Abstract
AIM: To develop a novel method for the rapid and efficient extraction of exosomes secreted by tumor cells.
METHODS: Unlike the traditional extraction method, the supernatants of cell cultures were concentrated, and the exosomes were isolated promptly and effectively using a novel nanomaterial called ExoQuick. Coomassie brilliant blue staining was used for protein quantification, and the morphology of the exosomes extracted by both methods was visualized by transmission electron microscopy. Exosome marker proteins were detected by Western blot analysis. Two potential hepatoma-associated proteins, tissue transglutaminase 2 (TGM2) and annexin A2, were analyzed.
RESULTS: The exosomes separated by the new extraction assay based on the nanomaterial were disc-shaped, intact vesicles with lipid bilayer membranes. They were approximately 30-100 nm in diameter, which is similar to the diameter of exosomes isolated by the traditional method. The protein concentration of exosomes extracted by the new method was approximately 780 μg/108 cells, and therefore, it was 19 times higher than that of exosomes extracted in the traditional manner. There were differences between the total proteins of Huh-7 cells and the exosomal proteins. Typical exosome proteins, such as the transmembrane protein CD63 and heat shock protein 70, were confirmed. Two potential hepatoma-associated proteins were also identified. TGM2 was first found to exist in the exosomes of human liver cancer cells, but annexin A2 was not secreted into exosomes.
CONCLUSION: The new extraction method based on the nanomaterial is quick and efficient. The cancer-associated protein TGM2 can be secreted through an exosome-mediated non-classical secretion pathway, and it may be a valuable tumor marker.
Keywords: Exosome, Membrane vesicles, Tissue transglutaminase 2, Annexin A2, Nanomaterials
Core tip: The traditional extraction assay of exosomes is usually complicated and time consuming. In this manuscript, we investigate a novel assay based on a nanomaterial to extract exosomes more quickly and efficiently compared with the traditional method. A hepatoma-associated protein, tissue transglutaminase 2, was first found to exist in the exosome of the human liver cancer cell line Huh-7. This protein can be secreted through an exosome-mediated non-classical secretion pathway and is a potential tumor marker.
INTRODUCTION
The term “exosome” was first used by Pan and Johnstone to describe the small vesicles secreted by reticulocytes during their differentiation into mature red blood cells; in this context, exosomes could carry away plasma membrane proteins that were not required by mature red blood cells, such as transferrin receptor and acetylcholinesterase[1]. In recent years, it has been demonstrated that exosomes can induce anti-tumor immune responses, and researchers have therefore devoted increasing attention to both basic research on exosomes and applied research regarding the use of exosomes for the development of anti-tumor vaccines. Recent studies have reported that many types of cultured cells can secrete exosomes, and exosomes can also be isolated from many types of body fluids, such as plasma[2], bile[3], urine[4], breast milk[5], saliva[6], pleural fluid[7], ascites[8], and bronchoalveolar lavage fluid[9-11]. Traditional exosome extraction methods involve ultracentrifugation or density gradient centrifugation; however, in this study, a new nanomaterial was utilized to isolate and purify exosomes secreted by hepatoma cells. This novel procedure was convenient and saved time and effort relative to traditional procedures. To determine whether this nanomaterial-based approach could quickly and efficiently purify exosomes, exosome morphology was observed by transmission electron microscopy, and biological marker proteins of the isolated exosomes were identified by Western blotting. These validation steps confirmed that the new isolation and purification method served as an efficacious approach for use in subsequent exosome research. Furthermore, in this study, exosomes were isolated and purified from the supernatant of cultured hepatoma cells, and Western blot analysis revealed the presence of tissue transglutaminase 2 (TGM2) in these exosomes. Therefore, these findings provide a theoretical foundation for the secretion of proteins through non-classical exosome-mediated pathways.
