Abstract
Emerging data demonstrate important roles for the TYRO3/AXL/MERTK receptor tyrosine kinase (TAM RTK) family in diverse cancers. We investigated the prognostic relevance of GAS6 expression, encoding the common TAM RTK ligand, in 270 adults (n=71 aged <60 years; n=199 aged ≥60 years) with de novo cytogenetically normal acute myeloid leukemia (CN-AML). Patients expressing GAS6 (GAS6+), especially those aged ≥60 years, more often failed to achieve a complete remission (CR). In all patients, GAS6+ patients had shorter disease-free (DFS) and overall (OS) survival than patients without GAS6 expression (GAS6−). After adjusting for other prognostic markers, GAS6+ predicted CR failure (P=0.02), shorter DFS (P=0.004) and OS (P=0.04). To gain further biologic insights, we derived a GAS6-associated gene-expression signature (P<0.001) that in GAS6+ patients included overexpressed BAALC and MN1, known to confer adverse prognosis in CN-AML, and overexpressed CXCL12, encoding stromal cell-derived factor, and its receptor genes, CXCR4 and CXCR7. This study reports for the first time that GAS6 expression is an adverse prognostic marker in CN-AML. Although GAS6 decoy receptors are not yet available in the clinic for GAS6+ CN-AML therapy, potential alternative therapies targeting GAS6+-associated pathways, e.g., CXCR4 antagonists may be considered for GAS6+ patients to sensitize them to chemotherapy.
Keywords: GAS6, acute myeloid leukemia, prognosis
INTRODUCTION
Constitutive activity of the receptor tyrosine kinase (RTK) family has been observed in malignant blasts from patients with acute myeloid leukemia (AML). Members of the RTK family include FLT3 and KIT, whose constitutive kinase activity can occur through several mechanisms, such as mutationally-induced autophosphorylation, receptor overexpression and/or aberrant expression of the receptors’ ligands.1-3 The constitutive activity of FLT3 and KIT has been associated with poor clinical outcomes, and therapeutic targeting of activated RTKs is currently an area of intense investigation.4-8
In cancer, another RTK family, the TAM RTKs (i.e., TYRO3, AXL and MERTK), has been shown to support survival, proliferation, migration, invasion, angiogenesis, metastasis and chemoresistance,9-12 and TAM RTK inhibitors are already in pre-clinical and clinical development for several solid tumors.11-13 Although TAM RTKs are aberrantly expressed in AML,11,14,15 to date, only AXL expression has been reported to adversely impact outcome in adults with cytogenetically normal AML (CN-AML).16
No AXL mutations have been described in AML, suggesting activation of AXL may occur at least in part via aberrant autocrine expression of GAS6, that binds AXL with high affinity and is also the common ligand for all three TAM RTKs.17 GAS6 was shown to be aberrantly expressed in AML cell lines.18 These data point to a possible role for the GAS6/TAM RTK signaling axis in AML and prompted us to test the clinical impact of GAS6 expression in a molecularly characterized cohort of chemotherapy-treated adults with de novo CN-AML.
METHODS
Patients
Available pretreatment bone marrow or blood samples were obtained from 270 patients with de novo CN-AML (aged 18 to 83 years; median, 66 years; n=71 aged <60 years; n=199 aged ≥60 years) enrolled on Cancer and Leukemia Group B (CALGB)/Alliance companion protocols 8461 (cytogenetic analyses), 20202 (molecular analyses) and 9665 (tissue banking). Patients were treated on CALGB/Alliance protocols 8525, 8923, 9420, 9720, 10201, or 19808.19-25 The treatment protocols included cytarabine/daunorubicin-based induction but differed with regard to consolidation therapy (for details see Supplemental Material). Per protocols, no patient received allogeneic stem-cell transplantation in first complete remission (CR). All protocols were in accordance with the Declaration of Helsinki and approved by institutional review boards at each center, and all patients provided written informed consent.
Cytogenetic and molecular analyses
For the patient’s karyotype to be considered normal, ≥20 metaphases from short-term cultures of the bone marrow specimens obtained at diagnosis had to have been analyzed and the normal result confirmed by central karyotype review.26 Tissue samples were cryopreserved after mononuclear cell enrichment through a Ficoll gradient. The presence or absence of FLT3 internal tandem duplication (FLT3-ITD),27 FLT3 tyrosine kinase domain mutations (FLT3-TKD),28 MLL partial tandem duplication (MLL-PTD),29,30 mutations in the NPM1,31 CEBPA,32 WT1,33 TET2,34 IDH1/2,35 RUNX1,36 ASXL136 and DNMT3A37 genes, and BAALC,38 ERG,38 and MN139 expression levels were assessed centrally as previously described. Patients were also categorized according to the European LeukemiaNet (ELN) reporting system.40 CN-AML patients with CEBPA mutation and/or NPM1 mutation without FLT3-ITD were classified in a Favorable genetic group and those with wild-type CEBPA, FLT3-ITD and/or NPM1 mutation, or wild-type NPM1 in an Intermediate-I genetic group
Expression analysis of GAS6 and TAM RTKs
GAS6, TYRO3, AXL and MERTK transcript expression levels measured with Affymetrix U133 plus 2.0 array (Affymetrix, Santa Clara, CA, USA) assays. The GeneAnnot chip definition file was used to derive a single expression value for each gene per patient sample.41 For array normalization and expression value computation, the robust multichip average method was implemented separately for samples from older and younger patients.42
Patients were categorized as either expressing GAS6 (yes or GAS6-positive, hereafter denoted GAS6+) if the probe-set fluorescence intensity (PFI) was greater than background fluorescence intensity (BFI) and not expressing GAS6 (no or GAS6-negative denoted GAS6−) if the GAS6 PFI was less than or equal to the BFI. Similarly, patients were categorized as either TYRO3+ or AXL+ (if the PFIs were greater than BFI) and TYRO3− or AXL− (if the PFIs were less than or equal to BFI). The MERTK PFI was above the BFI in all samples and, based on an optimal cutpoint analysis (see Supplemental Material),43 patients were grouped into high expressers (MERTK+) or lower expressers (MERTK−) if they were in the upper two tertiles or in the lowest tertile groups, respectively.
