SUMMARY
We previously demonstrated that the deletion of the poly(ADP-ribose)polymerase (Parp)-1 gene in mice enhances oxidative metabolism, thereby protecting against diet-induced obesity. However, the therapeutic use of PARP inhibitors to enhance mitochondrial function remains to be explored. Here, we show tight negative correlation between Parp-1 expression and energy expenditure in heterogeneous mouse populations, indicating that variations in PARP-1 activity have an impact on metabolic homeostasis. Notably, these genetic correlations can be translated into pharmacological applications. Long-term treatment with PARP inhibitors enhances fitness in mice by increasing the abundance of mitochondrial respiratory complexes and boosting mitochondrial respiratory capacity. Furthermore, PARP inhibitors reverse mitochondrial defects in primary myotubes of obese humans and attenuate genetic defects of mitochondrial metabolism in human fibroblasts and C. elegans. Overall, our work validates in worm, mouse and human models that PARP inhibition may be used to treat both genetic and acquired muscle dysfunction linked to defective mitochondrial function.
INTRODUCTION
Reductions in mitochondrial number and activity are a hallmark of several inherited mitochondrial diseases, as well as a number of age-related neurodegenerative and metabolic disorders (Andreux et al., 2013). The sirtuin enzymes have emerged as critical regulators of mitochondrial and oxidative metabolism (Chalkiadaki and Guarente, 2012). Several studies have shown that NAD+ levels can be rate-limiting for the deacetylase activity of SIRT1, the best-characterized sirtuin (Canto and Auwerx, 2012). Boosting intracellular NAD+ levels has hence become an attractive approach to activate SIRT1 and mitochondrial metabolism. In line with this, NAD+ availability might even determine circadian shifts in oxidative capacity (Peek et al., 2013).
One strategy to increase NAD+ levels consists of the inhibition of alternative NAD+ consuming enzymes, such as poly(ADP-ribose) polymerase-1 (PARP-1) (Bai and Canto, 2012), which is involved in DNA damage detection and repair. Upon activation, PARP-1 can deplete intracellular NAD+ levels by 80%. Genetic ablation of Parp-1 increases NAD+ availability and SIRT1 activity in tissues, such as skeletal muscle and brown adipose tissue (Bai et al., 2011). As a consequence, mitochondrial oxidative capacity is enhanced in muscle of Parp-1−/− mice, protecting them against high-fat diet (HFD)-induced insulin resistance. Therefore, PARP inhibition constitutes a plausible strategy to improve metabolic homeostasis. PARP inhibitors (Paribs) already exist and are clinically tested in the cancer field (Curtin and Szabo, 2013). The role of PARPs in genome maintenance, however, questions whether Paribs can be used therapeutically over longer periods of time to prevent the detrimental metabolic consequences of mitochondrial dysfunction. Here, we demonstrate that long-term Parib treatment is sustainable and improves mitochondrial function in skeletal muscle, enhancing endurance performance and protecting against HFD-induced metabolic complications. We furthermore show that Parp-1 expression is negatively correlated with energy expenditure in mouse genetic reference populations. Finally, we demonstrate how acquired and genetic mitochondrial defects can be improved by Paribs. Therefore, our work sets the stage for the possible clinical use of Paribs in situations of defective mitochondrial function.
RESULTS AND DISCUSSION
Characterization of Paribs in C2C12 myotubes
We initially compared how different Paribs, BSI-201, PJ-34, ABT-888, AZD-2881 and MRL-45696, could rescue NAD+ decline upon H2O2-induced PARP-1 activation in differentiated mammalian cell lines, such as C2C12 myotubes. MRL-45696, a dual PARP-1 and PARP-2 inhibitor derived from niraparib (Chinnaiyan et al., 2012; Jones et al., 2009) most efficiently rescued the H2O2-induced NAD+ decline, with a low nanomolar IC50 (Supplemental Table 1). MRL-45696 reduced the redox potential in C2C12 myotubes at concentrations as low as 1–10nM (Figure S1A), and increased mitochondrial membrane potential (MMP) (Figure S1B) and O2 consumption rates (OCR) at 10nM (Figure S1C). To certify that the effects of MRL-45696 are derived from PARP inhibition, we also used another Parib, AZD-2281 (Olaparib). In agreement, nanomolar concentrations of AZD-2281 decreased redox potential, while increasing MMP and OCR (Figures S1E–S1G). Notably, MRL-45696 or AZD-2281 were not toxic at these concentrations, as ATP content was unaffected (Figures S1D and S1H).
