adding 37 MBq/L 2-deoxy-d-[3H] glucose and 1 mM regular glucose as the final concentrations to cells. After 10 min, the glucose uptake was terminated by washing cells three times with cold phosphate buffered saline (PBS). Cells were lysed with NaOH (0.2 mM), and the radioactivity retained in the cell lysates was measured by a LS 6000 series liquid scintillation counter (Beckman Coulter, Inc., Fullerton, CA). The data were analyzed statistically using the Student’s t-test, by comparison of the data from the experimental samples with those from the vehicle control, and p <0.05 was set as the level of significant difference.
Supplementary Material
ACKNOWLEDGMENTS
This investigation was supported by an Administrative Supplement from NCCAM, NIH to grant P01 CA125066, funded by the National Cancer Institute, NIH, Bethesda, MD, and was also supported partially by the Edison Program of the State of Ohio and Ohio University Heritage College of Osteopathic Medicine. We are grateful to Dr. Hans T. Beck, formerly of the Institute of Economic Botany, The New York Botanical Garden, Bronx, New York, for the plant collection. We thank Dr. David Hart, Department of Chemistry and Biochemistry, The Ohio State University, for his suggestions regarding the Diels-Alder cycloaddition. We thank Dr. Judith C. Gallucci, Department of Chemistry and Biochemistry, The Ohio State University, for her helpful comments and suggestions about the crystal structure of bicaryanone D. We thank Dr. Kari Green-Church and Mr. Mark E. Apsega of the Mass Spectrometry and Proteomics of the Campus Chemical Instrument Center, The Ohio State University, for access to the MS measurements, and Dr. Craig A. McElroy of College of Pharmacy, The Ohio State University, for access to the NMR instrumentation used in this investigation.
Footnotes
ASSOCIATED CONTENT
Supporting Information. MS and NMR spectra of compounds 1 and 2 and the 1H- and 13C NMR and other analytical data of the known compounds isolated from C. rubra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Van der Werff H, Richter HG. Ann. Missouri Bot. Gard. 1996;83:409–418. [Google Scholar]
- 2.Fu X, Sévenet T, Remy F, Pais M, Hamid A, Hadi A, Zeng LM. J. Nat. Prod. 1993;56:1153–1163. doi: 10.1021/np50097a021. [DOI] [PubMed] [Google Scholar]
- 3.Dumontet V, Gaspard C, Van Hung N, Fahy J, Tchertanov L, Sévenet T, Guéritte F. Tetrahedron. 2001;57:6189–6196. [Google Scholar]
- 4.Dumontet V, Van Hung N, Adeline M-T, Riche C, Chiaroni A, Sévenet T, Guéritte F. J. Nat. Prod. 2004;67:858–862. doi: 10.1021/np030510h. [DOI] [PubMed] [Google Scholar]
- 5.Meragelman TL, Scudiero DA, Davis RE, Staudt LM, McCloud TG, Cardellina JH, II, Shoemaker RH. J. Nat. Prod. 2009;72:336–339. doi: 10.1021/np800350x. [DOI] [PubMed] [Google Scholar]
- 6.Kurniadewi F, Juliawaty LD, Syah YM, Achmad SA, Hakim EH, Koyama K, Kinoshita K, Takahashi K. J. Nat. Med. 2010;64:121–125. doi: 10.1007/s11418-009-0368-y. [DOI] [PubMed] [Google Scholar]
- 7.Chou T-H, Chen J-J, Lee S-J, Chiang MY, Yang C-W, Chen I-S. J. Nat. Prod. 2010;73:1470–1475. doi: 10.1021/np100014j. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y-C, Kung F-L, Tsai I-L, Chou T-H, Chen I-S, Guh J-H. J. Urol. 2010;183:2409–2418. doi: 10.1016/j.juro.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 9.Davis RA, Demirkiran O, Sykes ML, Avery VM, Suraweera L, Fechner GA, Quinn RJ. Bioorg. Med. Chem. Lett. 2010;20:4057–4059. doi: 10.1016/j.bmcl.2010.05.091. [DOI] [PubMed] [Google Scholar]