MATERIALS AND METHODS
Cell line, antibodies, and reagents
The human liver cancer cell line Huh-7 was obtained from the Human Science Research Resources Bank (Osaka, Japan). Amicon Ultra-15 centrifugal ultrafiltration units (3 kDa) and polyvinylidene difluoride (PVDF) membranes were purchased from Millipore (Bedford, MA, United States); ExoQuick Exosome precipitation solution was purchased from SBI (San Francisco, CA, United States). Rabbit anti-human CD63 and heat shock protein 70 (HSP70) polyclonal antibodies, rabbit anti-human annexin A2 monoclonal antibody, and mouse anti-human TGM2 monoclonal antibody were purchased from Santa Cruz (Santa Cruz Biotech., CA, United States). Mouse anti-human β-actin monoclonal antibody was purchased from Sigma-Aldrich (St. Louis, MO, United States). Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG and goat anti-mouse IgG were obtained from ZSGB-BIO (Beijing, China). An ultrasensitive enhanced chemiluminescence solution kit was obtained from Thermo (Waltham, MA, United States).
Cell culture and preparation of concentrated culture medium
Human hepatocellular carcinoma Huh-7 cells were cultured in DMEM medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 g/mL streptomycin at 37 °C with 5% CO2. When the cells reached approximately 90% confluence (in total, approximately 1.8 × 108 cells), they were washed three times with 20 mL of phosphate-buffered saline (1 × PBS) and were cultured for 24 h in serum-free DMEM medium. Approximately 100 mL of culture supernatant was collected and centrifuged at 3000 × g for 15 min at 4 °C (480 g for 5 min followed by 2000 g for 10 min) to remove intact cells and cell debris, and then, the remaining supernatant was concentrated to 1 mL using an Amicon Ultra-15 via centrifugation in a swing-out rotor at 4 °C and 4000 × g. An ultracel centrifugal filter device with a 3 kDa molecular weight cutoff (MWCO) was used.
Ultracentrifugation exosome isolation
The concentrated culture medium (CCM) was then subjected to high-speed centrifugation at 100000 × g (TLA-45 fixed angle, Beckman Coulter) at 4 °C for 2 h. The pellet was resuspended in 1 mL of PBS and washed by recentrifugation at 100000 × g for 3 h. The resulting exosome-enriched pellet was resuspended in 50 μL 1 × PBS and either used immediately or stored at -80 °C.
Nanomaterial exosome isolation
The CCM was added to an equal volume of ExoQuick exosome precipitation solution, and the resulting solution was mixed by inverting the tube and allowing it to stand overnight in a refrigerator. This mixture was then centrifuged at 1500 × g for 30 min. The supernatant was discarded, and the precipitate consisted of exosomes. A portion of the precipitate was then re-suspended in 1 × PBS for morphological observations via electron microscopy. Proteins were extracted from the remaining precipitate through resuspension in protein lysis buffer containing protease inhibitors, and the resulting solution was stored at -80 °C for future analysis.
Morphology analysis by transmission electron microscopy
One drop of the solution of exosomes resuspended in 1 × PBS was placed on a copper mesh with a diameter of 2 mm. Fluid was gently absorbed from the edges of the copper mesh with filter paper. A drop of 2% phosphotungstic acid solution was added to the sample, and negative staining was performed for 10 min at room temperature. After the negative staining solution was absorbed by the filter paper, the sample was dried for 2 min under incandescent light. The copper mesh was placed under a transmission electron microscope, and exosome morphology was observed and photographed at 80 kV.
Western blot analysis
Western blotting was used to analyze annexin A2 and TGM2 in the exosomes secreted by Huh-7 hepatoma cells as previously described, with slight modifications[4,12]. Briefly, approximately 30 μg of Huh-7 whole cell lysates and exosomal protein were separated, added to loading buffer, and heated at 95 °C for 10 min. The samples were subjected to electrophoresis using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in running buffer at constant 80 V at 4 °C for 3 h, and then, they were transferred to PVDF membranes. After blocking, the membranes were incubated with primary rabbit anti-CD63 (1:200), rabbit anti-HSP70 (1:200), rabbit anti-annexin A2 (1:500), or mouse anti-TGM2 (1:1000) at 4 °C overnight followed by incubation with the secondary antibody, HRP-labeled goat anti-rabbit IgG or goat anti-mouse IgG (1:3000), at room temperature for 1 h. The membranes were washed three times after each incubation for 5 min and visualized using the LAS4000 imaging system (Fujifilm).