Chronic PARP inhibition enhances energy expenditure and SIRT1 activity
As defects in mitochondrial metabolism are a hallmark for many diseases, Paribs could in principle also be used for these non-oncological indications. However, such use could be overshadowed by several concerns. First, chronic Parib treatment could induce genomic instability (Curtin and Szabo, 2013). Second, Paribs would not just affect PARP-1, but also PARP-2, and the combined reduction of both activities could be detrimental for long-term viability (Menissier de Murcia et al., 2003). Hence, we characterized the impact of long-term MRL-45696 treatment in mice. We initially determined that dietary admixture achieved higher MRL-45696 levels in plasma and muscle than oral gavage (Figures S2A and S2B), hence choosing this route for further studies.
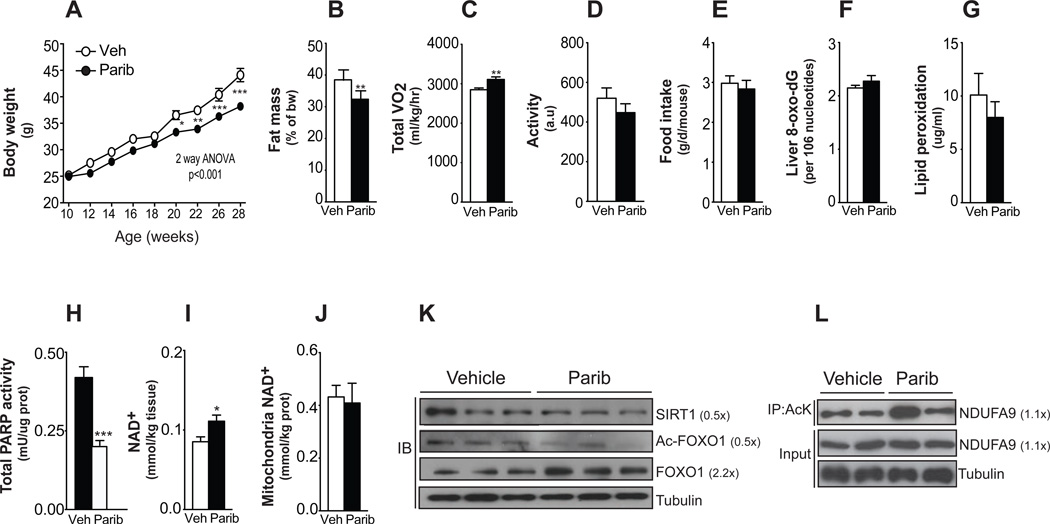
We next fed HFD admixed with MRL-45696 (50 mg/kg/day) to 10-wk old male C57BL/6J mice. MRL-45696 blunted HFD-induced body weight gain (Figure 1A) due to reduced fat accumulation (Figure 1B) and was associated with higher energy expenditure (Figure 1C), without affecting activity or food intake (Figures 1D and 1E). Although PARP-1 has been shown to impact genome stability and cell viability, no evidence for toxicity on genomic DNA or cellular damage was found, as liver 8-oxo-dG and muscle lipid peroxidation levels were similar between the groups (Figures 1F and 1G). This is in line with the fact that Parp-1−/− mice are viable and do not show signs of DNA damage unless challenged with cytotoxic stimuli (de Murcia et al., 1997).
Figure 1. Paribs protect from HFD-induced metabolic complications.
Ten-wk-old male C57Bl/6J mice were challenged with HFD supplemented with either vehicle (DMSO; Veh) or MRL-45696 (50 mg/kg/day) (n=10/group). (A) Body weight gain during 18wks of HFD. (B) Fat mass was measured using Echo-MRI. (C–D) A comprehensive laboratory animal monitoring system was used to evaluate VO2 (C) and activity (D) after 7wks of HFD. (E) Food intake measured by averaging weekly food consumption during HFD. (F) Liver 8-oxo-dG content, as indicator of DNA damage, (G) muscle lipid peroxidation-derived aldehyde, 4-hydroxy-2-nonenal, and (H) muscle total poly(ADP-ribose) (PAR) contents were measured in vehicle and MRL-45696-treated mice (n=7/group) (I–J) Total intracellular (I) and mitochondrial (J) NAD+ levels in gastrocnemius of refed vehicle and MRL-45696-treated mice (n=5–10/group). (K) SIRT1, acetylated FOXO1 and total FOXO1 protein levels were assessed in total homogenates from quadriceps of chow-diet fed mice. (L) The acetylation status of Ndufa9 immunoprecipates was tested as a marker of SIRT3 activity. Values are shown as mean+/−SEM. * indicates statistical significant difference vs. respective Veh group. *, p<0.05; **, p<0.01; ***, p<0.001. This figure is complemented by Figures S1 and S2, and Table S1.