Affymetrix-microarray gene expression profiling analysis
To establish a signature of genes differentially expressed between GAS6+ and GAS6− patients, we evaluated the aforementioned Affymetrix gene-expression profiles. Normalized expression values were compared between GAS6+ and GAS6− patients and a univariable significance level of P<0.001 was used to identify differentially expressed genes. A global test of significance based on a permutation procedure was performed to determine whether or not the number of differentially expressed genes was more than expected by chance. The false discovery rate (FDR) was used to assess multiple testing errors. A permutation test was computed based on 1 000 random permutations.
The Ingenuity Pathway Analysis tool (IPA Tool; Ingenuity H Systems, Redwood City, CA, USA; http://www.ingenuity.com) was used to identify enriched biological networks, global functions and functional pathways. Genes with altered expression profile associated with GAS6 expression status were imported into the IPA Tool. As a second means for identifying enriched ontologies, the web-based Database for Annotation, Visualization, and Integrated Discovery (DAVID) tool (DAVID Bioinformatics resources 6.7 http://david.abcc.ncifcrf.gov/) was used.
Clinical endpoints and statistical analyses
Baseline characteristics were compared between GAS6+ and GAS6− patients using the Fisher’s exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables. Definitions of clinical endpoints [i.e., CR, disease-free (DFS) and overall (OS) survival] and details of outcome analyses are provided in the Supplemental Material. Briefly, for time-to-event analyses, we calculated survival estimates using the Kaplan-Meier44 method, and compared groups by the log-rank test. We constructed age group-adjusted multivariable logistic regression models to analyze factors associated with the achievement of CR, and age group-adjusted multivariable Cox proportional hazards models45 for factors associated with survival endpoints. All analyses were performed by the Alliance for Clinical Trials in Oncology Statistics and Data Center.
RESULTS
Association of GAS6 expression status with clinical characteristics, TAM RTK expression status and molecular markers at diagnosis
Of the 270 patients, 26% of patients were GAS6+, (n=69) and 74% GAS6− (n=201). At diagnosis, GAS6+ patients had higher platelet counts (P=0.03), lower percentages of blood blasts (P=0.01), more often hepatomegaly (P=0.006) and co-expression of AXL (28% vs 5%, P<0.001) compared with GAS6− patients. There was no association between GAS6 and TYRO3 expression (P=0.74), whereas more GAS6− than GAS6+ patients were MERTK+ (P=0.02, Table 1). Compared with GAS6− patients, GAS6+ patients were more often wild-type for NPM1 (P<0.001) and CEBPA (P=0.02) and therefore more often in the ELN Intermediate-I Genetic Group (P<.001), and had mutations in RUNX1 (P<0.001) and ASXL1 (P=0.002), and high BAALC (P=0.02) and MN1 (P=0.05; Table 1) expression.
Table 1.
Comparison of clinical and molecular characteristics of de novo cytogenetically normal AML patients according to GAS6 expression status
| Variable | GAS6+a (n=69) |
GAS6−a (n=201) |
Pb |
|---|---|---|---|
| Age, years | 0.02 | ||
| Median | 68 | 65 | |
| Range | 37-81 | 18-83 | |
|
| |||
| Age group, n (%) | 0.35 | ||
| <60 years | 15 (22) | 56 (28) | |
| ≥60 years | 54 (78) | 145 (72) | |
|
| |||
| Male sex, n (%) | 35 (51) | 101 (50) | 1.00 |
|
| |||
| Race, n (%) | 0.81 | ||
| White | 63 (93) | 181 (91) | |
| Non-white | 5 (7) | 19 (9) | |
|
| |||
| Hemoglobin, g/dL | 0.56 | ||
| Median | 9.4 | 9.4 | |
| Range | 6.4-12.5 | 4.8-15.0 | |
|
| |||
| Platelet count, ×109/L | |||
| Median | 84 | 66 | |
| Range | 11-309 | 4-850 | |
|
| |||
| White blood cell count, ×109/L | |||
| Median | 21.1 | 26.5 | |
| Range | 1.0-434.1 | 1.0-450.0 | |
|
| |||
| Blood blasts (%) | 0.01 | ||
| Median | 40 | 59 | |
| Range | 0-96 | 0-99 | |
|
| |||
| Bone marrow blasts (%) | 0.87 | ||
| Median | 70 | 67 | |
| Range | 7-97 | 4-97 | |
|
| |||
| Extramedullary involvement, n (%) | 20 (29) | 45 (23) | 0.33 |
| Hepatomegaly | 8 (12) | 5 (3) | 0.006 |
|
| |||
| TYRO3 expression group a , n (%) | 0.74 | ||
| Positive | 16 (23) | 43 (21) | |
| Negative | 53 (77) | 158 (79) | |
|
| |||
| AXL expression group a , n (%) | <0.001 | ||
| Positive | 19 (28) | 11 (5) | |
| Negative | 50 (72) | 190 (95) | |
|
| |||
| MERTK expression c, n (%) | 0.02 | ||
| Positive | 38 (55) | 143 (71) | |
| Negative | 31 (45) | 58 (29) | |
|
| |||
| TYRO3/AXL dual receptor d , n (%) | 30 (43) | 51 (25) | 0.006 |
|
| |||
| NPM1, n (%) | <0.001 | ||
| Mutated | 20 (29) | 141 (70) | |
| Wild-type | 48 (71) | 60 (30) | |