RESULTS
Exosome morphology
The identity of the Huh-7 hepatocellular carcinoma cells was confirmed by short tandem repeats. Observations of the growth of the cells under an inverted phase contrast microscope revealed good adherence. Exosomes isolated from Huh-7 cells by the nanomaterial-based method and ultracentrifugation were viewed with a transmission electron microscope, which indicated the presence of many disc-shaped small vesicles of different sizes with lipid bilayer membranes. In particular, the diameters of these vesicles were between 30 and 100 nm (Figure 1). Therefore, the exosome morphology resulting from the new extraction assay based on the nanomaterial is similar to that of exosomes isolated by the traditional method.
Figure 1.
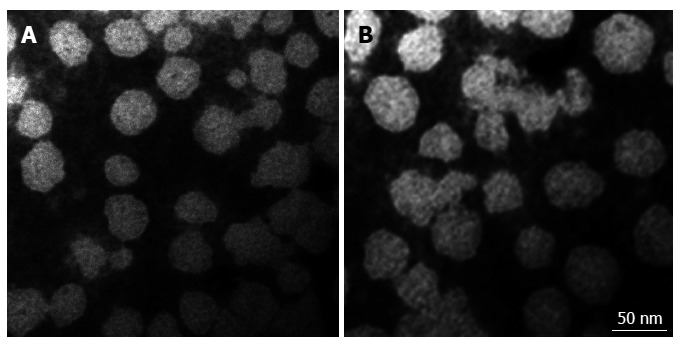
Characterization of the exosomes of Huh-7 cells extracted by two methods. Electron micrographs of exosomes isolated from Huh-7 cells in serum-free concentrated culture medium by the traditional method (A) and the new extraction method based on a nanomaterial (B). Negative-stained images show exosomes with a smooth, saucer-like morphology. The sizes are between 30-100 nm. The scale bar is 50 nm. Direct Mag: 100000 ×, HV = 80.0 kV.
Comparison of protein concentrations of exosomes isolated by two methods
Exosomal proteins from the supernatants of Huh-7 cells were quantified using the Bradford method. In particular, the actual protein concentrations were calculated using the standard curve equation and the appropriate dilutions. The concentration of exosomal proteins extracted by the new method based on the nanomaterial was approximately 780 μg/108 cells, which was more than 19 times higher than that of exosomes extracted in the traditional manner, which typically yields less than 40 μg/108 cells.
Exosomal protein identification and secretion mechanisms of human liver cancer-associated proteins
Approximately 30 μg of Huh-7 whole cell lysates and exosomal proteins were separated by SDS-PAGE. A Coomassie brilliant blue-stained gel obtained after SDS-PAGE demonstrated differences between the total proteins of Huh-7 cells and exosomal proteins secreted by Huh-7 cells (Figure 2). Exosomal proteins were separated by SDS-PAGE, electrotransferred, and probed with exosome markers (HSP70 and CD63). The membrane contained two bands of intense signal at 70 kDa and 53 kDa, which corresponded to the exosomal molecular biomarkers HSP70 and CD63, respectively (Figure 3). It is known that the hepatoma-associated proteins annexin A2 and TGM2 can be extracellularly secreted through non-classical secretory pathways, and the transport of proteins out of cells by exosomes is one of these non-classical secretory pathways. Therefore, we determined whether these proteins were localized to exosomes by Western blot analysis. The results indicated that exosomes secreted by Huh-7 cells contained TGM2 but not annexin A2 (Figure 3), confirming that TGM2 is extracellularly secreted through a non-classical exosome-mediated pathway.
Figure 2.
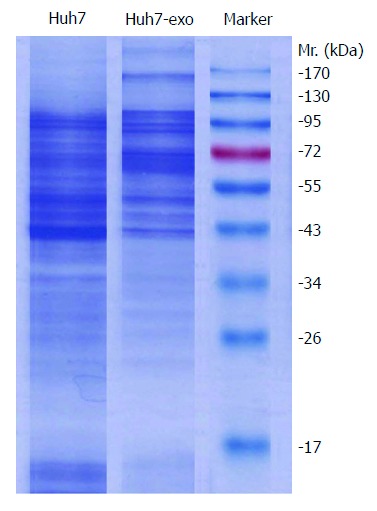
Detection of differences in Huh7 whole cell lysates and exosomal proteins. Approximately 30 μg of Huh-7 whole cell lysates and exosomal proteins isolated by the nanomaterial procedure were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were visualized by Coomassie brilliant blue staining. The marker is a prestained protein marker; Huh7-exo indicates exosomal proteins isolated from the supernatants of human hepatocellular carcinoma Huh-7 cells; Mr. stands for molecular weight.