MRL-45696 reduced total PARP activity in skeletal muscle (−50%; Figure 1H) and in others tissues tested, including brain and liver (Figure S2C). In line with PARP inhibition, intracellular NAD+ content was higher in muscle from treated mice (Figure 1I). The higher NAD+ levels were restricted to the nuclear and/or cytosolic compartments, since mitochondrial NAD+ content was similar between both groups (Figure 1J). Reductions in PARP activity translated also into SIRT1 activation, as reflected by lower FOXO1 acetylation, despite slightly lowered SIRT1 levels (Figure 1K). Conversely, the unaltered mitochondrial NAD+ levels did not change SIRT3 activity, as manifested in the acetylation of Ndufa9 (Figure 1L). These observations are in line with observations in Parp-1−/− mice, where SIRT1, but not SIRT3, activity was enhanced (Bai et al., 2011).
PARP inhibition enhances endurance and mitochondrial function
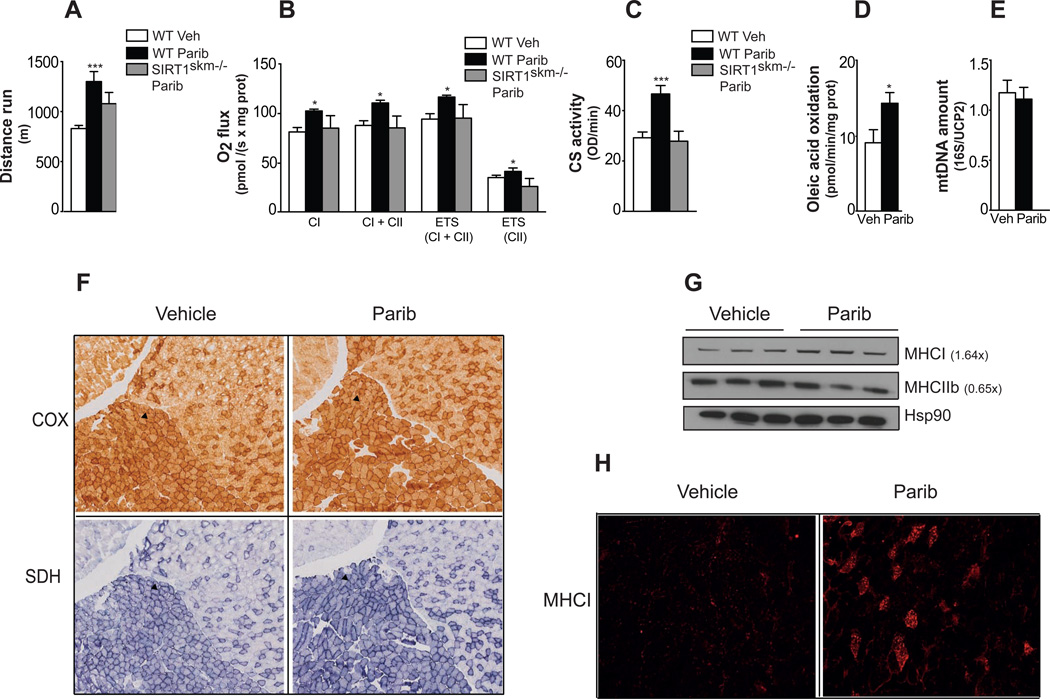
Given that MRL-45696 accumulates and inhibits PARP in the skeletal muscle (Figure 1H and S2C), we next examined the impact of Paribs on muscle performance and mitochondrial function in chow-fed mice. Despite similar body weight and composition (Figures S3A–S3C), MRL-45696-treated mice ran for a significantly longer distance (Figure 2A). Maximal O2 consumption capacity (VO2max), which critically determines endurance performance, was significantly higher with MRL-45696 (Figure S3D). As cardiovascular parameters, including ejection fraction, fractional shortening, ventricular mass and diameter (Figures S3E–S3I) were unchanged, the higher endurance was most likely due to improved muscle mitochondrial function. To substantiate this hypothesis, we performed respirometry tests in permeabilized EDL muscle fibers. Maximal respiration in the coupled state, with electron input through either complex I alone - using malate, pyruvate and glutamate - or complex I+II - using, in addition, succinate - was markedly higher in muscles after MRL-45696 (Figure 2B). Maximum electron transport system capacity was also higher after PARP inhibition, even after the addition of the complex I inhibitor rotenone (Figure 2B). These results are consistent with the marked increase in citrate synthase (CS) activity after MRL-45595 (Figure 2C). Finally, Paribs increased oleic acid oxidation rates by ~50% (Figure 2D). Mitochondrial DNA abundance was, however, not changed (Figure 2E), suggesting that MRL-45696 does not influence muscle mitochondrial function by increasing mitochondrial number.
Figure 2. Paribs enhance exercise capacity and muscle mitochondrial function.
