Abstract
Transient leukemia (TL) is evident in 5–10% of all neonates with Down syndrome (DS) and associated with N-terminal truncating GATA1-mutations (GATA1s). Here we report that TL cell clones generate abundant eosinophils in a substantial fraction of patients. Sorted eosinophils from patients with TL and eosinophilia carried the same GATA1s-mutation as sorted TL-blasts, consistent with their clonal origin. TL-blasts exhibited a genetic program characteristic of eosinophils and differentiated along the eosinophil lineage in vitro. Similarly, ectopic expression of Gata1s, but not Gata1, in wild-type CD34+-hematopoietic stem and progenitor cells induced hyperproliferation of eosinophil promyelocytes in vitro. While GATA1s retained the function of GATA1 to induce eosinophil genes by occupying their promoter regions, GATA1s was impaired in its ability to repress oncogenic MYC and the pro-proliferative E2F transcription network. ChIP-seq indicated reduced GATA1s occupancy at the MYC promoter. Knockdown of MYC, or the obligate E2F-cooperation partner DP1, rescued the GATA1s-induced hyperproliferative phenotype. In agreement, terminal eosinophil maturation was blocked in Gata1Δe2 knockin mice, exclusively expressing Gata1s, leading to accumulation of eosinophil precursors in blood and bone marrow. These data suggest a direct relationship between the N-terminal truncating mutations of GATA1 and clonal eosinophilia in DS patients.
Keywords: Down syndrome, eosinophilia, GATA1s, MYC, E2F
Introduction
Children with Down syndrome (DS) have an increased risk of developing acute leukemia (1;2). Approximately 10% of DS neonates present with transient leukemia (TL), which is characterized by the accumulation of immature megakaryoblasts. TL resolves spontaneously in the majority of cases (3). However, 20 to 30% of TL-patients progress to acute megakaryoblastic leukemia (AMKL) (3–7).
Acquired mutations in exon 2/3 of the hematopoietic transcription factor GATA1 are consistently present in DS-AMKL and TL (8). GATA1 is essential for the regulation of differentiation, proliferation and apoptosis during erythropoiesis and megakaryopoiesis (9–12). The exon 2/3 mutations in GATA1 that are associated with DS-AMKL and TL lead to the expression of a shorter isoform (GATA1s), which lacks the N-terminal domain (8). We previously showed that, compared to full-length GATA1, GATA1s fails to repress the E2F transcription network due to impaired interaction with E2F proteins during fetal megakaryocyte development (13). However, the molecular basis of impaired transcriptional repression of transcription factors, including Gata2, Ikaros, Sfpi1 (encoding PU.1), Myb and Myc, remained elusive (14–16).
GATA1 is essential not only for the formation of erythroid and megakaryocytic cells, but also for proper development of eosinophils, basophils and mast cells (17–21). Normal development of eosinophils is instructed by an interplay of the transcription factors GATA1, GATA2, CEBPα and PU.1 (22). Promoter regions of numerous eosinophil-specific genes encoding eosinophil granule proteins (PRG2 [eosinophil granule major basic protein], ECP [eosinophil cationic protein; RNASE3], EDN [eosinophil-derived neurotoxin; RNASE2], EPX [eosinophil peroxidase], CLC [Charcot-Leyden crystal protein]) and surface receptors (IL-5Rα [interleukin-5 receptor alpha] and CCR3 [chemokine (C-C motif) receptor 3]) include known or putative GATA1 binding sites (22;23). Consistent with these findings, enforced expression of Gata1 in committed mouse myelomonocytic progenitors converts them into eosinophils, erythrocytes and basophil-like cells (24). Similarly, enforced expression of GATA1 in human primary myeloid progenitors switches myelomonocytic to an eosinophil fate (18). Therefore, GATA1 plays an instructive and essential role in eosinophil development, which was proposed to be independent of its N-terminal domain (18). TL and AMKL blasts contain basophil-like cytoplasmic granules (25) and differentiate along the megakaryocytic lineage, but also along the basophil lineage (26;27).
Here we report differentiation of TL-blasts to the eosinophil lineage and show that eosinophils are part of the TL cell clone. We provide a molecular link between GATA1s expression and activation of eosinophil genes in TL-blasts, thereby accounting for the accompanying eosinophilia.
Materials and Methods
Cell lines and patient samples
Human cell lines were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Culturing and maintenance were performed according to the supplier's instructions. Patient samples were provided by the AML-'Berlin-Frankfurt-Münster' Study Group (AML-BFM-SG, Hannover, Germany) and ‘The Hospital for Sick Children’, Toronto. TL-blasts for in vitro studies were obtained from peripheral blood of 4 TL-patients by ficoll-paque density centrifugation followed by sorting for CD34+/CD41+/low blast population. Human CD34+-fetal liver cells were purchased from Novogenic Laboratories, LLC (LA, USA). All investigations were performed in accordance to the Declaration of Helsinki and informed consent was obtained according to local laws and regulations.
Cell culture and lentiviral infection
TL-blasts were cultured as described (13;28). Differentiation was induced by myeloid differentiation medium [RPMI, 20% FCS, 1% P/S, SCF (5ng/ml), GM-CSF (10ng/ml), G-SCF (10ng/ml), IL-3 (5ng/ml)]. CD34+-cells were cultured, lentivirally transduced and sorted on a FACSAria (BD, Heidelberg, Germany) as previously described (29).
Viral vectors and lentivirus production
cDNAs of either full length or short Gata1 were cloned into a modified LeGO-iG vector (30), where the mU6 was replaced by the hU6 promoter. shRNA against human GATA1, obtained from Open Biosystems (Schwerte, Germany) clone ID: TRCN0000019223), was cloned downstream of the hU6 promoter. shRNA sequences for DP1 and MYC are available upon request. For production of lentiviral supernatant, standard protocols were followed as previously described (29;31).
High-throughput chip cytometry
Single colonies were picked from methylcellulose cultures, washed and resuspended in PBS before applying to cell-adhesive microfluidic-chips. Chip cytometry (http://www.chipcytometry.com) was performed on a Zeiss Axio Imager M2 automated microscope using CellHopperX software as described (32). Raw data generalized Box-Cox transformation and heat map production were performed with R package versions 1.8.0. for BioConductor (32).
ChIP-Seq
Gene coding for biotinylation enzyme BirA has been introduced into human megakaryocytic cell line CMK, in which only N-truncated GATA1s is endogenously expressed, by stable transfection via electroporation (Amaxa Cell Line Nucleofector Kit C, Promega). Positive clones were subsequently transduced by retroviral vector encoding BIO-tagged GATA1 or GATA1s and cultured in the presence of 2 µg/ml puromycin and 4 µg/ml G418. Chromatin immunoprecipitation (ChIP) was performed using streptavidin-bound Dynabeads (Dynabeads MyOne Streptavidin T1; Invitrogen) as previously described (33;34). ChIP-seq analysis was performed using the Illumina™ HiSeq2000. 10–20 ng of ChIP DNA was processed for library generation using the ChIP-seq Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Raw ChIP-seq data were processed using the Illumina software pipeline and ChIP-seq reads aligned to exactly one location in the reference human genome (UCSC, hg18) as previously described (34).
In vitro differentiation from mouse ESCs
Eosinophil in vitro differentiation from knockin murine embryonic stem cells (ESCs) expressing the BirA enzyme (BirA) together with a biotinylated form of Gata1 (bioGata1) or Gata1s (bioGata1s) from its endogenous locus (35) was adapted from a previously described protocol (36). Briefly, 2.5 × 104 ESCs were seeded into a 10 cm dish containing an OP9 cell layer and differentiated in α-MEM supplemented with 20% FBS (PAA, Yeovil Somerset; UK). On day 5, ESC colonies were dissociated with 0.25% Trypsin, cells were incubated in culture medium for 45 minutes at 37°C to let the OP9 cells attach to the dish and 106 suspension cells were transferred onto a fresh OP9 layer in a 10 cm dish. From day 7, IL3 (10 ng/mL) and IL5 (10 ng/mL) were added to the culture. On day 10 and 15, the cells in suspension and slightly attached cells were harvested by vigorous pipetting and transferred onto a fresh OP9 layer. On day 15, cells in suspension were transferred to a new dish without OP9 cells. On day 20, cells were analyzed by FACS.
Mice
All studies involving mice were approved by the Hannover Medical School Institute for laboratory animal science and local authorities and performed in accordance with the relevant protocol. Blood counts were measured on the scil Vet abc blood counter (Horiba Medical, Europe). Flow cytometry of total bone marrow and peripheral blood was performed after red blood cell lysis (Pharm Lyse reagent; Becton Dickinson).
Statistical analysis
Statistical evaluation between the two groups was carried out using Student’s t-test and for more than two groups by 1-way ANOVA with Duncan’s or Bonferroni’s post-hoc analysis. The level of significance was set at P<0.05. Calculations were performed using GraphPad Prism 5 (STATCON, Witzenhausen, Germany).