Figure 3.
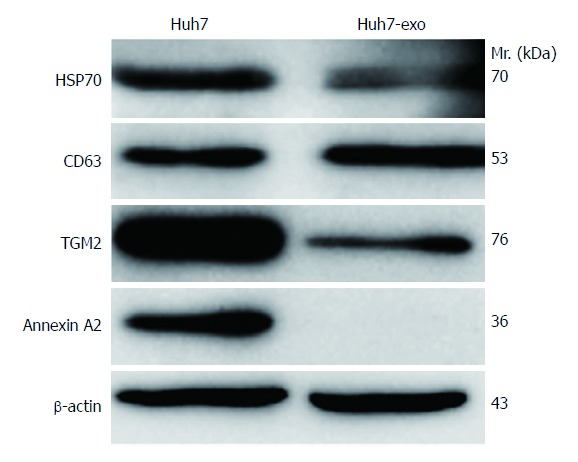
Exosome protein identification and secretion mechanisms of human liver cancer-associated proteins as analyzed by Western blotting. In total, 30 μg of proteins from Huh-7 whole cell lysates and exosomal proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred and incubated with antibodies against exosome protein markers (HSP70 and CD63) and hepatocellular carcinoma-associated proteins (TGM2 and annexin A2). β-actin was used as an internal control. Exosome protein markers (HSP70 and CD63) were identified on the surfaces of exosome membranes. Exosomes secreted by Huh-7 hepatocellular carcinoma cells contained the hepatoma-associated protein TGM2 but were negative for annexin A2. Huh7-exo indicates exosomal proteins isolated from the supernatants of human hepatocellular carcinoma Huh-7 cells; Mr. stands for molecular weight.
DISCUSSION
Exosomes are small, membranous vesicles with diameters between 30 and 100 nm that are extracellularly secreted by cells. Their surfaces contain large quantities of protein and lipid components that are closely associated with exosomal origins and functions, and microRNAs (miRNAs), messenger RNAs (mRNAs), and cytoplasmic proteins are contained within exosomes. The unique biological characteristics of exosomes have attracted great interest. For example, the membranous structure of exosomes can protect cytokines or other proteins carried by these vesicles from degradation by serum proteases, and exosomes therefore serve as important carriers that transmit information between cells in the body. Exosomes are derived from endosomes during endocytosis. In particular, during this process, the membranes of endosomes may bud inward into the endosomal lumen to form membrane-enclosed structures. The bases of these structures gradually separate from the endosomal membrane to form small vesicles within endosomes. Multiple small vesicles can be formed at once; these sets of many small vesicles are known as multivesicular bodies (MVBs)[13]. During exosome formation, cytoplasmic proteins and membrane proteins are selectively incorporated into exosomes. Because these proteins may include a variety of cell surface receptors and/or ligands, exosomes can act in either an autocrine or a paracrine manner and can therefore regulate their own functions or affect distant target cells[10,14]. However, the specific mechanisms of these interactions and regulatory functions remain unclear. For example, interactions between exosomes and target cells may rely on growth factors or bioactive lipids expressed on the surfaces of exosomes and cells, the transfer of membrane receptors, or direct stimulation through the delivery of proteins to target cells. The presence of mRNAs and miRNAs in exosomes suggests that the exchange of genetic material may be another aspect of exosome-mediated information transfer between cells[15]. Exosomes from tumor cells have been proposed to induce the cytotoxic T lymphocyte response, and many scientists initially had high hopes for the antitumor functions of exosomes. 5-Aza-2’-deoxycytidine, an inhibitor of DNA methyltransferase, can increase exosomes produced by hepatoma cells and the immune-associated protein component of exosomes[16]. However, recent studies have demonstrated that exosomes can downregulate immune responses or induce immune tolerance under certain conditions, leading researchers to re-examine the immunologic properties of exosomes[17]. The finding that glioblastoma tumor cell-derived exosomes contain mRNAs and microRNAs coupled with the detection of these exosomes in the serum of glioblastoma patients suggests that blood-based exosomes may provide valuable diagnostic information and aid in therapeutic decisions for cancer patients[18]. Exosomes were quantified by cytometry in the plasma of colorectal cancer patients to evaluate their potential as a tumor indicator. The fraction of exosomes in cancer patients was statistically higher than that in healthy controls[19]. To better understand the biological role of exosomes, it is important to work with highly purified material.