|
| |||
| FLT3-ITD, n (%) | 0.56 | ||
| Present | 26 (38) | 68 (34) | |
| Absent | 42 (62) | 133 (66) | |
|
| |||
| CEBPA, n (%) | 0.02 | ||
| Mutated | 4 (6) | 35 (17) | |
| Single mutated | 4 | 19 | |
| Double mutated | 0 | 16 | |
| Wild-type | 64 (94) | 166 (83) | |
|
| |||
| ELN Genetic Group e , n (%) | <0.001 | ||
| Favorable | 12 (18) | 115 (57) | |
| Intermediate-I | 56 (82) | 86 (43) | |
|
| |||
| FLT3-TKD, n (%) | 1.00 | ||
| Present | 7 (10) | 23 (11) | |
| Absent | 61 (90) | 178 (89) | |
|
| |||
| WT1, n (%) | 0.25 | ||
| Mutated | 2 (3) | 15 (7) | |
| Wild-type | 66 (97) | 186 (93) | |
|
| |||
| TET2, n (%) | 0.43 | ||
| Mutated | 15 (22) | 55 (28) | |
| Wild-type | 52 (78) | 143 (72) | |
|
| |||
| MLL-PTD, n (%) | 1.00 | ||
| Present | 4 (6) | 13 (7) | |
| Absent | 64 (94) | 182 (93) | |
|
| |||
| IDH1, n (%) | 0.83 | ||
| R132 | 5 (7) | 24 (12) | |
| V71I | 2 (3) | 0 (0) | |
| Wild-type | 60 (90) | 173 (88) | |
|
| |||
| IDH2, n (%) | 0.72 | ||
| IDH2 | 14 (21) | 37 (19) | |
| R140 | 8 | 33 | |
| R172 | 6 | 4 | |
| Wild-type | 53 (79) | 160 (81) | |
|
| |||
| RUNX1, n (%) | <0.001 | ||
| Mutated | 27 (44) | 9 (5) | |
| Wild-type | 35 (56) | 173 (95) | |
|
| |||
| ASXL1, n (%) | 0.002 | ||
| Mutated | 16 (24) | 16 (8) | |
| Wild-type | 51 (76) | 179 (92) | |
|
| |||
| DNMT3A, n (%) | 0.76 | ||
| Mutated | 21 (32) | 66 (35) | |
| R882 | 17 | 40 | |
| Non-R882 | 4 | 26 | |
| Wild-type | 44 (68) | 124 (65) | |
|
| |||
| ERG expression group f , n (%) | 0.33 | ||
| High | 30 (43) | 102 (51) | |
| Low | 39 (57) | 99 (49) | |
|
| |||
| BAALC expression group f , n (%) | 0.02 | ||
| High | 44 (64) | 93 (46) | |
| Low | 25 (36) | 108 (54) | |
|
| |||
| MN1 expression group f , n (%) | 0.05 | ||
| High | 34 (65) | 65 (48) | |
| Low | 18 (35) | 70 (52) | |
Abbreviations: FLT3-ITD, internal tandem duplication of the FLT3 gene; ELN, European LeukemiaNet; FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene.
All patients with GAS6 probe-set fluorescence intensity greater than the background fluorescence intensity (BFI) are defined as GAS6-positive (GAS6+) and those with GAS6 probe-set intensity less than or equal to the BFI as GAS6-negative (GAS6−). Similarly, patients were categorized as either TYRO3+ (TYRO3 expression greater than BFI) or TYRO3− (if TYRO3 expression was less than or equal to BFI) and AXL+ (expression greater than BFI) or AXL− (expression less than or equal to BFI).
P-values for categorical variables are from Fisher’s exact test, P-values for continuous variables are from Wilcoxon rank sum test.
All patients in the upper 2/3 of the values of MERTK are defined as MERTK+. All patients in the lower 1/3 of the values of MERTK are defined as MERTK−.
If patient has AXL+ and TYRO3+ expression, AXL+ and TYRO3− expression, or AXL− and TYRO3+ expression then TYRO3/AXL dual receptor is defined to be positive. If a patient has AXL− and TYRO3− expression then TYRO3/AXL dual receptor is defined to be negative.
According to the ELN recommendations,40 Favorable Genetic Group is defined as CEBPA-mutated or FLT3-ITD-negative and NPM1-mutated. Intermediate-I Genetic Group is defined as CEBPA wild-type and FLT3-ITD-positive and NPM1-mutated, FLT3-ITD-negative and NPM1-wild-type, or FLT3-ITD-positive and NPM1-wild-type.
The median expression value was used as the cutoff for high and low values.
Impact of GAS6 expression on clinical outcomes of de novo CN-AML patients
In age group-adjusted analyses, GAS6+ expression associated with lower odds of achieving CR (P<0.001; Table 2), with CR rates significantly different in patients ≥60 years of age [46% GAS6+ (n=54) vs 74% GAS6− (n=145); P<0.001]. None of the TAM RTKs impacted on CR (Table S1). To assess whether GAS6 expression independently affects clinical outcomes when other known clinical and molecular prognostic features are considered, we performed multivariable analyses (MVAs). For CR, GAS6+ status predicted lower probability of achieving CR [(P=0.02; odds ratio (OR), 0.46; 95% confidence interval (CI), 0.23-0.89)], after adjusting for the ELN CN-AML Genetic Group45 status, BAALC expression status, white blood cell (WBC) count and age group (Table 3).
Table 2.
Age group-adjusted analyses of outcomes by GAS6 positive expression versus no expression in de novo cytogenetically normal AML patients
| Outcome Endpoint | OR/HR (95% CI) | P |
|---|---|---|
| CR | 0.35 (0.20, 0.63) | <0.001 |
| DFS | 1.55 (1.05, 2.26) | 0.03 |
| OS | 1.55 (1.16, 2.09) | 0.004 |
Abbreviations: CR, complete remission; DFS, disease-free survival; OS, overall survival; OR, odds ratio; HR, hazard ratio; CI, confidence interval.
Note: ORs < 1.0 means a lower CR rate, and HRs > 1.0 mean higher risk
Table 3.