Results
A substantial proportion of TL patients have increased eosinophils in the peripheral blood carrying the mutated GATA1s allele
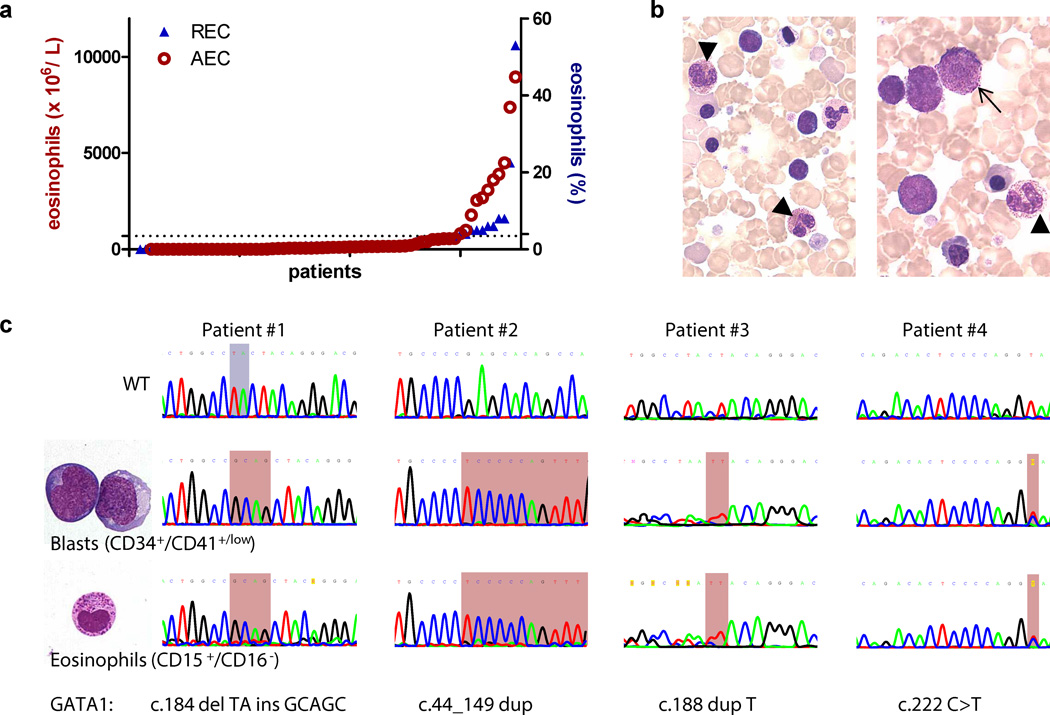
Between June 1998 and August 2011, 70 DS neonates were diagnosed with TL by morphological examination of blood smears in the central AML reference laboratory of the AML-BFM study group. The median age at diagnosis was 6.5 days (range: 2 – 56d) and GATA1s mutation was confirmed in 25/27 tested patients (93%). Interestingly, 11 patients (16%) presented with an elevated (more than 600 ×106/L) absolute eosinophil count (AEC; range: 820–8960 ×106/L, mean: 3655 ×106/L) or elevated (equal or more than 4%) relative eosinophil count (REC; range: 4–53%, mean: 11.5%; median age: 31 days; range: 1–65d; Figure 1a). Morphologically, most eosinophils in these patients appeared immature and contained basophilic granules (Figure 1b). Leukemic blasts with eosinophilic and basophilic granules were also seen (Figure 1b).
Figure 1. Abnormal eosinophils in a fraction of TL patients.
(a) Diagram showing the absolute eosinophil count (AEC; normal: less than 600 ×106/L) and relative eosinophil count (REC; normal: less than 4%) in n=70 DS neonates with TL as assessed by morphological examination of blood smears. (b) TL eosinophils (arrowhead) and a TL-blast (arrow) with eosinophilic granulation (MGG-staining; 1000× original magnification). (c) Microscopic images (left panel; MGG-staining; 1000× original magnification) and sequencing (right panel) of sorted CD15+/CD16+ TL-eosinophils and CD34+/CD41+/low blasts (n=4). The mutated regions are highlighted in red (duplication, insertion and point mutations) and blue (deletion). Patient #1 and #2 are male (GATA1 hemizygous), while #3 and #4 are female (GATA1 heterozygous).
All photomicrographs were taken using an Olympus BX41 Microscope.
GATA1 is involved in eosinophil differentiation. Since TL and DS-AMKL blasts have elevated levels of GATA1s (37), we sought to investigate whether immature eosinophils were intrinsic to the leukemic clone or reflected a secondary reaction to the TL. We sorted populations of blasts (CD34+/CD41+/low) and eosinophils (CD15+/CD52+/CD16−) from four TL patients with eosinophilia (Supplementary Figure 1a–b). Efficiency of sorting was confirmed by cytospins and qRT-PCR (Figure 1c and Supplementary Figure 1c). Morphologically sorted CD15+/CD52+/CD16− cells constituted a mixture of eosinophil myelocytes and metamyelocytes. Basophilic granules are characteristic for eosinophil precursors and disappear during normal eosinophil differentiation (Supplementary Figure 2).
Sequencing of the GATA1 locus from the sorted populations revealed the same mutation in the eosinophilic cells and megakaryoblasts (Figure 1c). Thus, the eosinophilic cells in TL can originate from the preleukemic clone.
TL-blasts can differentiate into eosinophils in vitro
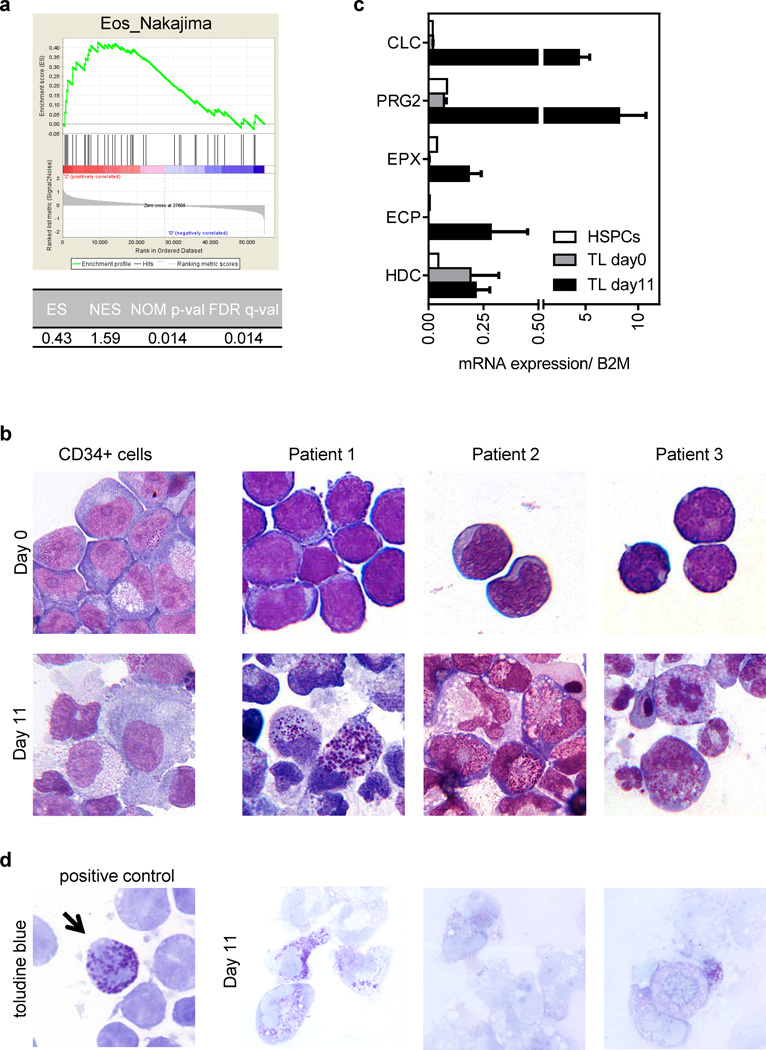
Next we performed gene set enrichment analysis (GSEA (38)) using previously published gene-expression profiles of sorted megakaryoblasts from TL patients without documented eosinophilia and non-DS-AMKL patients (13). This showed the enrichment of a set of specific eosinophil genes (39) in TL-blasts as compared to non-DS-AMKL-blasts (Figure 2a). Thus, TL-blasts express a gene program characteristic of differentiated eosinophils.
Figure 2. TL-blasts express an eosinophil program and can differentiate into eosinophils.
(a) GSEA of eosinophil genes (39) in sorted TL-blasts (n=6) compared to non-DS AMKL (n=5). (b) Microscopic images (MGG-staining, 1000× original magnification) of CD34+ cells (left) and TL-blasts (right; n=3) of DS patients on day 0 and on day 11 of culture in myeloid (SCF/ IL3/ G-CSF/ GM-CSF) differentiation medium. (c) Expression of eosinophil and basophil genes in undifferentiated (day 0) and differentiated TL-blasts (day 11) as well as HSPCs measured by qRT-PCR (n=3; mean±SD). B2M expression was used as an endogenous control. (d) Microscopic images (toluidine blue staining, 1000× original magnification) of TL-blasts of one representative patient on day 11 of culture in myeloid differentiation medium. Normal blood basophils were used as a positive staining control (left).
Photomicrographs were taken with a Keyence Biorevo BZ-9000 microscope and processed with the BZ-II viewer software.