To date, few studies have examined exosomes. One reason for this lack of research is that the traditional exosome extraction process is relatively complex and cumbersome, rendering exosome research difficult. In prior studies in which exosomes have been extracted using conventional methods, such as ultracentrifugation and density gradient centrifugation, the reported quantities of protein obtained from exosomes have generally been no higher than 40 μg/108 cells[20]. In contrast, our approach, which involves the use of a new type of nanomaterial to isolate and extract exosomes, yielded protein quantities of up to 780 μg/108 cells from exosomes secreted by hepatoma cells. In recent years, it has been reported that an immunomagnetic method can be used to purify the exosome proteins and RNA. The quality has been greatly improved[21], but there still are cumbersome and expensive shortcomings. This novel procedure is convenient and efficient, and it involves only a short period during which the samples are exposed in an open system; as a result, the samples are not readily contaminated by environmental or cell debris and can therefore readily fulfill experimental and clinical needs. We first utilized low-speed centrifugation to remove cells and cell debris from cell culture supernatant samples and then used ultrafiltration tubes with a 3 kDa MWCO to remove proteins smaller than 3 kDa and a large quantity of fluid from each sample. Subsequently, the nanomaterial was mixed with this concentrated sample solution, and the resulting mixture was incubated overnight in a refrigerator and centrifuged; following centrifugation, the exosomes precipitated at the bottom of the centrifuge tube. Observations of exosomal morphology using transmission electron microscopy revealed that the isolated and purified exosomes were circular discs with diameters of 30 to 100 nm, and these findings were consistent with the descriptions of exosomes in the published literature. In addition, the biomarker proteins HSP70 and CD63 were detected in the isolated exosomes by Western blot analysis[22]. In particular, Western blotting revealed clear bands at the theoretical molecular weights of both proteins, indicating that HSP70 and CD63 are expressed in exosomes and confirming that the bodies extracted using the nanomaterial-based approach were indeed exosomes. These results validate our novel method for rapidly and efficiently isolating and purifying exosomes for exosome-related research.
Annexin A2 and TGM2 are hepatoma-related proteins, and our previous study reported histological and serological findings validating annexin A2 and TGM2 as candidate markers for hepatocellular carcinoma (HCC)[23]. The results of immunohistochemical examinations of HCC tissues have revealed that annexin A2 is a highly expressed protein in HCC cells. TGM2 is a member of the transglutaminase protein family with Ca2+-dependent transamidation functions. In particular, TGM2 facilitates the crosslinking or polymerization of proteins by crosslinking glutamine residues to the ε-amino groups of lysine residues. TGM2 has very diverse functions, including roles in cell differentiation, apoptosis, adhesion, receptor-mediated endocytosis, cell movement, and cell migration. Notably, TGM2 has bidirectional roles in cellular apoptosis, and the specific functions of TGM2 in different cell types are dependent on various factors, such as the specific apoptotic pathways and types of stimuli that are involved, the subcellular localization of TGM2, and the conversions catalyzed by the various enzymatic activities of TGM2[24-27]. TGM2 and annexin A2 do not contain signal peptides or transmembrane domains. The extracellular secretion of these proteins is thought to occur through non-classical secretory pathways[28]. However, these proteins were not detected during the construction of the normal human plasma protein database as part of the Human Plasma Proteome Project (HPPP). Based on the characteristic expression patterns of these proteins and the results indicating that these proteins are potential markers for HCC, it is possible that these proteins could be secreted into the plasma by hepatoma cells. Therefore, Western blot analyses were performed to detect annexin A2 and TGM2 in the isolated and purified exosomes secreted by Huh-7 hepatoma cells. These analyses indicated that exosomes secreted from Huh-7 cells contained TGM2 but not annexin A2. These findings confirmed our hypothesis that TGM2 could be secreted by hepatoma cells and that exosomes constitute one method of TGM2 transport. In contrast, annexin A2 may be extracellularly secreted through other non-classical secretory pathways. Furthermore, an exosome database (ExoCarta) was searched to determine the sources of exosome samples in which annexin A2 and TGM2 had been detected[22]. This search revealed that these proteins had not previously been detected in exosome samples from human hepatoma cells, and therefore, our experimental results address this gap in previously reported data.