Chow fed male C57BL/6J and congenic SIRT1skm−/− mice (n=5–10/group) treated with either vehicle (DMSO; Veh) and/or MRL-45696 (50 mg/kg/day) were subjected to (A) endurance treadmill test after 13wks treatments, (B) respirometry analysis of permeabilized EDL muscle fibers (CI, complex I; CII, complex II; ETS, electron transport system) and (C) CS activity measurement. (D) Oleic acid oxidation rate in the muscles of Veh and MRL-45696-treated mice after 18wks treatment (n=5–7/group). (E) Mitochondria DNA abundance in quadriceps of Veh and MRL-45696-treated mice (n=8/group). Results are expressed as mitochondrial DNA amount (16S) relative to genomic DNA (UCP2). (F) Cytochrome c oxidase (COX), succinate dehydrogenase (SDH) staining in gastrocnemius of Veh and MRL-45696-treated mice. Soleus is indicated by an arrow. (G) Protein levels of MHCI and IIb were evaluated, using heat shock protein 90 (Hsp90) as loading control (H) Myosin heavy chain I staining (MHCI) of gastrocnemius of Veh and MRL-45696-treated mice. Values are shown as mean+/−SEM. * indicates statistical significant difference vs. respective Veh group. *, p<0.05; **, p<0.01; ***, p<0.001. This figure is complemented by Figure S3.
Histological analysis further certified that Paribs enhanced the oxidative profile of skeletal muscle, as testified by more intense cytochrome c oxidase and succinate dehydrogenase staining in gastrocnemius (Figure 2F). Remarkably, an increase in slow oxidative MHCI, a marker for type I fibers, was observed, whereas MHCIIb levels, a type II fiber marker, were decreased by MRL-45696 (Figure 2G). Furthermore, MHCI staining of gastrocnemius confirmed the higher type I fiber content after MRL-45696 (Figure 2H). The higher oxidative capacity after Paribs was accompanied by increased insulin sensitivity, as reflected by the higher glucose infusion rate during a hyperinsulinemic-euglycemic clamp (Figure S3J).
We next evaluated the possible contribution of SIRT1 in mediating the effects of Paribs by using a muscle-specific SIRT1 deficient mouse line (SIRT1skm−/− mice). In line with previous reports (Menzies et al., 2013; Philp et al., 2011), endurance or VO2max was not impaired in SIRT1skm−/− mice (data not shown). Interestingly, Paribs were unable to enhance endurance, respiratory capacity and CS activity in the SIRT1skm−/− mice (Figures 2A–2C). Of note, the expression of Sirt1, Tfam and c-myc was also increased in Parib treated muscles (Figure S3K), which indicates that a recently identified SIRT1-dependent pathway (Gomes et al., 2013) by which NAD+ controls metabolic health might also be at play. Therefore, SIRT1 seems key to the actions of Paribs on muscle metabolism.
MRL-45696 enhances mitochondrial protein translation and triggers the UPRmt
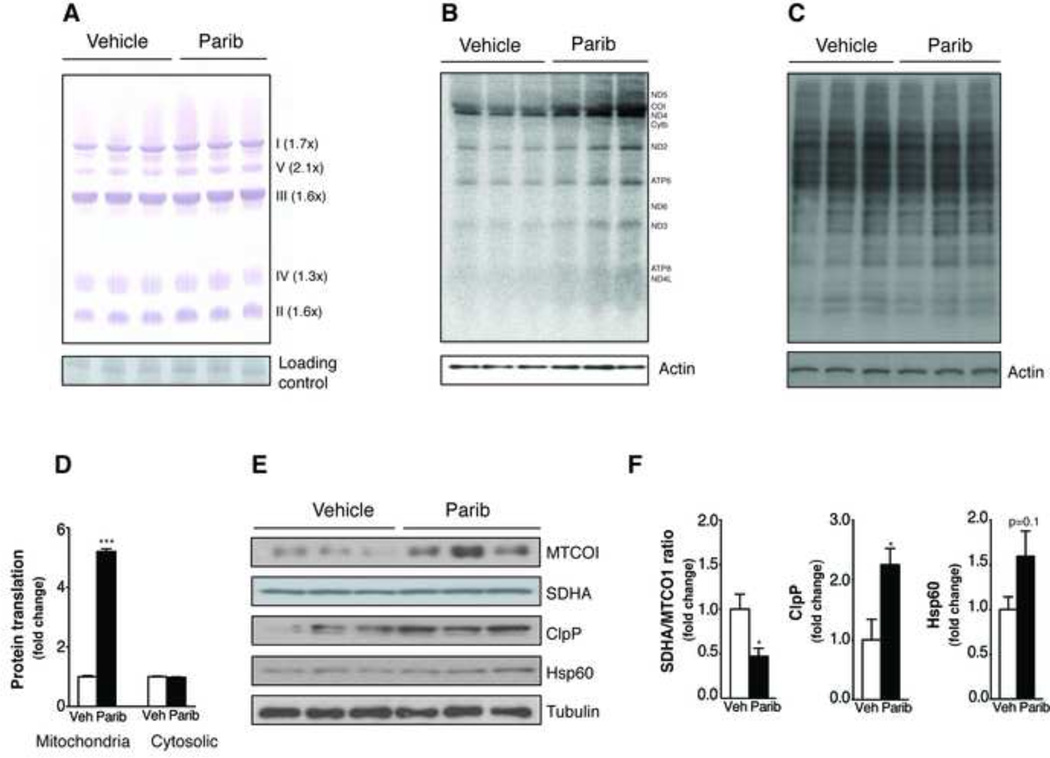
Paribs increase mitochondrial respiratory capacity in worms by triggering the mitochondrial unfolded protein response (UPRmt-) in a SIRT1-dependent manner (Mouchiroud et al., 2013). However, the possible translation of this finding into mammals has not been explored. We therefore performed blue native page analyses of mitochondrial complexes in muscles. MRL-45696 increased the abundance of mitochondrial complexes, but did not change their mobility (Figure 3A). Therefore, increased respiratory chain complex content, rather than changes in complex composition or stoichiometry, accounts for the enhanced mitochondrial function. This effect was also observed in cultured mouse embryonic fibroblast (MEFs) (Figure S4A). As MRL-45696 did not affect mRNA levels of mitochondrial complex subunits (Figure S4B), we speculated that the higher respiratory complex content could be due to enhanced mitochondrial protein translation. In line with this hypothesis, MRL-45696 specifically increased mitochondrial, but not cytosolic (Figures 3B–3D), protein translation rates in MEFs.
