Abstract
Optical trapping is a powerful manipulation and measurement technique widely employed in the biological and materials sciences1–8. Miniaturizing optical trap instruments onto optofluidic platforms holds promise for high throughput lab-on-chip applications9–16. However, a persistent challenge with existing optofluidic devices has been controlled and precise manipulation of trapped particles. Here we report a new class of on-chip optical trapping devices. Using photonic interference functionalities, an array of stable, three-dimensional on-chip optical traps is formed at the antinodes of a standing-wave evanescent field on a nanophotonic waveguide. By employing the thermo-optic effect via integrated electric microheaters, the traps can be repositioned at high speed (~ 30 kHz) with nanometer precision. We demonstrate sorting and manipulation of individual DNA molecules. In conjunction with laminar flows and fluorescence, we also show precise control of the chemical environment of a sample with simultaneous monitoring. Such a controllable trapping device has the potential for high-throughput precision measurements on chip.
A prominent example of the application of optical trapping techniques is the vibrant area of biophysics in which the mechanical behavior of biological molecules can be investigated at the single molecule level2–4. It is now possible to disrupt protein complexes with piconewton forces and track motor proteins with nanometer and millisecond resolution. However, conventional optical trapping instruments are only capable of manipulating one molecule at a time, which limits their throughput. Methods for generating multiple optical traps via time-sharing of a single laser beam17 or holographic modulation18 have the drawback of requiring proportionally increased laser power. For optical trapping to realize its full potential, a new platform is needed to enable manipulation with high resolution and high throughput.
Optical trapping based on photonic structures in an optofluidic platform presents a potential solution to these drawbacks. The strong gradient in the evanescent fields of these structures can trap and transport a large array of particles, even at a low optical power9–12,19–27. Despite recent advances, no device has achieved stable three-dimensional (3D) trapping with controllable repositioning, features that are essential for manipulation.
Here we present a platform enabling high-throughput 3D optical trapping with precision manipulation on chip. The core component of the device is a standing-wave interferometer (Figs. 1a and 1b), where light in a waveguide is split into two arms of equal light intensity and the two arms are then joined, leading to the interference of two counter-propagating waves and therefore the formation of a standing wave. Thus, in the portion of the waveguide exposed to fluid (Fig. 1c), stable 3D optical traps are formed by the evanescent field at the anti-nodes of the standing-wave. We refer to this type of device as a nanophotonic standing-wave array trap (nSWAT). In a conventional optical trap, a single laser beam typically traps only one particle, and thus the laser power must be proportionally increased with the number of traps. In contrast, in an nSWAT, the same laser beam is “recycled” to form an array of periodically spaced traps, so that a large number of traps can be formed, each with stiffness comparable to that of a conventional optical trap (Fig. S1), without the need for increasing the laser power.
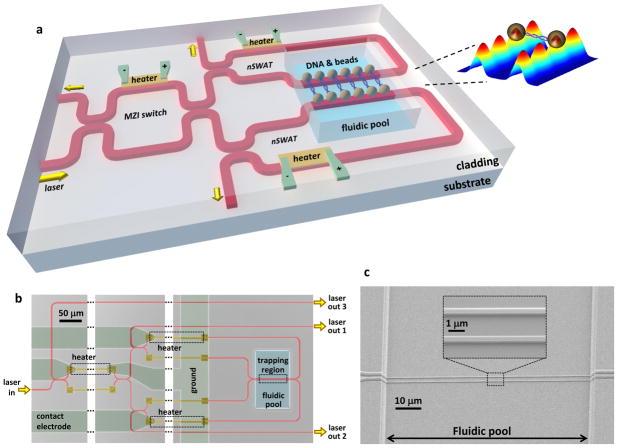
Figure 1. Design and fabrication of nanophotonic standing-wave array traps (nSWAT) device.