This study experimentally confirmed the feasibility of a novel approach utilizing a nanomaterial to rapidly and efficiently isolate and purify exosomes. In addition, annexin A2 and TGM2, which are candidate markers for HCC, were detected in exosomes secreted by hepatoma cells. The extracellular secretion of TGM2 must occur through non-classical secretory pathways because this protein possesses neither a signal peptide nor a transmembrane domain, and our results revealed that one such non-classical pathway for TGM2 secretion is the transport of this protein in exosomes. Together, these findings provide necessary theoretical foundations for future exosome-related research.
ACKNOWLEDGMENTS
We thank Professor Tao Zhou and Mr. Sa Zhang of the National Center of Biomedical Analysis of China for assistance with transmission electron microscopy analysis, and Dr. Liang Wang of Medical College of Wisconsin for his helpful advice on the manuscript.
COMMENTS
Background
Exosomes are small membranous vesicles with diameters of between 30 and 100 nm. They are released from different cell types under both normal and pathological conditions. Exosomes have been reported in many types of cultured cells and body fluids. MicroRNAs (miRNAs), messenger RNAs (mRNAs), and cytoplasmic proteins are contained within exosomes, and exosomes have diverse biological functions including immunomodulatory activity, intercellular communication abilities, and the transport of infectious cargo.
Research frontiers
The molecular composition of exosomes extracted from the cell culture medium from diverse cell types and various body fluids has been analyzed by proteomics. Proteomic studies with large data sets might contribute to the understanding of the biological functions of exosomes, such as the mechanism of regulating tumor development. Scientists try to develop exosomes as tools to target tumor cells. Exosomal proteins, as potential biomarkers, may be studied more frequently in the future.
Innovations and breakthroughs
Exosomes are membrane vesicles that are 40-100 nm in diameter and are of endocytic origin. They are released by most cell types upon the fusion of multivesicular bodies with the plasma membrane, presumably as a vehicle for intercellular communication. To better understand the biological role of exosomes, it is important to work with highly purified materials. However, the extraction assay of exosomes is normally complicated and time consuming. Here, the authors describe a novel assay based on a nanomaterial to extract exosomes more quickly and efficiently compared with the traditional method. A hepatoma-associated protein, tissue transglutaminase 2 (TGM2), was found to exist in the exosomes of the human liver cancer cell line Huh-7 for the first time. This protein can be secreted through an exosome-mediated non-classical secretion pathway as a potential tumor marker.
Applications
Improving conventional methods for purifying exosomes might contribute to studying exosome biological function, and an analysis of exosomal proteins may provide insights into the clinical monitoring of cancer patients.
Terminology
Exosomes are small membrane vesicles of endocytic origin secreted by various cell types, and they are thought to play important roles in intercellular communication. Cancer cell-derived exosomes play a role in immune escape, neovascularization, and metastasis. Although exosomes were originally described in 1983, interest in these vesicles has increased dramatically in recent years after the finding that they contain mRNAs and miRNAs.
Peer review
The authors describe a new extraction method for exosomes from hepatocellular carcinoma cells and show that the protein TGM2 can be secreted through an exosome-mediated non-classical secretion pathway. The methodology and the data reported are of great interest.