Multivariable models
| Complete remission | ||
| Variable | OR (95% CI) | P |
| GAS6 expression (+ vs −) | 0.46 (0.23-0.89) | 0.02 |
| ELN Genetic Group (Favorable vs Intermediate-I)a | 2.13 (1.07-4.23) | 0.03 |
| BAALC expression (high vs low) b | 0.26 (0.13-0.50) | <0.001 |
| WBC count (continuous, 50 unit increase) | 0.57 (0.42-0.77) | <0.001 |
| Age group (≥60 years vs <60 years) | 0.35 (0.16-0.75) | 0.007 |
| Disease-free survival | ||
| Variable | HR (95% CI) | P |
| GAS6 expression (+ vs −) | ||
| TYRO3/AXL dual receptor− patients | 2.12 (1.27-3.56) | 0.004 |
| TYRO3/AXL dual receptor+ patients | 0.73 (0.38-1.38) | 0.33 |
| Interaction between GAS6 expression status and TYRO3/AXL dual receptor status | 0.01 | |
| WT1 (mutated vs wild-type) | 2.62 (1.24-5.52) | 0.01 |
| DNMT3A (R882 mutated vs non-R882 mutated and wild-type) | 1.95 (1.29-2.93) | 0.001 |
| BAALC expression (high vs low)b | 1.58 (1.10-2.25) | 0.01 |
| Age group (≥60 years vs <60 years) | 2.62 (1.72-3.99) | <0.001 |
| Overall survival | ||
| Variable | HR (95% CI) | P |
| GAS6 expression (+ vs −) | ||
| TYRO3/AXL dual receptor− patients | 1.55 (1.01-2.38) | 0.04 |
| TYRO3/AXL dual receptor+ patients | 0.78 (0.47-1.30) | 0.34 |
| Interaction between GAS6 expression status and TYRO3/AXL dual receptor status | 0.04 | |
| ELN Genetic Group (Favorable vs Intermediate-I)a | 0.64 (0.46-0.89) | 0.008 |
| WT1 (mutated vs wild-type) | 2.59 (1.47-4.55) | 0.001 |
| DNMT3A (R882 mutated vs non-R882 mutated and wild-type) | 1.45 (1.02-2.05) | 0.04 |
| BAALC expression (high vs low)b | 1.74 (1.29-2.34) | <0.001 |
| WBC count (continuous, 50 unit increase) | 1.14 (1.04-1.25) | 0.008 |
| Age group (≥60 years vs <60 years) | 2.90 (2.00-4.22) | <0.001 |
Abbreviations: OR, odds ratio; CI, confidence interval; WBC, white blood cell; HR, hazard ratio.
According to the European LeukemiaNet (ELN) recommendations,40 Favorable Genetic Group is defined as CEBPA-mutated or FLT3-ITD-negative and NPM1-mutated. Intermediate-I Genetic Group is defined as CEBPA wild-type and FLT3-ITD-positive and NPM1-mutated, FLT3-ITD-negative and NPM1-wild-type, or FLT3-ITD-positive and NPM1-wild-type.
Median cut was used to determine whether patients were in the high or low expression group.
Note: ORs > (<) 1.0 mean a higher (lower) complete remission rate, and HRs > (<) 1.0 mean higher (lower) risk for the higher values of the continuous variables and the first category listed for the categorical variables. Variables considered were those significant at α=.20 in univariable models, ie, for complete remission, GAS6 (+ vs −), ELN (Favorable vs Intermediate-I), WT1 (mutated vs wild-type), ASXL1 (mutated vs wild-type), ERG (high vs low), BAALC (high vs low), platelet counts (50×109/L increase), WBC count (50×109/L increase), extramedullary involvement (present vs absent), age group (≥60 years vs <60 years); for disease-free survival, GAS6 (+ vs −), TYRO3/AXL dual receptor status (+ vs −),ELN (Favorable vs Intermediate-I), WT1 (mutated vs wild-type), MLL-PTD (present vs absent), DNMT3A (R882 mutated vs non-R882 mutated and wild-type), ERG (high vs low), BAALC (high vs low), age group (≥60 years vs <60 years); for overall survival, GAS6 (+ vs −), TYRO3/AXL dual receptor status (+ vs −), ELN (Favorable vs Intermediate-I), WT1 (mutated vs wild-type), MLL-PTD (present vs absent), RUNX1 (mutated vs wild-type), DNMT3A (R882 mutated vs non-R882 mutated and wild-type), ERG (high vs low), BAALC (high vs low), WBC count (50×109/L increase), age group (≥60 years vs <60 years).
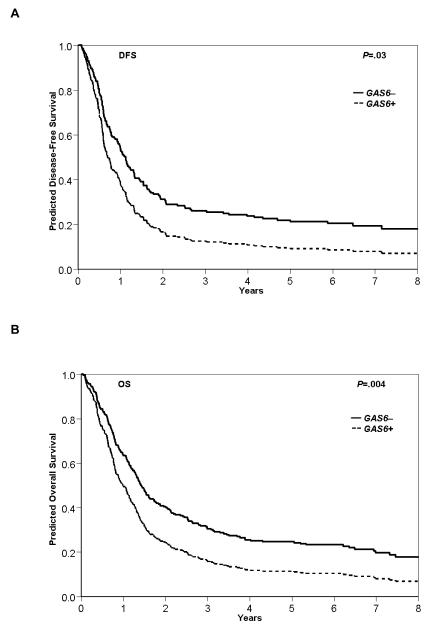
GAS6+ expression associated with shorter DFS (P=0.03) and OS (P=0.004) compared with GAS6− patients (Table 2 and Figure 1). As single markers, neither AXL nor MERTK influenced DFS or OS, whereas TYRO3 expression adversely impacted on both endpoints (Table S1). In multivariable modeling for DFS and OS, we noted a significant interaction (DFS, P=0.01; OS, P=0.04) between GAS6 expression and the combined TYRO3 and AXL expression status. In the dual receptor-positive patients, i.e., positive for one or both TYRO3 and AXL expression, GAS6 expression did not independently impact outcome, which may be reflective of the interplay between the GAS6 ligand and the TYRO3 and AXL receptors (Table 3). In the dual receptor-negative patients, i.e., negative for both TYRO3 and AXL expression, GAS6 expression remained an independent, adverse prognostic marker. Within the subgroup of dual receptor-negative patients, GAS6+ expression was a predictor of shorter DFS (P=0.004; hazard ratio (HR)=2.12; 95% CI, 1.27-3.56) and after adjusting for WT1 and DNMT3A R882 mutations, BAALC expression and age group; and shorter OS (P=0.04; HR=1.55; 95% CI, 1.01-2.38) after adjusting for ELN group, WT1 and DNMT3A R882 mutations, BAALC expression, WBC and age group.