a, A schematic illustration of the device design. nSWATs were implemented with silicon waveguides on a silicon-on-insulator (SOI) platform (Methods). Laser input to the waveguide is partitioned to two nSWATs using an MZI. nSWATs have a 50/50 waveguide beam splitter with connected output arms to create counter-propagating waves. There are three microheaters located above the waveguides: one in the MZI to control the partition of the laser into the two nSWATs, and two more to control the trap positions in each nSWAT. The microheaters and waveguides are buried in oxide, except for the exposed waveguides in the fluidic pool trapping region. The inset shows an array of traps with a DNA molecule suspended between two beads that are held by nSWATs. The colored 3-D plot shows the calculated energy density of standing waves on both waveguides (see also Figure S1).
b, An optical microscope image of the fabricated device (false colored). Each microheater is located on one of the two arms after a splitter while an unconnected strip of metal is located on the other arm to balance potential optical loss introduced by the metal in the proximity of a waveguide. The waveguides are made of silicon on a SOI wafer.
c, Scanning electron micrographs of the waveguides in the trapping region. The waveguides are 440 nm in width and 250 nm in height. Note that while all waveguides reside in the same plane, the protective layer of oxide outside the fluid channel gives the illusion that they do not by amplifying the structure of the underlying waveguides.
In an nSWAT, the entire trapping array can be precisely repositioned by controlling the phase difference of the counter-propagating waves. This phase difference is achieved via an integrated electric microheater which heats part of the waveguide, inducing a phase change due to the thermo-optic effect27,28 (Fig. S2). In order to suspend and manipulate single DNA molecules, we incorporated two copies of an nSWAT in a single device, each controlled independently by its own microheater. The power from an incoming laser beam is partitioned to the two nSWATs via a Mach-Zehnder interferometer (MZI) switch that is controlled by a third integrated electric microheater to allow for sorting.
An nSWAT is naturally stable because all optical elements creating the traps are on chip with a short path difference (~ 100 μm) between the counter-propagating waves. Such stable trapping is essential for precision measurements of molecular events. By contrast, in a conventional bench-top optical trap, drift is inevitable and must be minimized by employing elaborate measures29,30. To demonstrate the inherent stability of an nSWAT, we held a bead in a trap of an nSWAT and monitored its position over time relative to the waveguide via video tracking (Methods). Fig. 2a shows that the bead had no discernible drift relative to the waveguide for over 10 minutes. In addition to stability, we also demonstrate that nSWATs are exceptionally resistant to environmental noise and vibrations (Fig. S3). Thus, nSWATs are ideally suited for long-term, low noise measurements with no need for extensive drift reduction and vibration isolation.
Figure 2. Trapping stability and resolution.
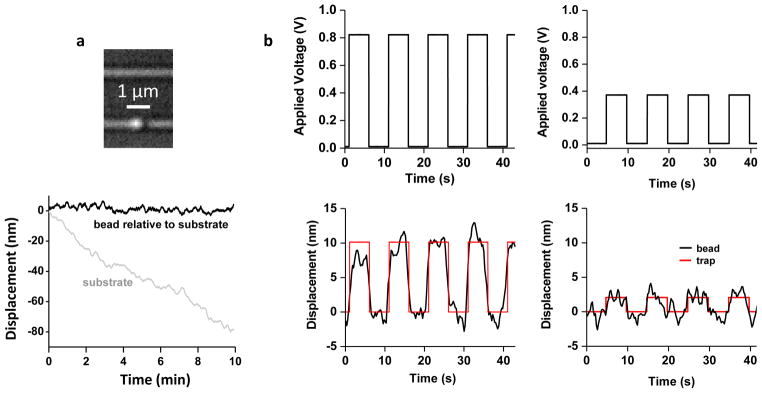
a, Trapping stability measurements. Top shows an optical microscope image of a bead (490 nm in diameter) held on the lower nSWAT. The bottom shows the bead position along the waveguide, relative to the waveguide, over 10 min. The oxide edge of the fluid region on the device was used as a fiducial marker.
b, Trapping control resolution. The position of a bead held by an nSWAT was measured as the trap was stepped in a square-wave fashion by the application of a voltage to the microheater to generate 9 nm (left) and 2 nm (right) steps. Red curves are fits to a periodic square wave function with the amplitude, period, and phase delay as fitting parameters.