To directly test whether TL-blasts can fulfill this intrinsic eosinophil program during differentiation, we cultured sorted TL-blasts from three patients (for GATA1s status see Supplementary Table 1) in medium containing G-CSF, GM-CSF, IL3 and SCF (40). Under these culture conditions, normal CD34+-hematopoietic stem and progenitor cells (HSPCs) differentiate into monocytes and granulocytes (Figure 2b and 3c–d) (41). Remarkably, despite the lack of documented eosinophilia in these patients in vivo, TL-blasts of all three patients differentiated into cells with densely granulated cytoplasm, resembling eosinophil precursors (Figure 2b). Eosinophil genes (ECP, EPX, CLC and PRG2) were significantly activated (Figure 2c). Expression of the basophil lineage gene HDC was only modestly affected (Figure 2c). The majority of cells were negative for toluidine blue, which heterochromatically stains heparin and histamine of mature basophils and mast cells (Figure 2d, see ‘positive control’). Thus, we propose that TL-blasts cannot only differentiate along the megakaryocytic lineage (26;27), but are also intrinsically fated to differentiate along the eosinophil lineage.
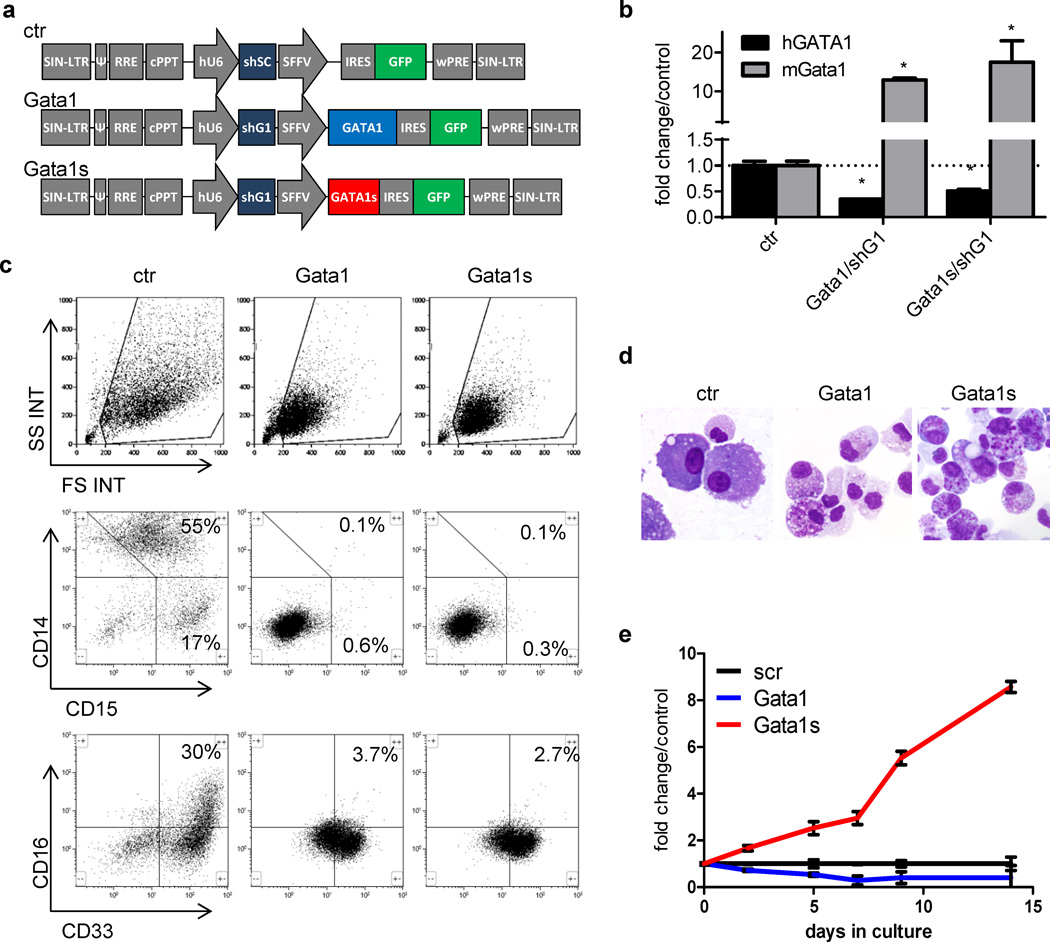
Figure 3. Overexpression of GATA1s perturbs normal myeloid differentiation of human fetal CD34+-HSPCs.
(a) Schematic illustration of the LeGO-iG-shSC (ctr), LeGO-iG-Gata1/shG1 (Gata1) and LeGO-iG-Gata1s/shG1 (Gata1s) vectors (30). (b) qRT-PCR of mouse Gata1 and human GATA1 using specific primers in LeGO-iG-shSC (ctr)-, LeGO-iG-Gata1/shG1 (Gata1/shG1)- and LeGO-iG-Gata1s/shG1 (Gata1s/shG1)-transduced fetal CD34+-HSPCs (mean±SD of replicates; *PANOVA<0.05 compared to ctr). (c) FACS analysis of Gata1-, Gata1s- and empty vector control (ctr)-transduced HSPCs grown for 14 days in myeloid differentiation medium. Representative dot plots of n=4 independent experiments are presented. (d) MGG staining of empty vector, Gata1- and Gata1s-transduced cells on day 14 of myeloid differentiation culture (1000× original magnification). Photomicrographs were taken with a Keyence Biorevo BZ-9000 microscope and processed with the BZ-II viewer software. (e) Number of Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal HSPCs grown in myeloid differentiation medium relative to the control. Data from n=4 independent experiments are presented as mean±SD. **PANOVA<0.01 compared to ctr.
Overexpression of Gata1s in human fetal HSPCs induces eosinophilic hyperproliferation
To investigate the role of GATA1-isoforms in TL-associated eosinophilia, we examined myeloid differentiation of human fetal CD34+-HSPCs ectopically expressing GATA1 or GATA1s. We used a modified lentiviral LeGO-iG vector (30) to express simultaneously murine Gata1s, or full length Gata1, and a shRNA directed to endogenous human GATA1 (LeGO-iG-Gata1/shG1 and LeGO-iG-Gata1s/shG1; Figure 3a). We achieved knock-down of endogenous GATA1 mRNA to 30% of the original level and stable expression of Gata1 and Gata1s mRNA and protein, respectively (Figure 3b and Supplementary Figure 3). This approach permitted investigation of the net effect of Gata1s in cells with a low level of endogenous GATA1. As a control, we used the modified LeGO-iG vector to express non-silencing shRNA (LeGO-iG-shSC; hereafter referred to as ‘empty vector control’).
Transduced cells were cultured in the same myeloid differentiation medium (containing G-CSF, GM-CSF, IL3 and SCF), which supports differentiation of normal CD34+-HSPCs to CD14+ monocytes and CD15+/CD16+ neutrophils (Figure 3c–d) (41), but supported eosinophil differentiation of GATA1s-mutated TL-blasts (Figure 2b). Overexpression of Gata1 or Gata1s induced formation of CD33mid/+ cells (Figure 3c) with highly granulated cytoplasm (Figure 3d), resembling eosinophil precursors (compare to Supplementary Figure 2). Some of those contained abnormal coarse basophilic granules, reminiscent of pro-eosinophilic granules found in AML FAB M4eo (Supplementary Figure 4). Expression of Gata1s blocked differentiation to CD14+ monocytes and CD15+/CD16+ neutrophils to the same extent as full length Gata1 (42–44) (Figure 3c). However, even though Gata1- and Gata1s-transduced cells displayed a similar immunophenotype and morphology (Figure 3c–d), only Gata1s-transduced eosinophil precursors were hyperproliferative in liquid culture in an IL3-dependent manner, maintaining viability for more than 45 days (Figure 3e and Supplementary Figure 5). In contrast, the proliferation rate of Gata1-transduced cells was reduced (Figure 3e). Of note, the presence of an extra copy of chromosome 21 in trisomy 21 fetal HSPCs did not have a significant additional impact on the observed Gata1s phenotype (Supplementary Figure 6).