Footnotes
Supported by Grants from the State Key Projects for Basic Research, No. 2011CB910703 and No. 2012ZX10002-017 (to Zhao XH); National High-tech R and D Program, No. 2013AA041201 (to Qian YM) and No. 2012AA020206 (to Zhao XH); National Natural Science Foundation of China, No. 81372591 and No. 81321091 (to Zhao XH); and the Research Foundation of the Center for Marine Medicine and Rescue of Tsinghua University and NGH (to Qian YM and Zhao XH)
P- Reviewers: Chetty R, Gentile V, Ishiguro H S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Wang CH
References
- 1.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 2.Dragovic RA, Southcombe JH, Tannetta DS, Redman CW, Sargent IL. Multicolor flow cytometry and nanoparticle tracking analysis of extracellular vesicles in the plasma of normal pregnant and pre-eclamptic women. Biol Reprod. 2013;89:151. doi: 10.1095/biolreprod.113.113266. [DOI] [PubMed] [Google Scholar]
- 3.Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, Splinter PL, LaRusso NF. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez S, Suazo C, Boltansky A, Ursu M, Carvajal D, Innocenti G, Vukusich A, Hurtado M, Villanueva S, Carreño JE, et al. Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplant Proc. 2013;45:3719–3723. doi: 10.1016/j.transproceed.2013.08.079. [DOI] [PubMed] [Google Scholar]
- 5.Torregrosa Paredes P, Gutzeit C, Johansson S, Admyre C, Stenius F, Alm J, Scheynius A, Gabrielsson S. Differences in exosome populations in human breast milk in relation to allergic sensitization and lifestyle. Allergy. 2014;69:463–471. doi: 10.1111/all.12357. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong H, Yang Y, Ma S, Xiu F, Cai Z, Zhao H, Du L. Induction of a tumour-specific CTL response by exosomes isolated from heat-treated malignant ascites of gastric cancer patients. Int J Hyperthermia. 2011;27:604–611. doi: 10.3109/02656736.2011.564598. [DOI] [PubMed] [Google Scholar]
- 9.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 10.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Levänen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, Sköld CM, Svartengren M, Grunewald J, Gabrielsson S, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131:894–903. doi: 10.1016/j.jaci.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Mi W, Cai J, Ying W, Liu F, Lu H, Qiao Y, Jia W, Bi X, Lu N, et al. Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J Proteome Res. 2008;7:3847–3859. doi: 10.1021/pr800153s. [DOI] [PubMed] [Google Scholar]
- 13.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 14.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao WH, Sanren GW, Zhu JH, Li QW, Kang HR, Wang RL, Song LP, Ye M. Effect of 5-aza-2’-deoxycytidine on immune-associated proteins in exosomes from hepatoma. World J Gastroenterol. 2010;16:2371–2377. doi: 10.3748/wjg.v16.i19.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, Garcia JM, Dominguez G, Campos-Martin Y, Cuevas J, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409–418. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 21.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Gao G, Cai J, Wang Y, Qu X, He L, Liu F, Zhang Y, Lin K, Ma S, et al. Annexin A2 is a discriminative serological candidate in early hepatocellular carcinoma. Carcinogenesis. 2013;34:595–604. doi: 10.1093/carcin/bgs372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beninati S, Facchiano F, Piacentini M. Transglutaminases: future perspectives. Amino Acids. 2013;44:1–9. doi: 10.1007/s00726-012-1431-7. [DOI] [PubMed] [Google Scholar]
- 25.Fésüs L, Szondy Z. Transglutaminase 2 in the balance of cell death and survival. FEBS Lett. 2005;579:3297–3302. doi: 10.1016/j.febslet.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 26.Király R, Barta E, Fésüs L. Polymorphism of transglutaminase 2: unusually low frequency of genomic variants with deficient functions. Amino Acids. 2013;44:215–225. doi: 10.1007/s00726-011-1194-6. [DOI] [PubMed] [Google Scholar]
- 27.Agnihotri N, Kumar S, Mehta K. Tissue transglutaminase as a central mediator in inflammation-induced progression of breast cancer. Breast Cancer Res. 2013;15:202. doi: 10.1186/bcr3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemskov EA, Mikhailenko I, Hsia RC, Zaritskaya L, Belkin AM. Unconventional secretion of tissue transglutaminase involves phospholipid-dependent delivery into recycling endosomes. PLoS One. 2011;6:e19414. doi: 10.1371/journal.pone.0019414. [DOI] [PMC free article] [PubMed] [Google Scholar]