Figure 1.
Clinical outcome by GAS6 expression status. Survival curves for (a) disease-free survival (DFS) and (b) overall survival (OS) are displayed for GAS6+ and GAS6− patient groups. Data were age adjusted (<60 years of age, n=71; ≥60 years of age, n=199).
A GAS6-associated gene expression signature in de novo CN-AML
To gain additional molecular insights into GAS6+ CN-AML, an Affymetrix microarray-based gene expression signature was derived. The signature contained 1 238 genes that were significantly differentially expressed between GAS6+ and GAS6− CN-AML blasts at diagnosis (Table S2). Within this signature, genes for which high expression level is an established adverse prognosticator in CN-AML were BAALC and MN1. These were overexpressed, respectively, 2.57-fold and 2.41-fold in the GAS6+ subgroup. Although hitherto not validated in large, independent patient sets, overexpression of the following four genes was reported to have an impact on outcome of AML patients. These were APP, which encodes the amyloid precursor protein and whose overexpression was associated with shorter survival than that of AML patients without APP overexpression46 (overexpressed 3.37-fold in GAS6+ patients); SETBP1, whose overexpression leads to PP2A inhibition, promoting proliferation of leukemic cells47 (overexpressed 1.73-fold); SPARC, which is overexpressed in patients with IDH2-R172 mutations and contributes to AML aggressiveness48 (overexpressed 2.05-fold); and CD74, whose lower surface protein levels associated with achievement of CR/partial CR in AML patients ages 60-75 years treated with bortezomib in combination with chemotherapy (overexpressed 2.51-fold).49 Moreover, overexpressed in GAS6+ CN-AML diagnostic samples were CXCL12 (3.2-fold), encoding for stromal cell-derived factor-1, and genes encoding both of its receptors, CXCR4 (1.52-fold) and CXCR7 (1.72-fold). Overexpression of CXCR4 has been previously associated with adverse clinical outcome in patients with CN-AML.50,51
CEBPA was among the 611 genes underexpressed in GAS6+ patients. Mutations in the CEBPA gene that encodes a transcription factor are associated with better outcome of AML patients (Table S2).32 CD33, encoding an immunotherapeutic target in AML, was also underexpressed (1.67-fold) in GAS6+ patients.
Gene expression signatures were not identified for any of the TAM RTKs (less than 10 genes, with FDRs of 10%; data not shown). Indeed, there was no apparent contribution from TAM RTKs in profiling analyses combining GAS6 with each of the TAM RTKs. This indicates that it is GAS6 expression status that drives the differential gene expression we observed.
Pathway analysis revealed that the GAS6-associated gene expression signature contained the following overrepresented molecular and cellular functions: a) cell cycle, b) cellular growth and proliferation, c) cell death and survival, d) cellular assembly and organization and, e) DNA replication, recombination and repair (Table 4). The top canonical pathways included a) IL-8 signaling, b) growth hormone signaling, c) mitotic roles of Polo-like kinase, d) CXCR4 signaling and e) Tec kinase signaling (Table 4). Of the top upstream regulators, colony stimulating factor 2 (granulocyte-macrophage), CSF2, was predicted by Ingenuity to be activated, while the cyclin dependent kinase inhibitor, CDKN1A, was predicted by Ingenuity to be inhibited (Table 4). A second analysis using DAVID also identified enriched clusters of genes, with the most highly enriched cluster (score of 9.66; Benjamini corrected P-values ranging from 2.8E-9 to 3.5E-6) containing genes involved in the cell cycle (data not shown).
Table 4.
Biological pathways over-represented in the GAS6 expression signature in cytogenetically normal AML
| Molecular and cellular functions (number of genes) a | P b |
| Cell cycle (202) | 7.75E-17 to 2.15E-03a |
| Cellular growth and proliferation (360) | 3.48E-15 to 2.41E-03 |
| Cell death and survival (363) | 5.28E-15 to 2.44E-03 |
| Cellular assembly and organization (195) | 2.76E-11 to 2.33E-03 |
| DNA replication, recombination and repair (136) | 2.76E-11 to 2.15E-03 |
|
| |
| Top canonical pathways a | |
| Interleukin-8 signaling | 5.43E-06 |
| Growth hormone signaling | 2.28E-05 |
| Mitotic roles of Polo-like kinase | 7.24E-05 |
| CXCR4 signaling | 1.23E-04 |
| Tec kinase signaling | 1.62E-04 |
|
| |
| Top upstream regulators a | |
| Colony stimulating factor 2 (granulocyte-macrophage), CSF2 | 9.17E-16 |
| Cyclin-dependent kinase inhibitor, CDKN1A | 2.55E-15 |
These data were obtained from Ingenuity’s Pathway Analysis program (see Methods)
Significance values shown indicate the range of P-values for each of the genes that were identified within each of the annotated functions listed
DISCUSSION
We report herein that GAS6 expressed by AML blasts is a marker of poor clinical outcomes in adults with CN-AML, independent of other established prognostic markers in this cytogenetic subset. Not only does GAS6 expression predict CR failure, albeit driven by older age, it has also a negative impact on DFS and OS in the studied cohort. The MVA revealed the negative prognostic impact of GAS6 was in patients whose leukemic blasts did not express TYRO3 and AXL. This suggests that GAS6 may contribute to a more aggressive disease through signaling mechanisms that are not dependent on AXL and TYRO3 expression within the AML cells. Perhaps it is the third GAS6 receptor, MERTK that together with GAS6 has a role within the TYRO3−/AXL− patient subgroup. Based on recently published data related to MERTK function in leukemia, MERTK, even when expressed at relatively low levels appears to contribute to a leukemic phenotype. 14 However, in the current study, too few numbers of patients prohibited a reliable GAS6/MERTK subgroup analyses.