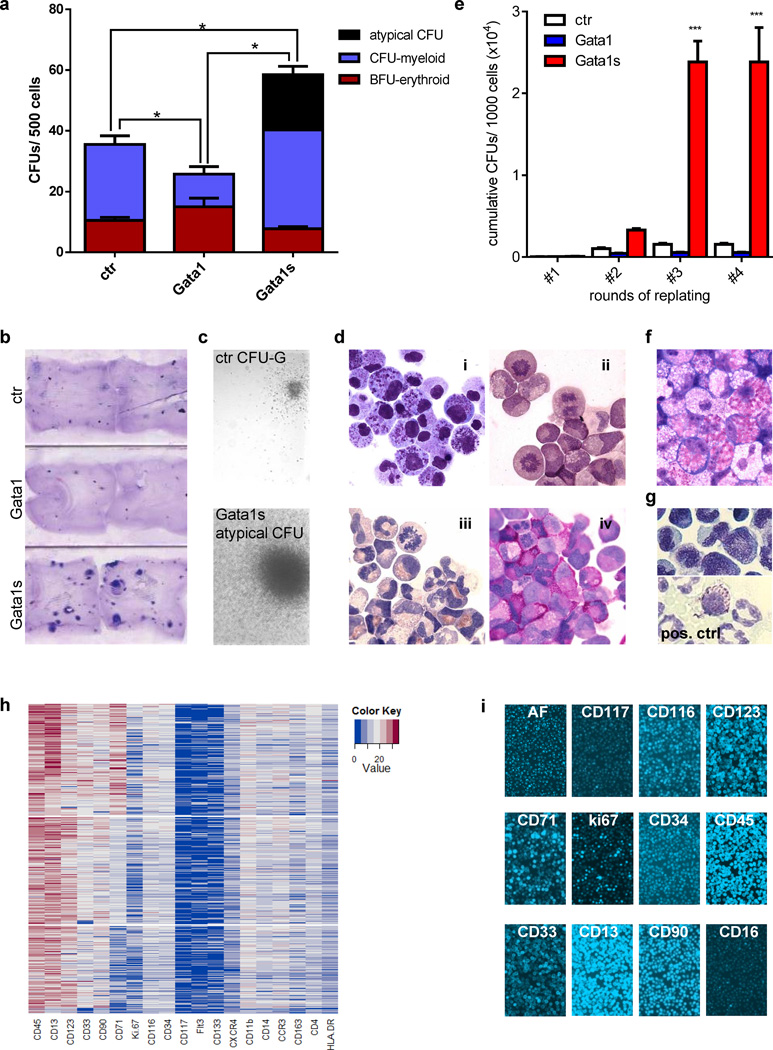
Gata1s induces formation of atypical eosinophil colonies
In the methylcellulose- and collagen-based myeloid colony-forming unit (CFU) assay, Gata1s increased the colony-forming capacity of transduced HSPCs (Figure 4a–b). Importantly, Gata1s-, but not Gata1- or empty vector-transduced cells, gave rise to atypical large myeloid colonies (Figure 4a and c). They were composed of eosinophil promyelocytes (Figure 4di and Supplementary Figure 7; compare to Supplementary Figure 2) with abnormal coarse pro-eosinophilic granules (compare to AML FAB M4eo in Supplementary Figure 4a). The cells were similar to those seen in liquid culture of transduced HSPCs (Figure 4di and 3d) or TL-blasts (Figure 2b). These atypical eosinophil colonies could be replated without changing morphology (Figure 4e–f), such that after the fourth round of replating, Gata1s-transduced cells had a 15-fold higher cumulative number of CFUs than empty vector-transduced control cells. Histochemical staining of cells from the atypical colonies were negative for toluidine blue (Figure 4g), alpha-naphtylacetate-esterase and weakly positive for myeloperoxidase (Figure 4di–iii) excluding basophil, monocytic or neutrophil lineage commitment, respectively. Many cells stained strongly positive for polysaccharides (PAS-staining; Figure 4div), consistent with their high proliferative rates as also indicated by the large number of metaphases (Figure 4dii–iv).
Figure 4. Gata1s induces the proliferation of eosinophil precursors.
(a) Number of CFUs per 500 Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal CD34+-HSPCs. Replicates from n=4 independent experiments are presented as mean±SD. (*PANOVA<0.05 of the total CFU count compared to ctr). (b) Photograph (1× original magnification) of collagen-based colony-forming assays of Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal CD34+-HSPCs. (c) Microscopic images (phase contrast, 50× original magnification) of a control CFU-G (top) and atypical Gata1s-CFUs (bottom). (d) Microscopic images of cells from an individual picked atypical Gata1s-CFU (1000× original magnification). (i) MGG-staining; (ii) esterase staining (EST); (iii) myeloperoxidase staining (MPO); (iv) periodic acid-Shiff staining (PAS). (e) Replating capacity of Gata1s-CFUs calculated as the cumulative CFU count per each round of replating compared to the Gata1- and empty vector-transduced HSPCs (***PANOVA<0.001 compared to ctr). (f) Morphology (MGG-staining; 1000× original magnification) of Gata1s-transduced HSPCs after the third round of replating. (g) Toluidine blue staining of Gata1s-transduced cells from a picked CFU (1000× original magnification). Peripheral blood basophils (bottom) were used as a positive control. (h–i) Expression of surface and intracellular markers of Gata1s-transduced cells from an individual picked Gata1s-CFU measured by chip cytometry. A clustered heatmap of transformed fluorescence intensity data (h) and raw image data (j) are presented.
Photomicrographs were taken with an Olympus BX41 Microscope and processed with AnalySIS software (Olympus Corporation).
Chip cytometry and gene expression profiling confirm eosinophil lineage affiliation of hyperproliferative Gata1s-transduced cells
To confirm that the Gata1s-transduced cells seen in the liquid cultures and CFU assays were indeed eosinophil precursors, we performed chip cytometry and global gene expression profiling.
High throughput chip cytometry (32), which allows more than 50 surface and intracellular antigens to be measured in small number of cells, was used to comprehensively analyze the immunophenotype of individual Gata1s-CFUs (Figure 4h–i). The cells exhibited bright autofluorescence, characteristic for granulated eosinophils, intermediate expression of the stem cell markers CD34 and CD90, and high levels of expression of the pan-leukocyte antigen CD45, early myeloid antigen CD13 and IL-3 receptor (CD123) (Figure 4h). Low CD117 expression excludes a mast cell origin of the hyperproliferative Gata1s-transduced cells (Figure 4h–i). Expression of mature monocytic (CD14) and neutrophilic markers (CD16) was low, consistent with the FACS data reflecting differentiation block imposed by enforced Gata1s expression.
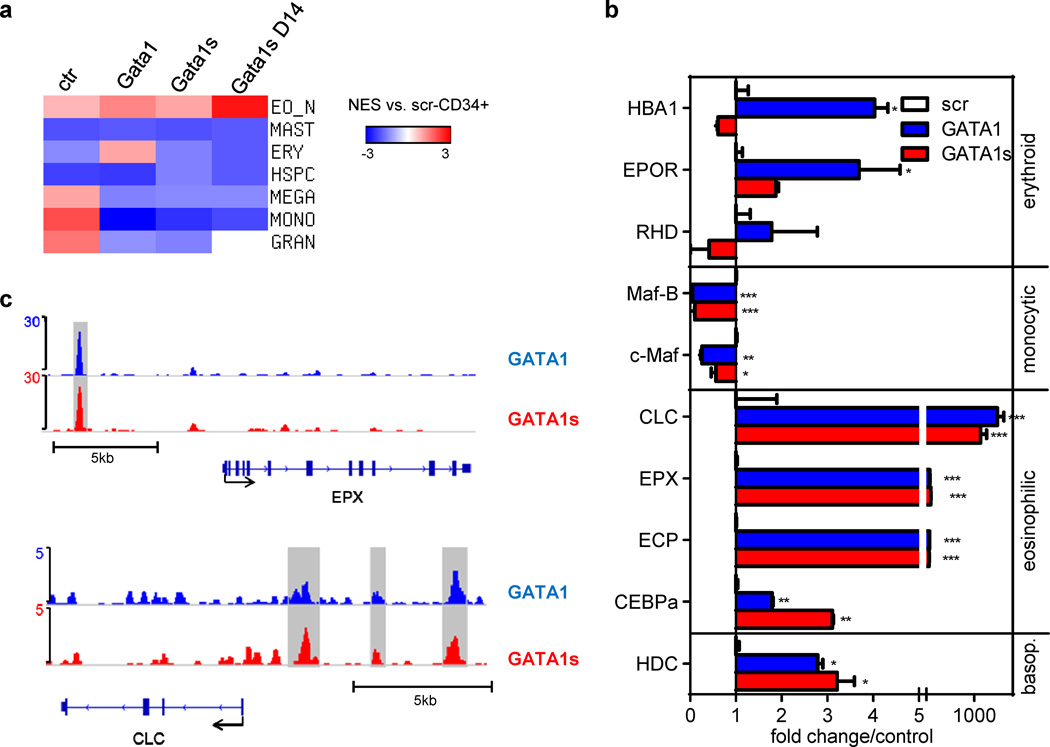
To determine the genetic program of Gata1s-, Gata1- and empty vector-transduced HSPCs in an unbiased manner, we performed global gene expression profiling followed by GSEA analysis (38). We used sets of genes, that are highly expressed (more than 2-fold, PANOVA<0.05) in human monocytes, granulocytes, megakaryocytes, erythroid cells, CD34+-HSPCs (S.E., J.H.K., unpublished data) compared to the other blood lineages. We also included sets of genes specific for eosinophils, as compared to basophils and mast cells (39;45). Using this approach, we first validated a significant (NOM p-val <0.05; FDR q-val <0.25) upregulation of monocytic and granulocytic genes and downregulation of erythroid and HSPC genes in the empty vector-transduced cells after four days of culture (Figure 5a and Supplementary Figure 8).
Figure 5. Expression of lineage markers and genes in Gata1-/ Gata1s-transduced HSPCs.
