Background: The mechanisms underlying differential regulation of gonadotropin subunit genes are not fully elucidated.
Results: Gonadotrope growth differentiation factor 9 (GDF9) expression, which is suppressed by GnRH, stimulates FSHβ expression.
Conclusion: Autocrine secretion of GDF9 contributes to FSH biosynthesis.
Significance: Regulation of FSH by GDF9 may contribute to gonadotrope function.
Keywords: Cell Signaling, Cell Surface Receptor, Endocrinology, Gene Regulation, Signal Transduction, GnRH Pulse Frequency, Growth Differentiation Factor 9, Follicle-stimulating Hormone β-Subunit, Gonadotropin-releasing Hormone, Incoherent Feed-forward Loop
Abstract
Gonadotropin-releasing hormone (GnRH) is secreted in brief pulses from the hypothalamus and regulates follicle-stimulating hormone β-subunit (FSHβ) gene expression in pituitary gonadotropes in a frequency-sensitive manner. The mechanisms underlying its preferential and paradoxical induction of FSHβ by low frequency GnRH pulses are incompletely understood. Here, we identify growth differentiation factor 9 (GDF9) as a GnRH-suppressed autocrine inducer of FSHβ gene expression. GDF9 gene transcription and expression were preferentially decreased by high frequency GnRH pulses. GnRH regulation of GDF9 was concentration-dependent and involved ERK and PKA. GDF9 knockdown or immunoneutralization reduced FSHβ mRNA expression. Conversely, exogenous GDF9 induced FSHβ expression in immortalized gonadotropes and in mouse primary pituitary cells. GDF9 exposure increased FSH secretion in rat primary pituitary cells. GDF9 induced Smad2/3 phosphorylation, which was impeded by ALK5 knockdown and by activin receptor-like kinase (ALK) receptor inhibitor SB-505124, which also suppressed FSHβ expression. Smad2/3 knockdown indicated that FSHβ induction by GDF9 involved Smad2 and Smad3. FSHβ mRNA induction by GDF9 and GnRH was synergistic. We hypothesized that GDF9 contributes to a regulatory loop that tunes the GnRH frequency-response characteristics of the FSHβ gene. To test this, we determined the effects of GDF9 knockdown on FSHβ induction at different GnRH pulse frequencies using a parallel perifusion system. Reduction of GDF9 shifted the characteristic pattern of GnRH pulse frequency sensitivity. These results identify GDF9 as contributing to an incoherent feed-forward loop, comprising both intracellular and secreted components, that regulates FSHβ expression in response to activation of cell surface GnRH receptors.
Introduction
Gonadotropin-releasing hormone (GnRH)2 plays an essential role in the regulation of mammalian reproductive function. Following release into the hypophyseal portal system in discrete pulses, GnRH complexes with its cell surface receptor on the anterior pituitary gonadotrope cells, where it stimulates the synthesis and secretion of the gonadotropins, luteinizing hormone (LH) and FSH. The gonadotropins regulate gonadal function, including sex steroid hormone synthesis, follicular growth and ovulation. LH and FSH have a common α subunit (CGA) and a specific β subunit (LHβ or FSHβ). Changes in the frequency of GnRH pulse secretion occur at various stages of the reproductive cycle (1). A high GnRH pulse frequency favors LH secretion during the late follicular phase of the menstrual cycle, whereas a low GnRH pulse frequency contributes to increased FSH secretion in the late luteal phase (2, 3). Unlike LH, which is stored and has a highly regulated secretion, FSH is largely constitutively secreted, and the major mechanism controlling its level is regulation of FSHβ gene expression (4).
FSH regulation shows a remarkable pattern of GnRH frequency sensitivity. In mouse gonadotropes, FSHβ gene expression is preferentially increased by brief, low concentration pulses of GnRH every 2 h. Paradoxically, doubling the amount of GnRH stimulation by increasing the pulse frequency to one pulse/hour leads to lower levels of FSHβ mRNA (5). This frequency-response pattern contributes to normal reproductive physiology. Alterations of GnRH frequency patterns are associated with reproductive disorders, such as polycystic ovary syndrome, and underlie the pharmacological treatment of gonadal hormone-dependent diseases, such as endometriosis and prostate cancer. Because of its biological and medical importance, the frequency system underlying gonadotropin regulation has been an area of study for decades, and several factors have been proposed to contribute to FSHβ frequency sensitivity (2, 5–11).