Given that GAS6 expression has an adverse impact on CR achievement, mainly in older patients, and on DFS and OS in all patients, this warrants development of novel, less toxic and perhaps more personalized therapies targeting GAS6. We recently reported that blocking the engagement of GAS6 to the AXL receptor with soluble AXL-Fc chimeric protein inhibits downstream signal transduction, inducing differentiation and apoptosis in human AML cell lines and patient samples with activated AXL.54 Consistent with our work,54 a recent study by Ben-Batalla et al,55 showed that pharmacologic inhibition of GAS6/AXL signaling induces leukemic cell death. We did not find an impact of AXL-positive expression as a sole marker on outcomes in our study, whereas AXL expression above the median was associated with shorter OS in the Ben-Batalla and colleagues study.55 This may be explained in part by therapy differences and differences in patient cohort characteristics. Their study exclusively analyzed adult patients ≤60 years of age, whereas 73% of patients in our study were 60 years of age or older.
As our study measured GAS6 transcript levels, the relationship between GAS6 mRNA, protein and secretion in AML is not yet clear. However, there are several studies of various solid cancers that report GAS6 mRNA and protein are present within the tumor cells. For example, Buehler and coworkers recently reported that GAS6 mRNA and the translated protein are both elevated within ovarian cancer.56 Additionally, GAS6 mRNA and protein levels were found in 81% and 74% of glioblastoma multiforme tissue samples, suggesting a close 1:1 relationship between transcription and translation of GAS6.57 As for secretion, Ben-Batalla, and co-workers performed immunohistochemistry for GAS6 on five AML patients’ bone marrows and concluded that stromal cells are the primary source of secreted GAS6 ligand (their Figure 1 and Supplement Table 3)55
Interaction of the GAS6/TAM RTK signaling axis with the stromal microenvironment in solid tumors has been associated with poor progression-free survival,58,59 A similar mechanism may be active in chemotherapy-resistant AML patients that express GAS6. This suggests a potential for novel therapies targeting GAS6+ leukemic blasts that could also simultaneously inhibit negative effects of GAS6 in the microenvironment, ultimately improving patient survival. Promising studies using decoy receptors showed significant activity against the growth of lung carcinoma cells in a xenograft model,13 and the absence of toxicity in normal murine tissues or hematopoiesis is encouraging.60 Given our current findings, GAS6-targeted therapeutic agents could lead to higher CR rates, particularly, as our data indicate, benefiting older patients, and possibly prolonging survival of the GAS6+ subset of CN-AML patients. Furthermore, while our results await independent validation, development of GAS6 decoy receptors, in addition to selective small molecule TAM inhibitors, appears warranted.
Meanwhile, in the short term, one possibility for improving outcomes of GAS6+ patients is alluded to by the GAS6-associated gene expression signature we identified. Overexpression of CXCR4 and its ligand was detected in pretreatment samples from patients expressing GAS6. In separate reports, overexpression of this signaling axis associated with increased risk of relapse and shorter overall survival in AML.50,51 CXCR4 is expressed on normal hematopoietic stem cells and regulates stem cell homing and retention in the BM when CXCL12, produced by BM stroma, engages the receptor.61 The CXCR4 antagonist, Plerixafor, currently has FDA approval in combination with G-CSF as a stem cell mobilizing agent for patients with multiple myeloma and non-Hodgkin lymphoma who undergo autologous hematopoietic stem cell transplantation.62 Uy and co-workers63 recently reported their results of a phase I/II clinical trial that demonstrated antagonizing the CXCL12/CXCR4 axis with Plerixafor induces chemosensitization of relapsed or refractory AML blasts. Thus, although GAS6 decoy receptors are not yet available in the clinic for GAS6+ CN-AML therapy, potential alternative therapies such as CXCR4 antagonists should be considered for GAS6+ patients to sensitize them to chemotherapy.
Supplementary Material
Acknowledgements
The authors express their sincere gratitude to Donna Bucci and the Alliance Leukemia Tissue Bank for sample processing and storage services, to Lisa J Sterling and Christine Finks for data management and to the OSU NCI-designated Comprehensive Cancer Center’s shared resources (Microarray and Nucleic Acid). We also thank Dan Sargent for critical review of the manuscript. The authors also acknowledge the physicians, nurses, clinical support staff and, most of all, patients for their invaluable contributions to these studies.
This work was supported in part by the National Cancer Institute (grants CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658 and CA129657), the Coleman Leukemia Research Foundation, the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.), the Pelotonia Fellowship Program (A.-K.E.), and the Conquer Cancer Foundation (J.H.M.).
Footnotes
Author contributions S.P.W., M.A.C., G.M., C.D.B designed the study and analyzed the data. S.P.W., J.K., K. Maharry, K. Mrózek, M.A.C., G.M. and C.D.B wrote the manuscript, and all authors agreed to the final version. J.K., K. Maharry, D.N. and S.V. performed statistical analyses. S.P.W., K.H.M., S.W., H.B., J.M., A.-K.E., A.J.C. and I-K.P. generated, compiled and interpreted lab data. B.L.P., T.H.C., M.R.B., J.E.K., R.M.S., M.A.C., G.M., C.D.B. were involved directly or indirectly in care of patients and/or sample procurement.
Supplemental material is available at the journal of Leukemia website. (http://www.nature.com/leu)
Conflicts of Interest statement: The authors do not have any conflicts of interest, including financial, to disclose related to this work.