Figure 3. Paribs induce UPRmt in muscle by increasing mitochondrial protein translation.
(A) Blue-Native Page using isolated mitochondria from quadriceps of vehicle (DMSO; Veh) and MRL-45696 treated mice. A non-specific band was used as loading control. (B) Mitochondrial and (C) cytosolic protein translation measured in MEFs after 100nM MRL-45696 treatment for 40hr. (D) Quantification of protein translation rates. (E) MTCOI, SDHA, ClpP and Hsp60 protein levels were evaluated in quadriceps from Veh and MRL-45696 mice, using tubulin as a loading control. (F) Quantification of mitonuclear and UPRmt markers. *, p<0.05 and ***<0.001. This figure is complemented by Figure S4.
Increased mitochondrial translation, without according changes in cytosolic translation rates, can lead to mitonuclear protein imbalance (Houtkooper et al., 2013), which was evidenced by altered protein ratios between MTCO1, which is encoded by mtDNA, and SDHA, encoded by nDNA, induced by MRL-45696 (Figures 3E and 3F). Such a mitonuclear protein imbalance could trigger UPRmt to restore optimal mitochondrial function (Houtkooper et al., 2013). In line with this premise, ClpP and Hsp60 levels, which reflect the presence of UPRmt, were increased in MRL-45696 treated muscles (Figures 3E and 3F). Altogether, these results illustrate the robust effects of MRL-45696 on mitochondrial translation, which culminate in the induction of UPRmt. While it is not clear how Paribs trigger mitonuclear imbalance and UPRmt in mammalian cells, results obtained in C2C12 cells suggest that SIRT1 is required for such an effect (Figure S4C), consistent with the mechanisms reported in worms (Mouchiroud et al., 2013). In addition, PARPs have been reported to silence translation during stress by binding and/or PARylating RNA-binding proteins that control the polyadenylation of mRNAs and protein translation rates (for review, see (Luo and Kraus, 2012)). Hence, the inhibition of PARP actions on translational processes could be a complementary mechanism to explain the induction of UPRmt.
Parp-1 expression negatively correlates with energy expenditure
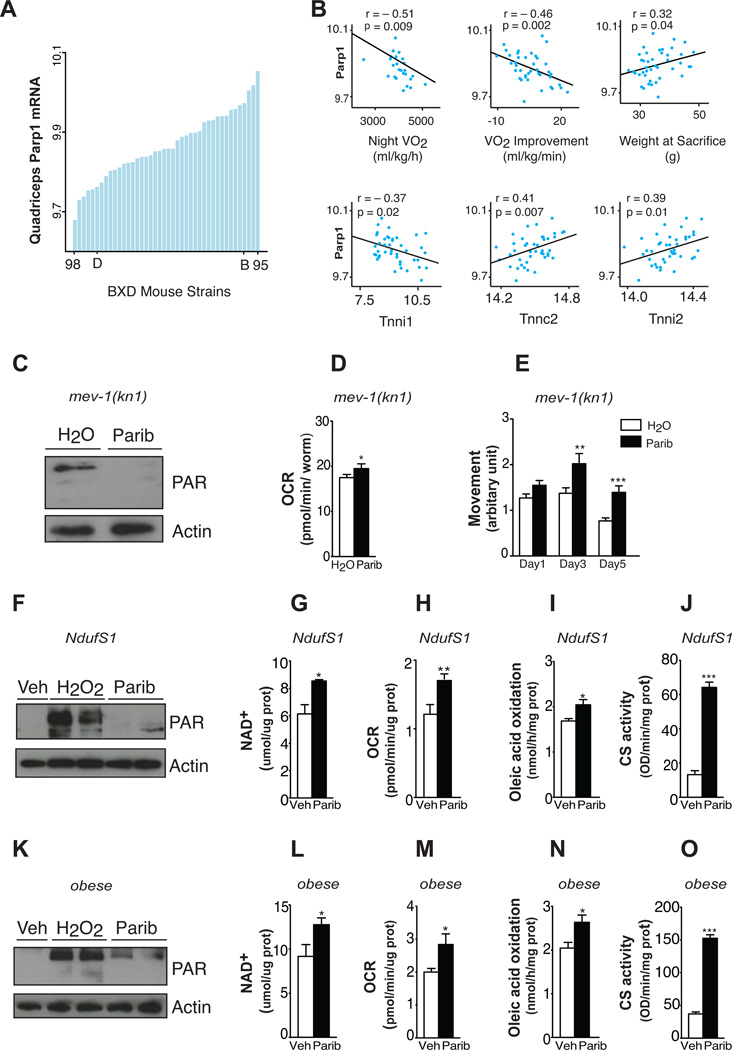
Given the effects of Paribs on mitochondrial function in the C57BL/6J mouse strain, we next aimed to elucidate whether Parp-1 could determine energy expenditure and oxidative capacity in genetically complex mouse populations such as the BXD mouse genetic population (Peirce et al., 2004). Parp-1 expression varied 1.3-fold in skeletal muscle of the BXD strains, enabling the study of gene expression correlations (Figure 4A). These changes in Parp-1 expression are not related to natural variations in fiber type composition, as Parp-1 expression is comparable in muscles having different fiber type content, such as gastrocnemius or soleus (data not shown). Parp-1 expression in muscles of the BXDs strains was negatively correlated with night VO2, VO2max improvement after 10 days training and with expression of the troponin I type 1 slow-twitch muscle isoform (Tnni1) (Figure 4B). In contrast, Parp-1 positively correlated with body weight, troponin C type 2 fast-twitch muscle isoform (Tnnc2) and troponin I type 2 fast-twitch muscle isoform (Tnni2) (Figure 4B). This robust genetic correlation suggests a physiological influence of PARP-1 activity on oxidative metabolism. Indeed, previous results indicated that PARP activity is increased upon HFD (Bai et al., 2011), likely favouring fat storage over burning (Erener et al., 2012). Consistent with this, PARP-1 activity was found to enhance adipogenesis (Erener et al., 2012). Oppositely, PARP activity was decreased in muscle upon fasting (Bai et al., 2011), when lipid oxidation is favored. Collectively, this indicates that physiological changes in PARP-1 activity might help balancing whole-body energy metabolism
Figure 4. Parp activity negatively correlates with energy expenditure in mouse populations and its inhibition improves mitochondrial function in worm and human models of mitochondria dysfunction.
(A) Expression of Parp-1 in the quadriceps of 37 strains of BXD mice. Each bar represents mRNA from a pool of ~5 animals per strain. Extreme strains and the parental strains are labelled. (B) Top: Correlation between quadriceps muscle Parp-1 expression and phenotypes collected from BXD mice. Night VO2 was measured though indirect calorimetry. VO2 improvement represents the increase in VO2max after 10 days of voluntary exercise. Bottom: Correlation between quadriceps muscle Parp-1 and muscle fiber type genes in the same dataset. (C) Total PAR content at day 2 of adulthood (n=~500 worms/sample) in mev-1(kn1) complex II-deficient C. elegans after water and 100nM MRL-45696 treatment. (D) Respiration at day 3 (n=10 worms/well, 19 wells/group) and (E) movement at day 1, 3 and 5 of adulthood (n=37–90 worms/group). (F) H2O2 (500µM)-induced PARylation after 48hr pretreatment with either vehicle (Veh) or 100nM MRL-45696 in NDUFS1 mutant human fibroblasts. (G–J) NAD+ levels (G), O2 consumption rates (H), oleic acid oxidation (I) and CS activity (J) in human NDUFS1 mutant fibroblasts after Veh and 100nM MRL-45696 for 48hr (n=3–12 samples/group). (K) H2O2 (500µM)-induced PAR content after 24hr pretreatment with either Veh or 10nM MRL-45696 in human primary myotubes. (L–O) NAD+ levels (L), O2 consumption rates (M), oleic acid oxidation (N) and CS activity (O) in human primary myotubes after Veh or 10nM MRL-45696 treatment for 72hr (n=6–12 samples/group). * indicates statistical significant difference vs. respective Veh group. *, p<0.05; **, p<0.01; ***, p<0.001.