(a) Heatmap of the normalized enrichment score (NES) obtained by GSEA of Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal HSPCs grown 4 days (and 14 days) in myeloid differentiation medium compared to empty vector control (ctr)-transduced cells on day 0 (n=2). Gene sets specific (more than 2-fold, PANOVA<0.05) for human monocytes, granulocytes, megakaryocytes, erythroid cells and CD34+-HSPCs (Emmrich and Klusmann, unpublished data), and specific for eosinophils and mast cells (39) were used. The corresponding enrichment plots and statistics are presented in Supplementary Figures 8–11. (b) The expression of key erythroid, monocytic, eosinophil and basophil genes was validated by qRT-PCR in replicates from another independent experiment (mean±SD of replicates; *PANOVA<0.05, **PANOVA<0.01, ***PANOVA<0.001 compared to ctr). (c) ChIP-seq density plots of Gata1 (blue) and Gata1s (red) enrichment on promoter regions of eosinophil genes EPX and CLC.
As expected, ectopic Gata1-expression led to increased expression of eosinophil and erythroid genes, while monocytic genes were reduced (Figure 5a and Supplementary Figure 9). In contrast, Gata1s-expression only caused an increase in the eosinophil gene set, reaching significance after 14 days (NES: 2.73; P<0.001; q<0.001) (Figure 5a and Supplementary Figures 10 and 11), while expression of monocytic, granulocytic, HSPC, erythroid and, importantly, mast cell/basophil (45) gene sets were all reduced (Figure 5a, Supplementary Figures 10–12). The results were validated by qRT-PCR analysis of representative genes for each gene set (Figure 5b).
Expression of critical eosinophil genes, such as CLC and EPX, was markedly increased in Gata1- and Gata1s-transduced cells (Figure 5b). To determine whether these genes are regulated directly by GATA1/GATA1s, we performed chromatin-immunoprecipitation followed by high throughput sequencing (ChIP-seq) in human GATA1s-mutated cell line CMK stably expressing biotinylated GATA1 or GATA1s (J.X., S.H.O., J.H.K., unpublished data). These analyses revealed that both GATA1 and GATA1s occupied promoter regions of the eosinophil genes EPX and CLC comparably (Figure 5c).
Thus, Gata1s instructs an eosinophil expression program in hyperproliferative Gata1s-transduced human fetal HSPCs (hereafter referred to as ‘Gata1s-eosinophil precursors’).
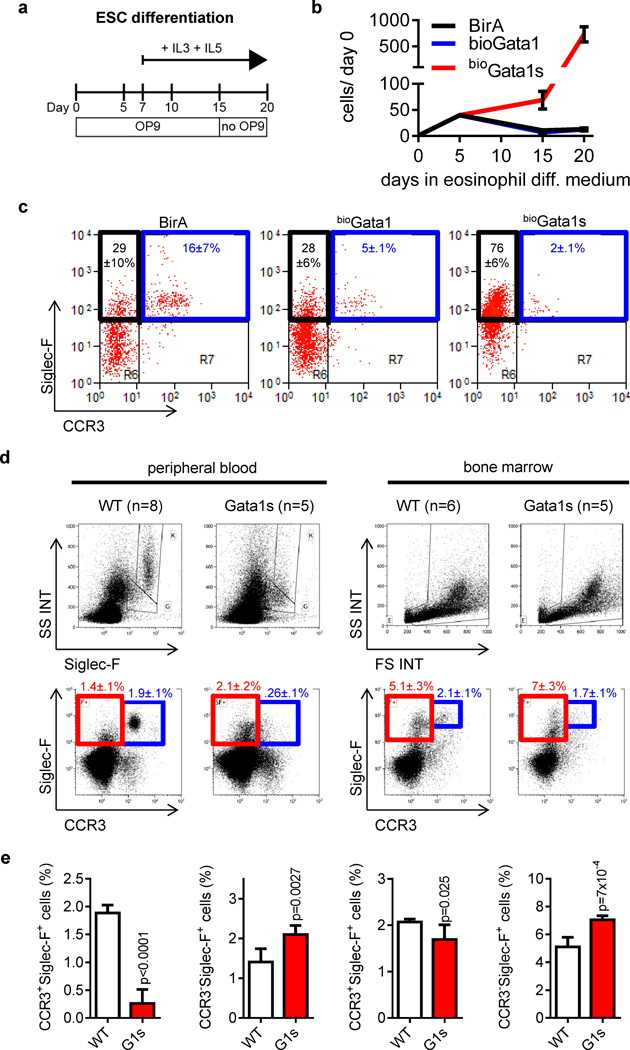
Eosinophil maturation is perturbed in Gata1s knockin mice
To evaluate the impact of Gata1s, expressed from its endogenous locus, on eosinophil differentiation, we investigated a knockin murine embryonic stem cell (ESC) model expressing the BirA enzyme (BirA) together with a biotinylated form of Gata1 (bioGata1) or Gata1s (bioGata1s) (35). During eosinophil in vitro differentiation, we also observed the hyperproliferation of Siglec-F+ eosinophils (46) only in bioGata1s-ESCs (Figure 6a–c). However, the percentage of Siglec-F+/CCR+ mature eosinophils (47) was markedly reduced (Figure 6c). Similarly, in the peripheral blood of Gata1Δe2-knockin mice (14), which exclusively express Gata1s, Siglec-F+/CCR+ or SSChigh/Siglec-F+ mature eosinophils were nearly absent (Figure 6d–e; for CBC see Supplementary Figure 13). However, the number of Siglec-F+/CCR− or SSClow/Siglec-F+ eosinophil precursors was elevated (Figure 6d–e). The accumulation of eosinophil precursors and the differentiation block was already evident in the bone marrow of Gata1Δe2-knockin mice (Figure 6d–e).
Figure 6. Gata1s perturbs eosinophilopoiesis in mice.
(a) Schematic illustration of the eosinophil ESC differentiation protocol. (b) Number of bioGata1-, bioGata1s- and BirA control murine ESCs grown in eosinophil differentiation medium relative to day 0. Data from n=3 independent experiments are presented as mean±SD. (c) FACS analysis on day 20 of eosinophil ESC differentiation. Mean±SD of n=3 independent experiments is indicated. (d–e) FACS analysis for eosinophils in peripheral blood and bone marrow of WT and Gata1Δe2-mice (14). (d) FACS-plots and mean±SEM. (e) Diagrams and statistics for the indicated populations in the peripheral blood (left) and bone marrow (right).
Thus, the N-terminal domain of Gata1 plays a previously unprecedented role in eosinophil development. Furthermore, these data exclude the possibility that the hyperproliferation of human fetal HSPCs is due to ectopic expression of Gata1s from a lentiviral vector.
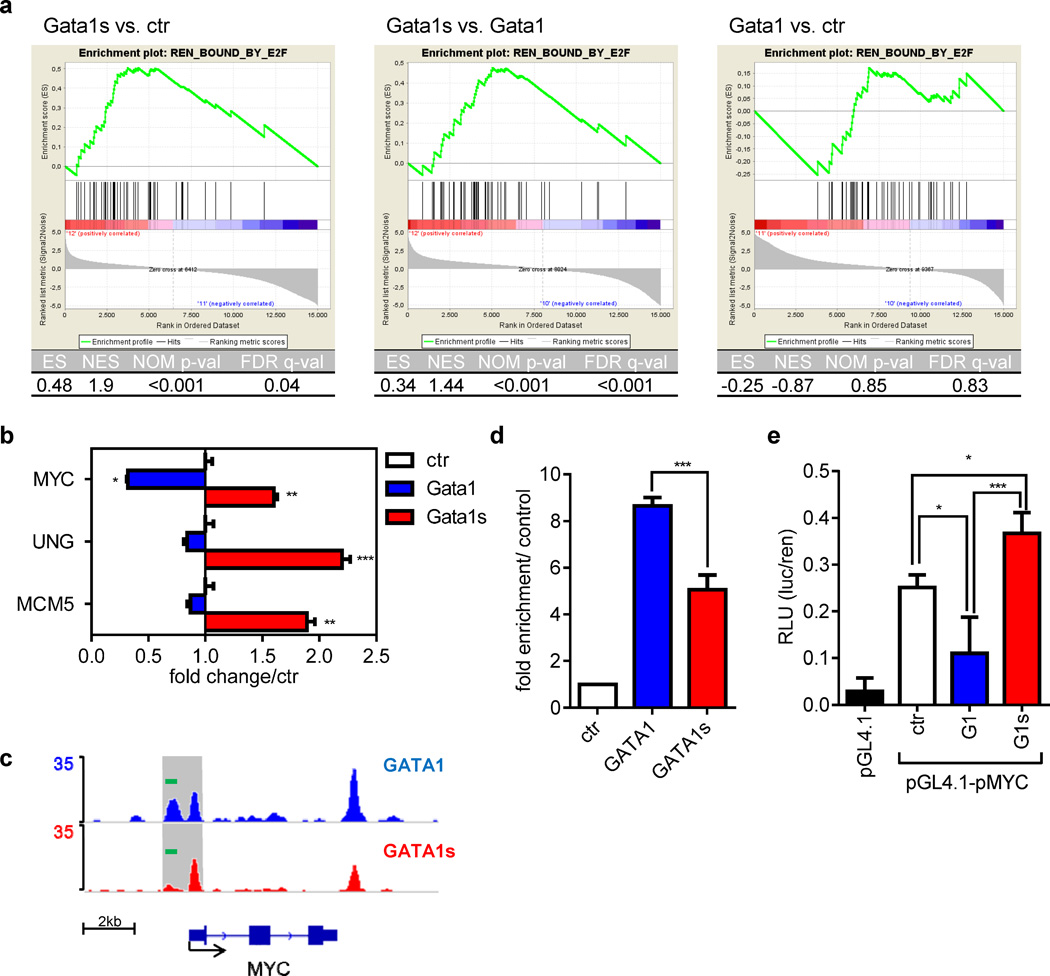
E2F-target genes are not repressed in Gata1s-eosinophil precursors
Next we assessed the expression of known E2F-bound target genes (48) in Gata1s-transduced fetal HSPCs by microarray (Figure 7a; Supplementary Figure 14). GATA1, but not GATA1s, represses the E2F transcription network by direct interaction with activating E2F transcription factors (13;49). The E2F transcription network is activated in cycling cells and repressed in terminally differentiating cells. As expected, E2F target genes were downregulated during myeloid differentiation in the empty vector transduced cells (Supplementary Figure 14a). Although ectopic expression of Gata1 perturbed myeloid differentiation, it was associated with downregulation of E2F target genes (Supplementary Figure 14b). In contrast, GATA1s failed to repress E2F target genes (Figure 7a, Supplementary Figure 14c). This was confirmed by qRT-PCR analysis of several representative genes from this gene set, including MYC, UNG and MCM5 (Figure 7b). The relative upregulation of E2F targets MCM5 and MYC was also evident in sorted TL-eosinophils (from three patients with eosinophilia) compared to eosinophils of a healthy donor (Supplementary Figure 15).