References
- 1.Masson K, Rönnstrand L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal. 2009;21:1717–1726. doi: 10.1016/j.cellsig.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Correll PH, Paulson RF, Wei X. Molecular regulation of receptor tyrosine kinases in hematopoietic malignancies. Gene. 2006;374:26–38. doi: 10.1016/j.gene.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Zheng R, Klang K, Gorin NC, Small D. Lack of KIT or FMS internal tandem duplications but co-expression with ligands in AML. Leuk Res. 2004;28:121–126. doi: 10.1016/s0145-2126(03)00184-x. [DOI] [PubMed] [Google Scholar]
- 4.Malaise M, Steinbach D, Corbacioglu S. Clinical implications of c-Kit mutations in acute myelogenous leukemia. Curr Hematol Malig Rep. 2009;4:77–82. doi: 10.1007/s11899-009-0011-8. [DOI] [PubMed] [Google Scholar]
- 5.Sritana N, Auewarakul CU. KIT and FLT3 receptor tyrosine kinase mutations in acute myeloid leukemia with favorable cytogenetics: two novel mutations and selective occurrence in leukemia subtypes and age groups. Exp Mol Pathol. 2008;85:227–231. doi: 10.1016/j.yexmp.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Nanri T, Matsuno N, Kawakita T, Suzushima H, Kawano F, Mitsuya H, et al. Mutations in the receptor tyrosine kinase pathway are associated with clinical outcome in patients with acute myeloblastic leukemia harboring t(8;21)(q22;q22) Leukemia. 2005;19:1361–1366. doi: 10.1038/sj.leu.2403803. [DOI] [PubMed] [Google Scholar]
- 7.Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 8.Fröhling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol. 2005;23:6285–6295. doi: 10.1200/JCO.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Hafizi S, Dahlbäck B. Signalling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev. 2006;17:295–304. doi: 10.1016/j.cytogfr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Verma A, Warner SL, Vankayalapati H, Bearss DJ, Sharma S. Targeting Axl and Mer kinases in cancer. Mol Cancer Ther. 2011;10:1763–1773. doi: 10.1158/1535-7163.MCT-11-0116. [DOI] [PubMed] [Google Scholar]
- 11.Brandão L, Migdall-Wilson J, Eisenman K, Graham DK. TAM receptors in leukemia: expression, signaling, and therapeutic implications. Crit Rev Oncog. 2011;16:47–63. doi: 10.1615/critrevoncog.v16.i1-2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suárez RM, Chevot F, Cavagnino A, Saettel N, Radvanyi F, Piguel S, et al. Inhibitors of the TAM subfamily of tyrosine kinases: synthesis and biological evaluation. Eur J Med Chem. 2013;61:2–25. doi: 10.1016/j.ejmech.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Ye X, Li Y, Stawicki S, Couto S, Eastham-Anderson J, Kallop D, et al. An anti-Axl monoclonal antibody attenuates xenograft tumor growth and enhances the effect of multiple anticancer therapies. Oncogene. 2010;29:5254–5264. doi: 10.1038/onc.2010.268. [DOI] [PubMed] [Google Scholar]
- 14.Lee-Sherick AB, Eisenman KM, Sather S, McGranahan A, Armistead PM, McGary CS, et al. Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia. Oncogene. 2013 doi: 10.1038/onc.2013.40. epub ahead of print March 11 2013; doi: 10.1038/onc.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosier PS, Hall LR, Vitas MR, Lewis PM, Crosier KE. Identification of a novel receptor tyrosine kinase expressed in acute myeloid leukemic blasts. Leuk Lymphoma. 1995;18:443–449. doi: 10.3109/10428199509059643. [DOI] [PubMed] [Google Scholar]
- 16.Rochlitz C, Lohri A, Bacchi M, Schmidt M, Nagel S, Fopp M, et al. Axl expression is associated with adverse prognosis and with expression of Bcl-2 and CD34 in de novo acute myeloid leukemia (AML): results from a multicenter trial of the Swiss Group for Clinical Cancer Research (SAKK) Leukemia. 1999;13:1352–1358. doi: 10.1038/sj.leu.2401484. [DOI] [PubMed] [Google Scholar]
- 17.Manfioletti G, Brancolini C, Avanzi G, Schneider C. The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K-dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol Cell Biol. 1993;13:4976–4985. doi: 10.1128/mcb.13.8.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirks W, Rome D, Ringel F, Jäger K, MacLeod RA, Drexler HG. Expression of the growth arrest-specific gene 6 (GAS6) in leukemia and lymphoma cell lines. Leuk Res. 1999;23:643–651. doi: 10.1016/s0145-2126(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 19.Mayer RJ, Davis RB, Schiffer CA, Berg DT, Powell BL, Schulman P, et al. Intensive postremission chemotherapy in adults with acute myeloid leukemia. N Engl J Med. 1994;331:896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 20.Stone RM, Berg DT, George SL, Dodge RK, Paciucci PA, Schulman P, et al. Granulocyte-macrophage colony-stimulating factor after initial chemotherapy for elderly patients with primary acute myelogenous leukemia. N Engl J Med. 1995;332:1671–1677. doi: 10.1056/NEJM199506223322503. [DOI] [PubMed] [Google Scholar]
- 21.Lee EJ, George SL, Caligiuri M, Szatrowski TP, Powell BL, Lemke S, et al. Parallel phase I studies of daunorubicin given with cytarabine and etoposide with or without the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age or older with acute myeloid leukemia: results of Cancer and Leukemia Group B study 9420. J Clin Oncol. 1999;17:2831–2839. doi: 10.1200/JCO.1999.17.9.2831. [DOI] [PubMed] [Google Scholar]
- 22.Baer MR, George SL, Caligiuri MA, Sanford BL, Bothun SM, Mrózek K, et al. Low-dose interleukin-2 immunotherapy does not improve outcome of patients age 60 years and older with acute myeloid leukemia in first complete remission: Cancer and Leukemia Group B study 9720. J Clin Oncol. 2008;26:4934–4939. doi: 10.1200/JCO.2008.17.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baer MR, George SL, Sanford BL, Mrózek K, Kolitz JE, Moore JO, et al. Escalation of daunorubicin and addition of etoposide in the ADE regimen in acute myeloid leukemia patients aged 60 years and older: Cancer and Leukemia Group B study 9720. Leukemia. 2011;25:800–807. doi: 10.1038/leu.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcucci G, Moser B, Blum W, Stock W, Wetzler M, Kolitz JE, et al. A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a proapoptotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients >60 years old [abstract] J Clin Oncol. 2007;25(suppl):360s. (abstract 7012) [Google Scholar]