We demonstrate nanometer resolution control of positioning of trapped beads using an nSWAT. To determine how well a trapped bead can be positioned, we applied a square-wave voltage to the microheater while simultaneously measuring the trapped bead’s position. As shown in Fig. 2b, 9 nm steps are readily resolved, and even 2 nm steps are discernible, indicating the potential of this device for detecting molecular events that occur at the nanometer scale31.
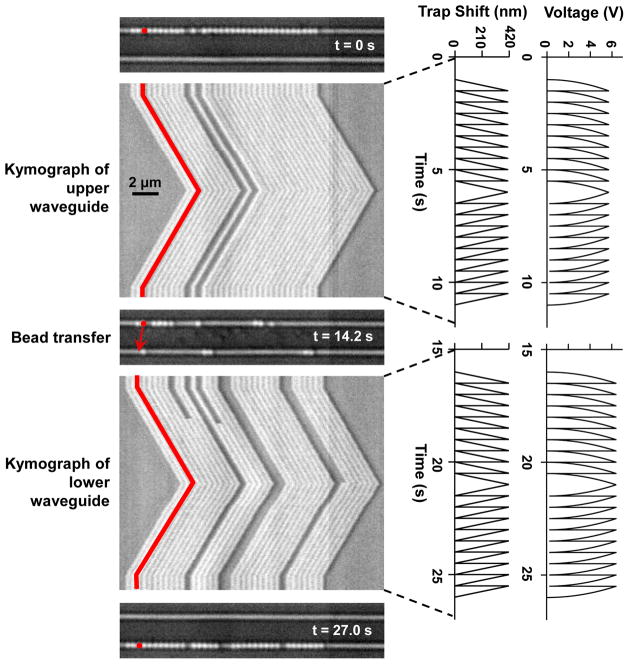
We also demonstrate a method for precision transport of trapped particles over many microns using an nSWAT without applying damagingly high voltage to the microheater (Fig. 3). To do this, we took advantage of the periodic spacing of the traps. The positioning of the nSWATs was precisely calibrated (Fig. S2) such that by applying a non-linear increasing voltage ramp to the microheater of an nSWAT (Fig. 3, a linear ramp in the trap array position was generated to steadily displace the array by one spatial period of the standing wave (~ 430 nm; Methods). Then the voltage was reset to zero, and the ramping process was repeated, resulting in a sawtooth-pattern in the trap array position. Because the trap array was reset much faster (~ 30 kHz, Fig. S4) than the corner frequency of a trapped bead (150 Hz, Fig. S1), a trapped bead could not respond to the sudden trap position reset and instead behaved as if the microheater voltage was ramped continuously (Methods). This resulted in a steady movement of trapped beads at a constant controlled speed with the transport distance limited only by the size of the trapping region. Fig. 3 shows an example of this long-distance transport of an array of beads (see also Video S1). This array was initially trapped on the upper waveguide and was transported at a constant speed over several microns, in both directions. Then the laser power was switched from the upper to the lower waveguide using the MZI switch microheater. When this took place, a modest fluid flow was applied to direct motion of the beads downward, and the array of trapped beads was shifted to the lower waveguide. The beads were then transported along the lower waveguide. Note that the spacing of the beads in this packed array corresponds well with the expected periodicity of the standing wave (Fig. S5).
Figure 3. Controlled long range transportation by an nSWAT.
An array of beads (356 nm in diameter, one false-colored) were initially trapped on the upper waveguide and transported in a controlled manner along the waveguide in both directions (Video S1). Subsequently, laser power was switched from the upper to the lower waveguide using the MZI switch and beads were then trapped and transported along the lower waveguide. Each kymograph shows a line scan of an image of the active waveguide (horizontal axis) versus time (vertical axis), with the corresponding voltage applied to the microheater and the resulting phase shift of the standing wave plotted on the right.