Figure 7. Gata1s fails to repress E2F target genes in eosinophilic precursors.
(a) GSEAs of E2F target genes previously identified by ChIP-chip analysis (48) in Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal HSPCs grown for 4 days in the myeloid differentiation medium: Gata1s vs. ctr (left), Gata1s vs. Gata1 (middle) and Gata1 vs. ctr (right). Enrichment plots (top) and statistics (table below) are provided. (b) qRT-PCR of key E2F target genes in Gata1-, Gata1s- and empty vector control (ctr)-transduced fetal HSPCs grown for 7 days in the myeloid differentiation medium from another independent experiment (mean±SD of replicates; *PANOVA<0.05, **PANOVA<0.01, ***PANOVA<0.001 compared to ctr). (c) ChIP-seq density plots of Gata1 and Gata1s on MYC genomic region and (d) ChIP followed by qRT-PCR (mean±SD of replicates; *PANOVA<0.05, **PANOVA<0.01, ***PANOVA<0.001 compared to ctr). The PCR amplicon is shown as green bar in Figure 7c. (e) Luciferase reporter assay using the MYC promoter in GATA1/GATA1s transfected CMK cells. pGL4.1 was used as a negative control (mean relative luminescence unit±SD of n=2 independent experiments; *PANOVA<0.05, **PANOVA<0.01, ***PANOVA<0.001 compared to empty vector ctr).
Taken together, our data suggest that GATA1s fails to repress pro-proliferative genes during differentiation, leading to hyperproliferation of eosinophil precursors.
GATA1s occupancy at the MYC promoter is reduced
MYC is a direct target gene of E2F1. MYC is also a direct target of GATA1 and is repressed by GATA1 during erythroid differentiation (10). To explore whether relative overexpression of MYC in GATA1s-transduced cells is due to incomplete repression of an E2F1 transcription network or direct transcriptional regulation by GATA1s, we analyzed occupancy of GATA1 and GATA1s at the MYC promoter by ChIP-seq analysis in CMK cells stably expressing biotinylated GATA1 or GATA1s (Figure 7c). We identified one distal (−900 to −600nt relative to MYC transcription start site [TSS]) and one proximal (−100 to +100nt relative to MYC TSS) ChIP-seq peak for GATA1. Occupancy of GATA1s was equal at the proximal region but modestly reduced at the distal region compared to that of GATA1 (Figure 7c; for quantitative ChIP-PCR analysis see Figure 7d). The distal region harbors three putative GATA1 binding sites (Supplementary Figure 16; ‘WGATAR motif’ (50;51). In contrast, the proximal peak contains only one putative GATA1 binding site located around the peak summit. The flanking DNA sequences of each GATA motif are different.
Reduced occupancy by GATA1s to the MYC promoter suggests that GATA1s may be less potent in repressing MYC expression. To test this, we performed a transient reporter assay using MYC promoter (P1 and P2 promoter) in CMK cells. While expression of GATA1 resulted in downregulation of luciferase reporter activity, GATA1s was unable to repress the activity of MYC promoter in this assay (Figure 7e). Our data suggest a dual role of GATA1s in the hyperproliferation of GATA1s-transduced promyelocytes. GATA1s fails to repress E2F transcription network by direct interaction with E2F1 (13), leading to overexpression of MYC; moreover, GATA1s is impaired in its capacity to repress MYC expression, most likely due to impaired occupancy at the MYC promoter.
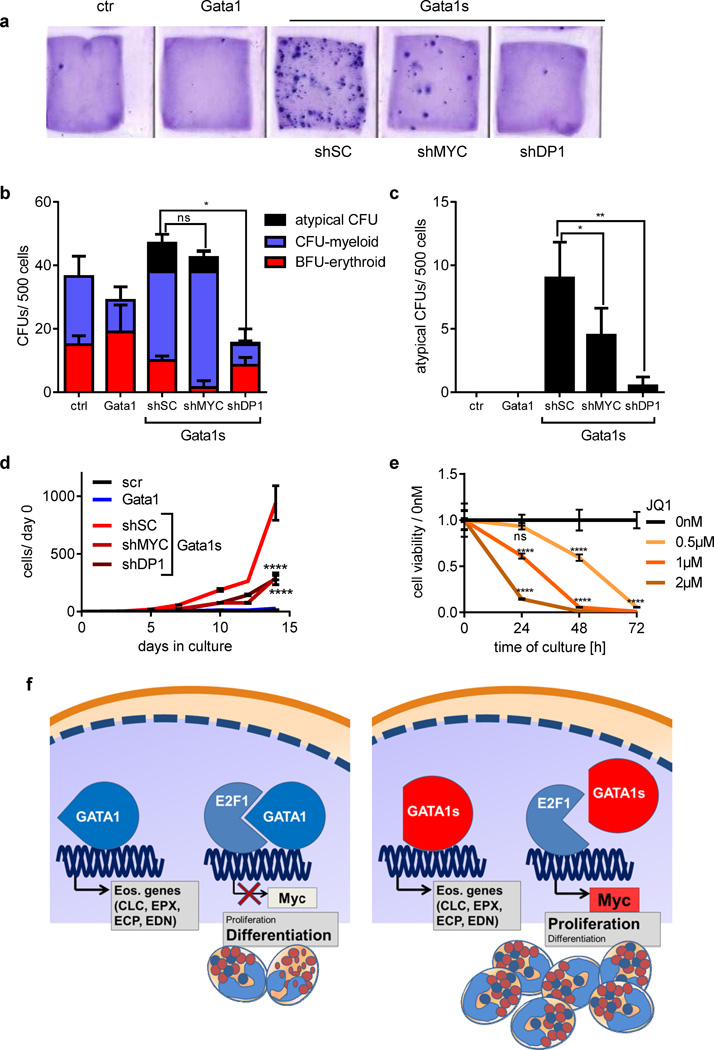
Knockdown of MYC or DP1 reverts the GATA1s-hyperproliferative phenotype of eosinophil promyelocytes
To test the hypothesis that the hyperproliferative phenotype associated with GATA1s is due to incomplete repression of an E2F transcription network and MYC, we performed a rescue experiment. MYC or DP1, an obligate E2F partner, were downregulated by an shRNA-approach and GATA1s expression in human fetal HSPCs was enforced simultaneously (Supplementary Figure 17a). Reduction of MYC (40% compared to control) or DP1 (60% compared to control; see Supplementary Figure 17b–c) was associated with the formation of significantly fewer colonies in collagen-based CFU assays (Figure 8a). In methocellulose-based CFU assays, DP1 knockdown led to a reduced number of total CFUs (Figure 8b). Of note, downregulation of either MYC or DP1 significantly decreased the formation of atypical eosinophil colonies (Figure 8c). In the case of MYC knockdown, these colonies were not only less abundant, but also smaller in size (Supplementary Figure 17d). In liquid culture, the proliferation was reduced upon DP1 or MYC knockdown, but the monocytic/neutrophilic differentiation block was retained (Figure 8d Supplementary Figure 17e). Moreover, treatment of Gata1s-eosinophil precursors with JQ1 led to marked dose-dependent decrease of viable cells (Figure 8e). JQ1 is a BRD4 inhibitor that was shown to selectively reduce MYC expression (52).
Figure 8. Knockdown of either MYC or DP1 partially rescues GATA1s-induced hyperproliferative phenotype of eosinophil precursor cells.