- 10.Chou T-H, Chen J-J, Peng C-F, Cheng M-J, Chen I-S. Chem. Biodivers. 2011;8:2015–2024. doi: 10.1002/cbdv.201000367. [DOI] [PubMed] [Google Scholar]
- 11.Feng R, Guo ZK, Yan CM, Li EG, Tan RX, Ge HM. Phytochemistry. 2012;76:98–105. doi: 10.1016/j.phytochem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Feng R, Wang T, Wei W, Tan RX, Ge HM. Phytochemistry. 2013;90:147–153. doi: 10.1016/j.phytochem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Kinghorn AD, Carcache de Blanco EJ, Chai H-B, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häberlein H, Tschiersch KP. Phytochemistry. 1994;35:765–768. [Google Scholar]
- 15.Ma X-M, Liu Y, Shi Y-P. Chem. Biodivers. 2007;4:2172–2181. doi: 10.1002/cbdv.200790174. [DOI] [PubMed] [Google Scholar]
- 16.Bertelli D, Papotti G, Bortolotti L, Marcazzan GL, Plessi M. Phytochem. Anal. 2012;23:260–266. doi: 10.1002/pca.1352. [DOI] [PubMed] [Google Scholar]
- 17.Lotti C, Piccinelli AL, Arevalo C, Ruiz I, Migliani De Castro GM, Figueira Reis De Sá L, Tessis AC, Ferreira-Pereira A, Rastrelli L. J. Agric. Food Chem. 2012;60:10540–10545. doi: 10.1021/jf302578r. [DOI] [PubMed] [Google Scholar]
- 18.Allard P-M, Dau ETH, Eydoux C, Guillemot J-C, Dumontet V, Poullain C, Canard B, Guéritte F, Litaudon M. J. Nat. Prod. 2011;74:2446–2453. doi: 10.1021/np200715v. [DOI] [PubMed] [Google Scholar]
- 19.Ren Y, Muñoz Acuña U, Jiménez F, García R, Mejía M, Chai H, Gallucci JC, Farnsworth NR, Soejarto DD, Carcache de Blanco EJ, Kinghorn AD. Tetrahedron. 2012;68:2671–2678. doi: 10.1016/j.tet.2012.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Fujimoto Y. Phytochemistry. 1993;33:151–153. [Google Scholar]
- 21.Si C-L, Kim J-K, Bae Y-S, Li S-M. Planta Med. 2009;75:1165–1167. doi: 10.1055/s-0029-1185476. [DOI] [PubMed] [Google Scholar]
- 22.Laville R, Thomas OP, Berrue F, Reyes F, Amade P. Eur. J. Org. Chem. 2008:121–125. [Google Scholar]
- 23.Nakamura N, Yamamoto T. Acta Cryst. 1994;C50:946–948. [Google Scholar]
- 24.Gottlieb HE, Kotlyar V, Nudelman A. J. Org. Chem. 1997;62:7512–7515. doi: 10.1021/jo971176v. [DOI] [PubMed] [Google Scholar]
- 25.Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, McCabe P, Pidcock E, Rodriguez-Monge L, Taylor R, van de Streek J, Wood PA. J. Appl. Cryst. 2008;41:466–470. [Google Scholar]
- 26.Pudukulathan Z, Manna S, Hwang S-W, Khanapure SP, Lawson JA, FitzGerald GA, Rokach J. J. Am. Chem. Soc. 1998;120:11953–11961. [Google Scholar]
- 27.Snider BB. Acc. Chem. Res. 1980;13:426–432. [Google Scholar]
- 28.Zhang W, Liu Y, Chen X, Bergmeier SC. Bioorg. Med. Chem. Lett. 2010;20:2191–2194. doi: 10.1016/j.bmcl.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Zhang W, Cao Y, Liu Y, Bergmeier S, Chen X. Cancer Lett. 2010;298:176–185. doi: 10.1016/j.canlet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, Colvin R, Ding J, Tong L, Wu S, Hines J, Chen X. Mol. Cancer Ther. 2012;11:1672–1682. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 31.Melstrom LG, Salabat MR, Ding X-Z, Milam BM, Strouch M, Pelling JC, Bentrem DJ. Pancreas. 2008;37:426–431. doi: 10.1097/MPA.0b013e3181735ccb. [DOI] [PubMed] [Google Scholar]
- 32.Melstrom LG, Salabat MR, Ding X-Z, Strouch MJ, Grippo PJ, Mirzoeva S, Pelling JC, Bentrem DJ. J. Surg. Res. 2011;167:173–181. doi: 10.1016/j.jss.2010.10.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.