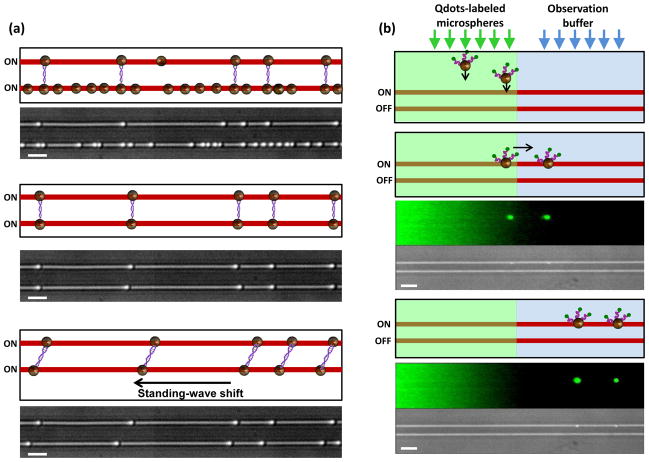
As an application of nSWAT to biomolecules, we demonstrate sorting of DNA dumbbells –single molecules of DNA with a bead attached at each end32 – from a mixture of other bead and DNA species (Methods), and the subsequent manipulation of the DNA dumbbells. Sorting is often a critical priming step for single molecule measurements. Here sorting was achieved by switching off the optical trapping force in one waveguide while applying a modest flow to direct motion of the released particles away from the other waveguide (Fig. 4a; Video S2). Trapping by the upper waveguide in the presence of downward fluid flow revealed DNA dumbbells. Subsequent trapping by the lower waveguide in the presence of upward flow retained DNA dumbbells and removed all other trapped bead species. This process may be repeated for further enrichment of DNA dumbbells. Subsequently, the array of DNA molecules was extended by moving the traps of the lower nSWAT relative to those of the upper nSWAT. We envision that this sorting and stretching ability would be advantageous in multiple situations. For example, DNA may be suspended between the two waveguides for visualization of motor protein movement along the DNA, complexes bound to the DNA may be disrupted by stretching the DNA, or enriched DNA dumbbells may be transported to a different region for a chemical or biomolecular reaction.
Figure 4. Manipulation, transport, and change of chemical environment of biomolecules.
a, Sorting and manipulation of individual DNA molecules. Cartoons and corresponding video frames (Video S2) explain the steps in the process. DNA dumbbells were formed using beads of 490 nm diameter and sorted by a combination of trapping and fluid flow forces. The sorted array of DNA dumbbells was extended by moving the traps of the lower nSWAT relative to those of the upper nSWAT. Scale bars are 2 μm.
b. On-chip changes of chemical environment of biomolecules with simultaneous fluorescence monitoring. Cartoons and corresponding video frames (Video S3) explain the steps in the process. Each video frame is composed of two panels with the upper panel showing a fluorescence image and the lower panel showing the corresponding bright field image. Free Qdots and Qdots-labelled beads were introduced by the left laminar flow. The cartoons show only beads (brown) labeled with Qdots (green) via DNA linkers (purple). Beads were trapped by the upper nSWAT and subsequently transported to and held in the right laminar flow (observation buffer) which had a different chemical environment. During the experiment, the lower nSWAT was off. Scale bars are 4 μm.
As another application of nSWAT to biochemical experiments, we demonstrate how a bead-bound sample can be transported from one chemical environment to another, potentially for a sequence of chemical reactions and analyses, all on chip (Figure 4b; Video S3; Methods). We formed two adjacent laminar flows over the waveguides with one laminar flow containing a mixture of free quantum dots (Qdots) and Qdots-labeled beads, and the other containing observation buffer. Only beads were trapped by an nSWAT. They were then transported to the adjacent laminar flow (observation buffer) and held briefly to simulate a potential reaction and/or measurement before release. We envision this technology will enable many potential applications for on-chip sorting, sampling, and monitoring of biochemical reactions.