(a) Photograph (1× original magnification) of collagen-based colony-forming assays of an empty vector control (ctr), Gata1-, Gata1s-, shMYC/Gata1s and shDP1/Gata1s-transduced fetal CD34+-HSPCs. Number of CFUs (b) and atypical eosinophilic CFUs (c) per 500 empty vector (ctr), Gata1-, Gata1s-, shMYC/Gata1s and shDP1/Gata1s-transduced fetal CD34+-HSPCs. Replicates from n=2 independent experiments are presented as mean±SD (*PANOVA<0.05 of the total CFU count compared to ctr). (d) Number of empty vector control (ctr), Gata1-, Gata1s-, shMYC/Gata1s and shDP1/Gata1s -transduced fetal CD34+-HSPCs grown in myeloid differentiation medium relative to day 0. Data from n=2 independent experiments are presented as mean±SD. ****PANOVA<0.0001 compared to Gata1s. (e) Number of Gata1s-eosinophil precursors (after 20 days of culture) treated with MYC inhibitor JQ1 (52) relative to the untreated control. Data from n=2 independent experiments are presented as mean±SD. ****PANOVA<0.0001. (f) Model of GATA1s-mediated proliferation of eosinophil progenitors. Both GATA1 and GATA1s bind promoters of eosinophil genes therefore activate eosinophil differentiation program. GATA1 represses E2F transcription network and transcription of MYC, therefore works as “molecular brake” to restrict proliferation of eosinophil progenitor cells. GATA1s fails to repress E2F transcription network and MYC and thus causes hyperproliferation of eosinophil progenitors in DS-TL patients.
These findings provide evidence that incomplete repression of the E2F transcription network, particularly the E2F and GATA1 target gene MYC, accounts in large measure for the hyperproliferative phenotype associated with GATA1s.
Discussion
Here we report that eosinophils are derived from a TL cell clone in a substantial fraction of DS patients with TL. TL-blasts display an expression program characteristic of eosinophils and can differentiate along the eosinophil lineage, even in the absence of documented eosinophilia. TL is consistently associated with N-terminal truncating GATA1-mutations (GATA1s) and the eosinophils in TL with eosinophilia harbor N-terminal truncating GATA1 mutations. This suggests that GATA1s plays an instructive role, which drives an intrinsic propensity of TL-blasts to differentiate along the eosinophil lineage. In fact we demonstrated that, as with wild-type GATA1, GATA1s promoted differentiation of fetal hematopoietic progenitors along the eosinophil lineage through occupancy and transactivation of eosinophil genes. Since GATA1s was unable to repress a subset of GATA1 target genes, most notably MYC and the pro-proliferative E2F transcription network, the balance between cell differentiation and proliferation is perturbed in the presence of GATA1s (Figure 8f), leading to hyperproliferation of eosinophil precursors. Strikingly, the accumulation of eosinophil precursors and the block in terminal eosinophil differentiation were also evident in Gata1s-knockin mouse ESCs and Gata1Δe2 mice in vivo.
Previously, we and others demonstrated that GATA1 serves as a molecular brake to restrict proliferation of megakaryocytic and erythroid progenitors through repression of E2F target genes (13;49). The repression is mediated in part by direct protein interaction with E2Fs (13;49). This function of GATA1 is impaired in GATA1s. Our study suggests that this function of GATA1 extends to eosinophil precursors and is not restricted to the megakaryocytic and erythroid lineages. The central role of E2F in GATA1s-induced hyperproliferation of eosinophilic precursors is underscored by abrogation of this phenotype upon knockdown of the essential E2F interaction partner DP1. Importantly, we also identified MYC, a direct target gene of E2Fs and GATA1 (10;53), as an effector of GATA1s-mediated transformation. MYC activates the E2F transcription network either directly (54) or indirectly (55). Here we demonstrated that the occupancy of GATA1s at the MYC promoter is reduced. It remains unclear whether the presence of multiple putative GATA1 binding sites, the DNA sequence flanking the GATA1 motif and/or the failure to interact with bound cofactors contribute to the differential binding affinity of GATA1 and GATA1s to this genomic region. However, our results suggest a model in which GATA1s is defective in repressing MYC expression through impaired promoter association, leading to perturbation of the MYC-E2F loop in TL eosinophil precursors. Whether pharmacologic inhibition of MYC using JQ1 might also be a therapeutic option in DS-AMKL or clinically relevant TL, warrants consideration.
These data suggest that GATA1s may exert a dual role in TL. On one hand, it contributes to unrestricted proliferation of leukemic blasts through incomplete repression of MYC and the E2F transcription network. On the other hand, GATA1s transactivates an eosinophil expression program, thereby promoting the differentiation of the TL-blasts into this lineage. We demonstrated that at least in some instances of TL, eosinophilia is intrinsic to the leukemic clone, as the same mutation in GATA1 is present in both eosinophils and megakaryoblasts. Eosinophilia may have been overlooked previously in TL either because eosinophil counts were performed only at initial diagnosis, before blasts differentiated, or, in other cases, the mutant GATA1 clone is small thereby masking the enhanced eosinophil differentiation. Similarly, the accumulation of eosinophil precursors and the blocked terminal eosinophil differentiation in Gata1Δe2 mice was previously unrecognized. Thus, Gata1Δe2 mice may represent a useful tool to study eosinophilopoiesis and the role of eosinophils in allergic responses in vivo. It would also be interesting to investigate whether individuals with inherited GATA1s-mutations without trisomy 21 (56) have defective eosinophilopoiesis.
Miyauchi et al. (26) also demonstrated differentiation of TL megakaryoblasts into granulocytic cells with coarse basophilic granules. Based on electron microscopy, they described a hybrid basophilic/mast cell nature of the cells. Suda et al. (27) and Worth et al. (57) also reported infants that developed TL with extreme basophilia. Basophils and eosinophils share common surface antigens (CCR3, IL3R, IL5R, CD116) and some granule proteins (CLC, HDC) (45). Although distinct progenitors of mast cells, basophils and eosinophils have been identified, these cells share many features (58). The Gata1/Gata1s-derived eosinophil precursors and TL-eosinophils contained eosinophilic and coarse basophilic (pro-eosinophilic) granules. These pro-eosinophilic granules are otherwise only described in MDS and AML FAB M4eo. The observed eosinophilia and basophilia suggest that TL-blasts may give rise to both lineages. However, due to their abnormal coarse basophilic granules, the eosinophil precursors may also be misinterpreted as basophils. Our comprehensive evaluation of Gata1s-transduced hyperproliferative progenitors, however, clearly supports the eosinophil over other lineages.
In conclusion, our work provides novel insight into molecular mechanisms underlying TL and associated clonal eosinophilia. Our findings illustrate the pleiotropic effects of GATA1 in the hematopoietic system in a GATA1-associated hematologic disorder. Moreover, our observations have clinical implications. We propose that in addition to TL patients, the status of GATA1 should also be assessed in all DS neonates that present with abnormal eosinophils in blood. Such patients should be monitored for the development of subsequent DS-AKML.
Supplementary Material
Acknowledgements
The authors thank J. Strouboulis and E. Karkoulia for providing BioGata1/Gata1s ESCs. J. Schoening for general lab support; Drs. K. Weber and B. Fehse for providing plasmids; Dr. A. Mirenska for chip cytometry data analysis; Dr. R. Geffers for microarray analysis. A.M., L.S., S.E. and T.D. were supported by the Hannover Biomedical Research School. J.H.K is a fellow of the Emmy Noether-Programme from the German National Academic Foundation (KL-2374/2-1). This work was supported by a grant to J.H.K. from the German Research Foundation (KL-2374/1-1).
Footnotes
Conflict of interest
C.H. has filed patents regarding chip cytometry. The other authors declare no competing financial interests.
Author contribution
A.M., K.R., and T.D. performed experiments; L.S., S.E., J.X., Z.S. and G.J. performed experiments and analyzed results. A.M. and J.H.K. analyzed and interpreted results, prepared the figures and wrote the manuscript; J.H.K. designed the research. D.R. and J.H. provided patient material and revised the manuscript. I.R., C.H., K.R., P.V., Z.L., and S.O. analyzed and interpreted results and revised the manuscript. G.H. provided access and support to laboratory equipment and revised the manuscript.
Supplementary Information is available at Leukemia’s website.