- 25.Kolitz JE, George SL, Marcucci G, Vij R, Powell BL, Allen SL, et al. P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients under age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–1421. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mrózek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 27.Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 28.Whitman SP, Ruppert AS, Radmacher MD, Mrózek K, Paschka P, Langer C, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caligiuri MA, Strout MP, Schichman SA, Mrózek K, Arthur DC, Herzig GP, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 30.Whitman SP, Ruppert AS, Marcucci G, Mrózek K, Paschka P, Langer C, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: a Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrózek K, Maharry K, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzeler KH, Maharry K, Radmacher MD, Mrózek K, Margeson D, Becker H, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcucci G, Maharry K, Wu Y-Z, Radmacher MD, Mrózek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrózek K, et al. Age related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwind S, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Holland KB, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116:5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2009;27:3198–3204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, et al. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8:446. doi: 10.1186/1471-2105-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. Springer; New York, NY: 2003. [Google Scholar]
- 44.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 45.Cox DR. Regression models and life-tables. J Royal Stat Soc Series B (Methodological) 1972;34:187–220. [Google Scholar]
- 46.Jiang L, Yu G, Meng W, Wang Z, Meng F, Ma W. Overexpression of amyloid precursor protein in acute myeloid leukemia enhances extramedullary infiltration by MMP-2. Tumor Biol. 2013;34:629–636. doi: 10.1007/s13277-012-0589-7. [DOI] [PubMed] [Google Scholar]
- 47.Cristóbal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 48.Alachkar H, Maharry K, Santhanam R, Neviani P, Volinia S, Kohlschmidt J, et al. SPARC contributes to leukemia growth and aggressive disease in acute myeloid leukemia (AML) Blood. 2012;120 (abstract 773) [Google Scholar]
- 49.Attar EC, Johnson JL, Amrein PC, Lozanski G, Wadleigh M, DeAngelo DJ, et al. Bortezomib added to daunorubicin and cytarabine during induction therapy and to intermediate-dose cytarabine for consolidation in patients with previously untreated acute myeloid leukemia age 60 to 75 years: CALGB (Alliance) study 10502. J Clin Oncol. 2013;31:923–929. doi: 10.1200/JCO.2012.45.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn JY, Seo K, Weinberg OK, Arber DA. The prognostic value of CXCR4 in acute myeloid leukemia. Appl Immunohistochem Mol Morphol. 2013;21:79–84. doi: 10.1097/PAI.0b013e3182606f4d. [DOI] [PubMed] [Google Scholar]
- 51.Konoplev S, Rassidakis GZ, Estey E, Kantarjian H, Liakou CI, Huang X, et al. Overexpression of CXCR4 predicts adverse overall and event-free survival in patients with unmutated FLT3 acute myeloid leukemia with normal karyotype. Cancer. 2007;109:1152–1156. doi: 10.1002/cncr.22510. [DOI] [PubMed] [Google Scholar]
- 52.Deschler B, de Witte T, Mertelsmann R, Lübbert M. Treatment decision-making for older patients with high-risk myelodysplastic syndrome or acute myeloid leukemia: problems and approaches. Haematologica. 2006;91:1513–1522. [PubMed] [Google Scholar]
- 53.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 54.Park IK, Mishra A, Chandler J, Whitman SP, Marcucci G, Caligiuri MA, et al. Inhibition of the receptor tyrosine kinase Axl impedes activation of the FLT3 internal tandem duplication in human acute myeloid leukemia: implications for Axl as a potential therapeutic target. Blood. 2013;121:2064–2073. doi: 10.1182/blood-2012-07-444018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Batalla I, Schultze A, Wroblewski M, Erdmann R, Heuser M, Waizenegger JS, et al. Axl, a prognostic and therapeutic target in acute myeloid leukemia mediates paracrine cross-talk of leukemia cells with bone marrow stroma. Blood. 2013;122:2443–2452. doi: 10.1182/blood-2013-03-491431. [DOI] [PubMed] [Google Scholar]
- 56.Buehler M, Tse B, Leboucq A, Jacob F, Caduff R, Fink D, et al. Meta-analysis of microarray data identifies GAS6 expression as an independent predictor of poor survival in ovarian cancer. Biomed.Res. Int. doi: 10.1155/2013/238284. Epub 2013 Jun 27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutterer M, Knyazev P, Abate A, Reschke M, Maier H, Stefanova N, Knyazeva T, et al. Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:130–138. doi: 10.1158/1078-0432.CCR-07-0862. [DOI] [PubMed] [Google Scholar]
- 58.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt T, Ben-Batalla I, Schultze A, Loges S. Macrophage-tumor crosstalk: role of TAMR tyrosine kinase receptors and of their ligands. Cell Mol Life Sci. 2012;69:1391–1414. doi: 10.1007/s00018-011-0863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rankin EB, Fuh KC, Taylor TE, Krieg AJ, Musser M, Yuan J, et al. AXL is an essential factor and therapeutic target for metastatic ovarian cancer. Cancer Res. 2010;70:7570–7579. doi: 10.1158/0008-5472.CAN-10-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hattermann K, Mentlein R. An infernal trio: the chemokine CXCL12 and its receptors CXCR4 and CXCR7 in tumor biology. Ann Anat. 2013;195:103–110. doi: 10.1016/j.aanat.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Micallef IN, Stiff PJ, Stadtmauer EA, Bolwell BJ, Nademanee AP, Maziarz RT, et al. Safety and efficacy of upfront plerixafor + G-CSF vs. placebo + G-CSF for mobilization of CD34+ hematopoietic progenitor cells in patients ≥60 and <60 years of age with non-Hodgkin’s lymphoma or multiple myeloma. Am J Hematol. 2013 doi: 10.1002/ajh.23561. epub ahead of print August 1 2013; doi: 10.1002/ajh.23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119:3917–3924. doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.