In conclusion, we have demonstrated a new platform for the next generation of optical trapping instruments. This platform has a number of advantages. First, it enables high throughput, as tens or potentially hundreds of trapped particles can be monitored and manipulated at the same time. Second, this multiplexing is achieved using laser power comparable to that of a single optical trap, in contrast to conventional microscope-based multiplexing approaches. Third, the on-chip nature of our device makes it inherently stable, eliminating the need for complicated drift-reduction techniques. Finally, the horizontal geometry and ultra-fast trap control are well-suited for integration with fluorescence and laminar flow channels. While in this work, the nSWAT device was implemented with silicon waveguides, it can be realized with different optical materials and at different wavelength ranges. We envision that our instrument will enable routine high-throughput optical trapping experiments for biophysical and biochemical analysis. Thus nSWATs establish parallel processing and promise to make manipulation and precision measurements broadly available.
Methods
Device fabrication and characterization
The device fabrication process is similar to, and modified from, the one we have previously described27. The details can be found in Supplementary Section 1. For the device characterization, a tunable laser (at 1550 nm, Ando AD4321Q) after being amplified by a laser amplifier (IPG Photonics) was coupled to the chip using an optical tapered lensed fiber. A fiber polarization controller adjusted the light polarization to the transverse magnetic (TM) polarization mode of the waveguide. The transmitted light through the chip was collected from the opposite end of the chip using an additional tapered lensed optical fiber and sent to a photodetector (Thorlabs, PDB150C). A microprobe (GBB Industries), for applying electric voltage, was connected to the contact electrodes of the microheater (with a measured resistance of ~ 300 Ω). The optimal operational wavelength range is when minimum light is transmitted through the output port of the nSWAT to the photodetector. This means that a standing-wave made of counter-propagating waves with nearly equal amplitudes has been formed in the trapping region. In all the experiments shown in this paper, the laser power in the trapping region was 30 to 40 mW for all measurements except for calibrations.
Trapping methods
To prevent beads from sticking to the device surfaces, we coated the sample chamber with 1 mg/ML 1, 2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) in 100 mM NaCl and 10 mM Tris pH 8.0.
For trapping experiments, polystyrene beads (Polysciences) with diameters of 356 nm or 490 nm in 20 mM Tris-HCl pH 8.0 were flowed into the microfluidic channel using syringe pumps (Harvard PhD 2000). Using image tracking (see below), the spacing between traps in the nSWAT was experimentally determined to be 423 nm (Figs. 3 and S5).
To ensure continuous long distance transport as shown in Fig. 3, a trapped bead should not move substantially during the brief reset of the microheater voltage (~ 30 μs, Fig. S4). For simplicity, assume that the bead undergoes diffusion during the voltage reset, then the distance it diffuses during the reset will be ~ 15 nm, smaller than the amplitude of its Brownian motion in a stationary trap (~ 20 nm) (Fig. S1). Even if the bead is subjected to an external force during the reset (e.g., 1 pN), its biased motion is still limited (~ 9 nm displacement along the direction of the force). Therefore, because the voltage reset is rapid, the bead effectively remains stationary during the reset and is transported smoothly without interruption.
Imaging and tracking of trapped beads
The sample plane was imaged by a 100X, 1.3 NA oil-immersion objective (Nikon) using a CCD camera (JAI RM-6740GE, 7.4 μm pixel). Bead positions were determined by fitting a circular Gaussian spot to the image of each bead in each frame33. This allowed us to localize a bead to better than 10 nm using one frame, and higher resolution was achieved by averaging over multiple frames. For stability measurements (Fig. 2a), images were acquired at 10 frames per second (FPS) and then data were averaged to 0.1 FPS, while stepping measurements (Fig. 2b) were acquired at 540 FPS and averaged to 10.8 FPS. In order to correct for drift of the sample relative to the camera, we also tracked the position of one edge of the fluidic pool, and subtracted this displacement from all measured bead displacements.
Preparation of DNA
The 10 kbp DNA template used in the experiments was prepared using methods similar to those previously described34,35. Briefly, a 10 kbp plasmid (available upon request) was cut with SapI (NEB) to produce nonpalindromic overhangs. A fill-in reaction with Klenow fragment (NEB), digoxigenin-11-dUTP and biotin-11-dATP (Roche) was used to label the linearized plasmid DNA. This resulted in one end of the DNA labeled with digoxigenin and the other end labeled with biotin. A similar method was used to prepare the 30 bp DNA used for Qdot-labeling.