References
- 1.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355(9199):165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 2.Hasle H, Abrahamsson J, Arola M, Karow A, O'Marcaigh A, Reinhardt D, et al. Myeloid leukemia in children 4 years or older with Down syndrome often lacks GATA1 mutation and cytogenetics and risk of relapse are more akin to sporadic AML. Leukemia. 2008;22(7):1428–1430. doi: 10.1038/sj.leu.2405060. [DOI] [PubMed] [Google Scholar]
- 3.Klusmann JH, Creutzig U, Zimmermann M, Dworzak M, Jorch N, Langebrake C, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111(6):2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lightfoot J, Hitzler JK, Zipursky A, Albert M, Macgregor PF. Distinct gene signatures of transient and acute megakaryoblastic leukemia in Down syndrome. Leukemia. 2004;18(10):1617–1623. doi: 10.1038/sj.leu.2403466. [DOI] [PubMed] [Google Scholar]
- 5.Blink M, Buitenkamp TD, van den Heuvel-Eibrink MM, Danen-van Oorschot AA, de HV, Reinhardt D, et al. Frequency and prognostic implications of JAK 1-3 aberrations in Down syndrome acute lymphoblastic and myeloid leukemia. Leukemia. 2011;25(8):1365–1368. doi: 10.1038/leu.2011.86. [DOI] [PubMed] [Google Scholar]
- 6.Klusmann JH, Reinhardt D, Hasle H, Kaspers GJ, Creutzig U, Hahlen K, et al. Janus kinase mutations in the development of acute megakaryoblastic leukemia in children with and without Down's syndrome. Leukemia. 2007;21(7):1584–1587. doi: 10.1038/sj.leu.2404694. [DOI] [PubMed] [Google Scholar]
- 7.Kiyoi H, Yamaji S, Kojima S, Naoe T. JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia. 2007;21(3):574–576. doi: 10.1038/sj.leu.2404527. [DOI] [PubMed] [Google Scholar]
- 8.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 9.Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2):231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, et al. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23(14):5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23(2):99–107. [PubMed] [Google Scholar]
- 12.Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24(7):1249–1257. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- 13.Klusmann JH, Godinho FJ, Heitmann K, Maroz A, Koch ML, Reinhardt D, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24(15):1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37(6):613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 15.Ge Y, LaFiura KM, Dombkowski AA, Chen Q, Payton SG, Buck SA, et al. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia. 2008;22(3):521–529. doi: 10.1038/sj.leu.2405066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stankiewicz MJ, Crispino JD. AKT collaborates with ERG and Gata1s to dysregulate megakaryopoiesis and promote AMKL. Leukemia. 2013;27(6):1339–1347. doi: 10.1038/leu.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harigae H, Takahashi S, Suwabe N, Ohtsu H, Gu L, Yang Z, et al. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells. 1998;3(1):39–50. doi: 10.1046/j.1365-2443.1998.00166.x. [DOI] [PubMed] [Google Scholar]
- 18.Hirasawa R, Shimizu R, Takahashi S, Osawa M, Takayanagi S, Kato Y, et al. Essential and instructive roles of GATA factors in eosinophil development. J Exp Med. 2002;195(11):1379–1386. doi: 10.1084/jem.20020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi S, Komeno T, Suwabe N, Yoh K, Nakajima O, Nishimura S, et al. Role of GATA-1 in proliferation and differentiation of definitive erythroid and megakaryocytic cells in vivo. Blood. 1998;92(2):434–442. [PubMed] [Google Scholar]
- 20.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei Y, Obata-Ninomiya K, Tsutsui H, Ishiwata K, Miyasaka M, Matsumoto K, et al. GATA-1 regulates the generation and function of basophils. Proc Natl Acad Sci U S A. 2013;110(46):18620–18625. doi: 10.1073/pnas.1311668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, et al. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002;277(45):43481–43494. doi: 10.1074/jbc.M204777200. [DOI] [PubMed] [Google Scholar]
- 23.Uhm TG, Kim BS, Chung IY. Eosinophil development, regulation of eosinophil-specific genes, and role of eosinophils in the pathogenesis of asthma. Allergy Asthma Immunol Res. 2012;4(2):68–79. doi: 10.4168/aair.2012.4.2.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyworth C, Pearson S, May G, Enver T. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 2002;21(14):3770–3781. doi: 10.1093/emboj/cdf368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eguchi M, Sakaibara H, Suda J, Ozawa T, Hayashi Y, Sato T, et al. Ultrastructural and ultracytochemical differences between transient myeloproliferative disorder and megakaryoblastic leukaemia in Down's syndrome. Br J Haematol. 1989;73(3):315–322. doi: 10.1111/j.1365-2141.1989.tb07746.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyauchi J, Ito Y, Tsukamoto K, Takahashi H, Ishikura K, Sugita K, et al. Blasts in transient leukaemia in neonates with Down syndrome differentiate into basophil/mast-cell and megakaryocyte lineages in vitro in association with down-regulation of truncated form of GATA1. Br J Haematol. 2010;148(6):898–909. doi: 10.1111/j.1365-2141.2009.08038.x. [DOI] [PubMed] [Google Scholar]
- 27.Suda T, Suda J, Miura Y, Hayashi Y, Eguchi M, Tadokoro K, et al. Clonal analysis of basophil differentiation in bone marrow cultures from a Down's syndrome patient with megakaryoblastic leukemia. Blood. 1985;66(6):1278–1283. [PubMed] [Google Scholar]
- 28.Stankov MV, El Khatib M, Kumar TB, Heitmann K, Panayotova-Dimitrova D, Schoening J, et al. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy. Leukemia. 2013 doi: 10.1038/leu.2013.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmrich S, Henke K, Hegermann J, Ochs M, Reinhardt D, Klusmann JH. miRNAs can increase the efficiency of ex vivo platelet generation. Ann Hematol. 2012 doi: 10.1007/s00277-012-1517-z. [DOI] [PubMed] [Google Scholar]
- 30.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16(4):698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 31.Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24(5):478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennig C, Adams N, Hansen G. A versatile platform for comprehensive chip-based explorative cytometry. Cytometry A. 2009;75(4):362–370. doi: 10.1002/cyto.a.20668. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4(4):506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23(4):796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez P, Braun H, Kolodziej KE, de Boer E, Campbell J, Bonte E, et al. Isolation of transcription factor complexes by in vivo biotinylation tagging and direct binding to streptavidin beads. Methods Mol Biol. 2006;338:305–323. doi: 10.1385/1-59745-097-9:305. [DOI] [PubMed] [Google Scholar]
- 36.Hamaguchi-Tsuru E, Nobumoto A, Hirose N, Kataoka S, Fujikawa-Adachi K, Furuya M, et al. Development and functional analysis of eosinophils from murine embryonic stem cells. Br J Haematol. 2004;124(6):819–827. doi: 10.1111/j.1365-2141.2004.04850.x. [DOI] [PubMed] [Google Scholar]
- 37.Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A. 2006;103(9):3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakajima T, Matsumoto K, Suto H, Tanaka K, Ebisawa M, Tomita H, et al. Gene expression screening of human mast cells and eosinophils using high-density oligonucleotide probe arrays: abundant expression of major basic protein in mast cells. Blood. 2001;98(4):1127–1134. doi: 10.1182/blood.v98.4.1127. [DOI] [PubMed] [Google Scholar]
- 40.Toki T, Kanezaki R, Adachi S, Fujino H, Xu G, Sato T, et al. The key role of stem cell factor/KIT signaling in the proliferation of blast cells from Down syndrome-related leukemia. Leukemia. 2009;23(1):95–103. doi: 10.1038/leu.2008.267. [DOI] [PubMed] [Google Scholar]
- 41.Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schaffer AA, et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med. 2007;13(1):38–45. doi: 10.1038/nm1528. [DOI] [PubMed] [Google Scholar]
- 42.Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9(10):1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka H, Matsumura I, Nakajima K, Daino H, Sonoyama J, Yoshida H, et al. GATA-1 blocks IL-6-induced macrophage differentiation and apoptosis through the sustained expression of cyclin D1 and bcl-2 in a murine myeloid cell line M1. Blood. 2000;95(4):1264–1273. [PubMed] [Google Scholar]
- 44.Yamaguchi Y, Zon LI, Ackerman SJ, Yamamoto M, Suda T. Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 1998;91(2):450–457. [PubMed] [Google Scholar]
- 45.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20(3):267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 46.Dyer KD, Garcia-Crespo KE, Percopo CM, Sturm EM, Rosenberg HF. Protocols for identifying, enumerating, and assessing mouse eosinophils. Methods Mol Biol. 2013;1032:59–77. doi: 10.1007/978-1-62703-496-8_5. [DOI] [PubMed] [Google Scholar]
- 47.Voehringer D, Van Rooijen N, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007;81(6):1434–1444. doi: 10.1189/jlb.1106686. [DOI] [PubMed] [Google Scholar]
- 48.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadri Z, Shimizu R, Ohneda O, Maouche-Chretien L, Gisselbrecht S, Yamamoto M, et al. Direct binding of pRb/E2F-2 to GATA-1 regulates maturation and terminal cell division during erythropoiesis. PLoS Biol. 2009;7(6):e1000123. doi: 10.1371/journal.pbio.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36(4):682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thalmeier K, Synovzik H, Mertz R, Winnacker EL, Lipp M. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev. 1989;3(4):527–536. doi: 10.1101/gad.3.4.527. [DOI] [PubMed] [Google Scholar]
- 54.Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17(9):5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19(7):4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hollanda LM, Lima CS, Cunha AF, Albuquerque DM, Vassallo J, Ozelo MC, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38(7):807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 57.Worth LL, Zipursky A, Christensen H, Tubergen D. Transient leukemia with extreme basophilia in a phenotypically normal infant with blast cells containing a pseudodiploid clone, 46,XY i(21)(q10) J Pediatr Hematol Oncol. 1999;21(1):63–66. [PubMed] [Google Scholar]
- 58.Boyce JA, Friend D, Matsumoto R, Austen KF, Owen WF. Differentiation in vitro of hybrid eosinophil/basophil granulocytes: autocrine function of an eosinophil developmental intermediate. J Exp Med. 1995;182(1):49–57. doi: 10.1084/jem.182.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.