Paribs improve oxidative capacity in worm and human models of reduced mitochondrial function
To elucidate whether Paribs could recover mitochondrial function in models with genetically determined mitochondrial defects, we first used the C. elegans mev-1 strain, which contains a missense mutation in cyt-1, the worm homolog of mammalian succinate dehydrogenase cytochrome b, a component of complex II (Senoo-Matsuda et al., 2001). The ability of complex II to catalyse electron transport from succinate to ubiquinone is compromised in mev-1 mutants, leading to increased superoxide levels and premature aging. MRL-45696 effectively inhibited PARP activity (Figure 4C) and OCR in mev-1 mutants (Figure 4D), as previously noticed in wild-type worms using other Paribs (Mouchiroud et al., 2013). The increased respiration also translated into effective prevention against age-related physiological decline, as old mev-1 mutants maintained youthful motility (Figure 4E).
To test whether these observations could also apply in human models, we used skin fibroblasts from a patient with a mutation in the nuclear NDUFS1 gene, which reduced complex I activity by 87% (Bugiani et al., 2004). MRL-45696 efficiently prevented H2O2-induced PARP activation (Figure 4F) and increased NAD+ (Figure 4G) in NDUFS1 mutant fibroblasts. In line with observations in mouse tissues, MRL-45696 also increased OCR (Figures 4H–4J), lipid oxidation and CS activity in these cells, testifying for enhanced mitochondrial function.
Finally, we evaluated whether Paribs could improve acquired defects in mitochondrial function, such as the decreased mitochondrial activity observed in skeletal muscle cells of obese humans (Patti and Corvera, 2010). MRL-45696 effectively abrogated PARP activation, enhanced NAD+ levels (Figures 4K and 4L) and stimulated OCR (Figure 4M) in primary myotubes from obese patients (Timmers et al., 2012). In addition, MRL-45696 granted a higher ability to oxidize lipids (Figure 4N), probably by enhancing mitochondrial function, as suggested by the higher CS activity (Figure 4O). Our data therefore illustrate that Paribs enhance mitochondrial function in both acquired and genetically determined mitochondrial alterations. Hence, the interrelation between PARP activity and mitochondrial function is well conserved across the evolutionary spectrum from worms to mice and humans.
In sum, this work provides evidence that drugs used at present in the cancer field could be retooled to treat metabolic dysfunction linked to impaired mitochondrial activity, even in a long-term fashion. Paribs are currently being tested for oncology indications, with positive outcomes in tumors with dysfunctional homologous DNA recombination repair (Curtin and Szabo, 2013). The success of Paribs is thought to rely on the high dependency of these cancer cells on PARP enzymes for DNA repair (Curtin and Szabo, 2013), but could also involve their capacity to reprogram cells towards oxidative metabolism (Bai et al., 2011). Notably, Paribs are in general well tolerated by patients with few reported side effects (Audeh et al., 2010; Bundred et al., 2013; Tutt et al., 2010). Within the time-frame of this study, and in line with findings in Parp-1−/− mice (Bai et al., 2011) and in patients treated with Olaparib (Audeh et al., 2010; Bundred et al., 2013; Tutt et al., 2010), no toxicity or genomic instability was seen in MRL-45696-treated mice. In contrast, marked metabolic effects were observed, such as improved mitochondrial function and protection against diet-induced obesity. Importantly, PARP inhibition rescued mitochondrial activity in situations of genetically determined mitochondrial dysfunction. Therefore, our results open the path of using Paribs to improve invalidating diseases caused by impaired mitochondrial function. While this could also set the stage for Paribs to impinge on other metabolic complex diseases, such as seen during type 2 diabetes, further work must be done to ensure the safety and feasibility of these treatments in non-life threatening diseases.
EXPERIMENTAL PROCEDURES
Materials
BSI-201, ABT-888 and AZD-2281 were purchased from Selleck Chemicals and PJ-34 from Sigma. MRL-45696, kindly provided by Thomas Vogt from Merck Research Laboratories, is closely related to the potent Parib, niraparib or 2-phenyl-2H-indazole-7-carboxamide (Jones et al., 2009). MRL-45696 is a dual PARP-1/2 inhibitor with IC50 of 0.8 and 0.3nM, respectively. In contrast, it displays an IC50 in the micromolar range against PARP-3, vPARP, and Tankyrase. C2C12 cells were obtained from ATCC. Myoblasts from obese patients and NDUFS1 mutant fibroblasts were provided by Drs. Schrauwen (Timmers et al., 2012) and Zeviani (Bugiani et al., 2004).
Cell culture and molecular biology studies
Methods for cellular and molecular assays are provided in the Supplemental Procedures.