Preparation of DNA-tethered beads
To form DNA dumbbells, 10 kbp DNA was mixed with both 490 nm streptavidin-coated beads and anti-digoxigenin-coated beads at molar ratios 3:1:1 following a previously described protocol34.
Preparation of Qdots-labeled beads
To form Qdots-labeled beads, Qdot® 525 streptavidin conjugate (Invitrogen) was mixed with 490 nm anti-digoxigenin-coated polystyrene beads and a 30 bp DNA with biotin-label at one end and digoxigenin label at the other end, following previously described protocols34.
Laminar flow was established by multiple inlets to our flow cells. The relative size of each flow was controlled by adjusting the inlet pressures. The outlet flow rate was controlled by a syringe pump. Bright field and fluorescence imaging were interlaced onto a single cooled EM-CCD camera (Andor Ixon3 897). Fluorescence was excited at 488 nm by an Argon ion laser (Lexel Laser Inc.) and controlled by a mechanical shutter. The bright field was illuminated by a TTL-controlled LED (Thorlabs).
Supplementary Material
Acknowledgments
We thank members of the Wang Lab and the Lipson Lab for critical comments on this work. We wish to acknowledge postdoctoral support to R.A.F. from the American Cancer Society (125126-PF-13-205-01-DMC), graduate traineeship support to S.N.S from Cornell’s Molecular Biophysics Training Grant funded by National Institutes of Health (T32GM008267) and a National Science Foundation Graduate Research Fellowship under Grant No. (DGE-1144153), and support to M.D.W. by the National Institutes of Health (GM059849) and the National Science Foundation (MCB-0820293). This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECCS-0335765).
References
- 1.Chu S. The manipulation of neutral particles. Reviews of Modern Physics. 1998;70:685–706. [Google Scholar]
- 2.Forth S, Sheinin M, Inman J, Wang M. Torque Measurement at the Single-Molecule Level. Annu Rev Biophys. 2013 doi: 10.1146/annurev-biophys-083012-130412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 4.Hilario J, Kowalczykowski SC. Visualizing protein-DNA interactions at the single-molecule level. Curr Opin Chem Biol. 2010;14:15–22. doi: 10.1016/j.cbpa.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dholakia K, Cizmar T. Shaping the future of manipulation. Nature Photonics. 2011;5:335–342. [Google Scholar]
- 6.Padgett M, Bowman R. Tweezers with a twist. Nature Photonics. 2011;5:343–348. [Google Scholar]
- 7.Jannasch A, Demirors AF, van Oostrum PDJ, van Blaaderen A, Schaffer E. Nanonewton optical force trap employing anti-reflection coated, high-refractive-index titania microspheres. Nature Photonics. 2012;6:469–473. [Google Scholar]
- 8.Grier DG. A revolution in optical manipulation. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 9.Fainman Y. Optofluidics : fundamentals, devices, and applications. McGraw-Hill; New York: 2010. p. xvii. 510 p., 16 p. of plates. [Google Scholar]
- 10.Erickson D, Serey X, Chen YF, Mandal S. Nanomanipulation using near field photonics. Lab on a Chip. 2011;11:995–1009. doi: 10.1039/c0lc00482k. [DOI] [PubMed] [Google Scholar]
- 11.Erickson D, Rockwood T, Emery T, Scherer A, Psaltis D. Nanofluidic tuning of photonic crystal circuits. Optics Letters. 2006;31:59–61. doi: 10.1364/ol.31.000059. [DOI] [PubMed] [Google Scholar]
- 12.Diehl L, et al. Microfluidic tuning of distributed feedback quantum cascade lasers. Optics Express. 2006;14:11660–11667. doi: 10.1364/oe.14.011660. [DOI] [PubMed] [Google Scholar]
- 13.Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–6. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 14.Juan ML, Righini M, Quidant R. Plasmon nano-optical tweezers. Nature Photonics. 2011;5:349–356. [Google Scholar]
- 15.Schmidt H, Hawkins AR. The photonic integration of non-solid media using optofluidics. Nature Photonics. 2011;5:598–604. [Google Scholar]
- 16.Fan XD, White IM. Optofluidic microsystems for chemical and biological analysis. Nature Photonics. 2011;5:591–597. doi: 10.1038/nphoton.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378:209–12. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 18.Farre A, van der Horst A, Blab GA, Downing BPB, Forde NR. Stretching single DNA molecules to demonstrate high-force capabilities of holographic optical tweezers. Journal of Biophotonics. 2010;3:224–233. doi: 10.1002/jbio.200900107. [DOI] [PubMed] [Google Scholar]
- 19.Yang AHJ, et al. Optical manipulation of nanoparticles and biomolecules in sub-wavelength slot waveguides. Nature. 2009;457:71–75. doi: 10.1038/nature07593. [DOI] [PubMed] [Google Scholar]
- 20.Lin SY, Crozier KB. Planar silicon microrings as wavelength-multiplexed optical traps for storing and sensing particles. Lab on a Chip. 2011;11:4047–4051. doi: 10.1039/c1lc20574a. [DOI] [PubMed] [Google Scholar]
- 21.Lin SY, Crozier KB. An integrated microparticle sorting system based on near-field optical forces and a structural perturbation. Optics Express. 2012;20:3367–3374. doi: 10.1364/OE.20.003367. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt BS, Yang AHJ, Erickson D, Lipson M. Optofluidic trapping and transport on solid core waveguides within a microfluidic device. Optics Express. 2007;15:14322–14334. doi: 10.1364/oe.15.014322. [DOI] [PubMed] [Google Scholar]
- 23.Mandal S, Serey X, Erickson D. Nanomanipulation using silicon photonic crystal resonators. Nano Letters. 2010;10:99–104. doi: 10.1021/nl9029225. [DOI] [PubMed] [Google Scholar]
- 24.Renaut C, et al. On chip shapeable optical tweezers. Scientific Reports. 2013;3 doi: 10.1038/srep02290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaquay E, Martinez LJ, Mejia CA, Povinelli ML. Light-Assisted, Templated Self-Assembly Using a Photonic-Crystal Slab. Nano Letters. 2013;13:2290–2294. doi: 10.1021/nl400918x. [DOI] [PubMed] [Google Scholar]
- 26.Lei T, Poon AW. Silicon-on-insulator multimode-interference waveguide-based arrayed optical tweezers (SMART) for two-dimensional microparticle trapping and manipulation. Optics Express. 2013;21:1520–1530. doi: 10.1364/OE.21.001520. [DOI] [PubMed] [Google Scholar]
- 27.Soltani M, Inman JT, Lipson M, Wang MD. Electro-optofluidics: achieving dynamic control on-chip. Optics Express. 2012;20:22314–22326. doi: 10.1364/OE.20.022314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atabaki AH, Hosseini ES, Eftekhar AA, Yegnanarayanan S, Adibi A. Optimization of metallic microheaters for high-speed reconfigurable silicon photonics. Optics Express. 2010;18:18312–18323. doi: 10.1364/OE.18.018312. [DOI] [PubMed] [Google Scholar]
- 29.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun B, et al. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–5. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forget AL, Dombrowski CC, Amitani I, Kowalczykowski SC. Exploring protein-DNA interactions in 3D using in situ construction, manipulation and visualization of individual DNA dumbbells with optical traps, microfluidics and fluorescence microscopy. Nat Protoc. 2013;8:525–38. doi: 10.1038/nprot.2013.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–3. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 34.Fuller DN, et al. A general method for manipulating DNA sequences from any organism with optical tweezers. Nucleic Acids Research. 2006;34 doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laib S, Robertson RM, Smith DE. Preparation and characterization of a set of linear DNA molecules for polymer physics and rheology studies. Macromolecules. 2006;39:4115–4119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.