In vivo and ex vivo studies
To compare administration routes, male C57BL/6J mice were given MRL-45696 by gavage or food admix at a dose of 50 mg/kg/d for 5 days. Mice were sacrificed either 6hr after the last gavage or in the random fed state, respectively. MRL-45696 concentration in plasma and muscle was measured by mass spectrometry. For the long-term animal studies, 10-week-old male C57BL/6J mice (Charles River) or SIRT1skm−/− mice were fed pellets containing either vehicle and/or PARP inhibitor (50 mg/g/kg) for 18wks. The generation of SIRT1skm−/− mice is described in the Supplemental Procedures. During the experiment, mice were housed under a 12hr dark-light cycle and had ad libitum access to water and food. Clinical tests were carried out according to standard operational procedures (SOPs). Mitochondrial function in permeabilized EDL muscle fibers was evaluated using the Oxygraph-2k respirometer (Oroboros, Austria). Correlations in the BXD mouse genetic reference population were identified using the GeneNetwork database (http://www.genenetwork.org). Details are in the Supplemental Procedures.
C. elegans studies
Worm strains were cultured at 20°C on nematode growth media agar plates seeded with E. coli strain OP50. Mev-1(kn1) mutant worms were provided by the Caenorhabditis Genetics Center (University of Minnesota). Protocols used for western blotting and worm phenotyping are described in the Supplemental Procedures.
Statistical methods
Statistical analyses were performed with Prism software (GraphPad). Differences between two groups were analysed using Student’s t-test (two-tailed) and multiple comparisons were analyzed by ANOVA with a Bonferroni post-hoc test. A p value less than 0.05 was considered significant. Data are expressed as means±SEM.
Supplementary Material
HIGHLIGHTS.
Inhibition of poly(ADP-ribose) polymerases enhances endurance performance
Inhibition of PARPs improves mitochondrial function in skeletal muscle
Parp-1 correlates with energy expenditure in heterogeneous mouse populations
Genetic and acquired mitochondrial defects can be rescued by PARP inhibition
ACKNOWLEDGEMENTS
We thank the members of the Auwerx lab and M. Hottiger for discussions and P. Cettour-Rose, B. Rochat, D. Dahlmans, N. Ziegler and S.J. Sturla for technical help. EP was funded by the Academy of Finland, the Saastamoinen Foundation, the Finnish Cultural Foundation and the Finnish Diabetes Foundation. CC is an employee of the Nestlé Institute of Health Sciences. JA is the Nestlé Chair in Energy Metabolism and a founder and scientific advisor to Mitokyne. AS has intellectual property in the field of NAD enhancers. This work is supported by the Ecole Polytechnique Fédérale de Lausanne, the EU Ideas program (AdG-23138 and AdG-322424), the NIH (R01HL106511-01A and R01AG043930), and the Swiss National Science Foundation (31003A-124713 and 31003A-125487 and CSRII3-1362.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. Oral poly(ADP-ribose) polymerase inhibitor Olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, Carrara F, Moroni I, Farina L, Spada M, Donati MA, et al. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta. 2004;1659:136–147. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Bundred N, Gardovskis J, Jaskiewicz J, Eglitis J, Paramonov V, McCormack P, Swaisland H, Cavallin M, Parry T, Carmichael J, et al. Evaluation of the pharmacodynamics and pharmacokinetics of the PARP inhibitor Olaparib: A phase I multicentre trial in patients scheduled for elective breast cancer surgery. Invest. New Drugs. 2013;31:949–958. doi: 10.1007/s10637-012-9922-7. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: All you need is NAD+? Pharmacol. Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan A, Brenner J, Yocum A. PARP1 targeted therapy. US2012035244. Patent number. 2012
- Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: Anticancer therapy and beyond. Molecul. Aspects Med. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, Donath MY, Richards PJ, Hottiger MO. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J. 2012;26:2631–2638. doi: 10.1096/fj.11-200212. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C, Lamartina S, Monteagudo E, Ontoria JM, Orsale MV, et al. Discovery of 2-{4-[(3s)-piperidin-3-yl]phenyl}-2h-indazole-7-carboxamide (MK-4827): A novel oral poly(ADP-ribose) polymerase (PARP) inhibitor efficacious in BRCA-1 and-2 mutant tumors. J. Med. Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies KJ, Singh K, Saleem A, Hood DA. Sirtuin 1-mediated effects of exercise and resveratrol on mitochondrial biogenesis. J. Biol. Chem. 2013;288:6968–6979. doi: 10.1074/jbc.M112.431155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, et al. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J. Biol. Chem. 2011;286:30561–30570. doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, Nakazawa H, Hartman PS, Ishii N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in caenorhabditis elegans. J. Biol. Chem. 2001;276:41553–41558. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- Timmers S, Nabben M, Bosma M, van Bree B, Lenaers E, van Beurden D, Schaart G, Westerterp-Plantenga MS, Langhans W, Hesselink MK, et al. Augmenting muscle diacylglycerol and triacylglycerol content by blocking fatty acid oxidation does not impede insulin sensitivity. Proc. Nat. Acad. Sci. USA. 2012;109:11711–11716. doi: 10.1073/pnas.1206868109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor Olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.