The major information processing properties of molecular signaling networks are largely determined by simple network motifs, and specific feedback or feed-forward circuits are necessary to create a robust low frequency detector (12–14). The components and network motifs underlying the paradoxical preferential induction of FSHβ by low frequency GnRH pulses are not well understood.
Most studies of the signaling pathways connecting the cell surface GnRH receptor and the nuclear FSHβ gene have been directed at intracellular signaling components and transcriptional regulators. We recently identified inhibin α, a secreted extracellular autocrine factor, as contributing to FSHβ regulation in the gonadotrope (5). This finding suggested that important regulatory network motifs might be constructed from intracellular pathways and extracellular secreted regulatory factors. The identification of novel secreted FSHβ regulators might help elucidate the fundamental mechanisms underlying frequency sensitivity. Growth differentiation factor 9 (GDF9), a member of the transforming growth factor β (TGFβ) superfamily that is expressed in primary pituitary cultures and gonadotrope cell lines (15), was one candidate selected for study. We found that GDF9 is a strong autocrine inducer of FSHβ that is suppressed by GnRH and forms a network motif contributing to the GnRH frequency sensitivity of FSHβ regulation.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources: GnRH, Bachem (Torrance, CA); cholera toxin (CTX), Calbiochem; ERK inhibitor U0126, JNK inhibitor SP600125, p38 inhibitor SB203580, Src inhibitor PP2, and PKA inhibitor H-89, Calbiochem; GDF9, ALK5, Smad2/3, and inhibin α siRNAs, Dharmacon (On-Target plus siRNA SMARTpool, Denver, CO); GDF9, R&D Systems; actinomycin D, Sigma; anti-GDF9 (c-18) and GAPDH antibodies, Santa Cruz Biotechnology; anti-Smad2/3 and anti-phospho-Smad2/3 antibodies, Cell Signaling Technology; anti-GDF9 antibody used in immunoneutralization experiments, Biorbyt (Cambridge, UK); activin A human recombinant, VWR (Lutterworth, UK); SB-505124, Tocris Bioscience (Bristol, UK).
Cell Culture
LβT2 cells were obtained from Professor Pamela Mellon (University of California, San Diego, CA). Cells were cultured as described previously (5).
Mouse pituitary glands were harvested from 13–15-week-old CD-1 male mice. Male mice were used as a source of cells because cells from randomly cycling female mice result in more abundant multiresponsive and polyhormonal anterior pituitary cells than males (16). Briefly, pituitaries were dispersed by enzymatic digestion with collagenase and pancreatin (5). Undigested tissue and cell aggregates were filtered out using a 52-μm pore nylon mesh. Primary pituitary cells were then seeded in 96-well tissue culture plates at 100,000 cells/well in M-199 complete medium (Invitrogen) containing 10% FBS and 1% penicillin/Fungizone and incubated for 2 days at 37 °C in a humidified air atmosphere of 5% CO2. Cells were then pretreated with follistatin overnight in serum-free medium to antagonize the impact of endogenous activin on FSHβ expression (17).
Rat pituitary glands were harvested from adult, female ovariectomized rats and prepared for cell culture as described previously (18). Briefly, pituitaries were enzymatically dispersed with a trypsin solution (19). Pituitary cells were plated in 48-well tissue culture plates at 1.8 × 106cells/ml per well in minimal essential medium (Invitrogen) containing 10% FBS that had been charcoal-stripped to remove endogenous steroids, particularly glucocorticoids, estrogens, and progesterone present in fetal serum. Cells were incubated for 2–3 h at 37 °C in a humidified air atmosphere of 5% CO2.
siRNA Interference
LβT2 cells were transfected using the Amaxa Cell Line Nucleofector Kit L (Lonza Walkersville, Walkersville, MD), as described previously (5). Immediately after transfection, cells were seeded in a cell culture plate containing DMEM + 10% FBS and incubated at 37 °C for 24 h. Next, culture medium was replaced with serum free-DMEM, and cells were incubated overnight at 37 °C for serum starvation. Cells were then stimulated with GnRH or GDF9, harvested by trypsinization, and pelleted by centrifugation.
Perifusion Experiment
The laminar flow microprocessor-controlled parallel perifusion system was designed and built in the laboratory, as reported previously (5). Cells were subjected to 2 nm GnRH pulses of 5-min duration at varying frequencies of GnRH stimulation for 8 h, then harvested 30 min after the last pulse for RNA extraction.
Quantitative Real-time PCR
Quantitative real-time PCR (qPCR) experiments were carried out as described previously (5). Primer sequences were as follows: GDF9/sense, 5′-CATGGGGGCCACTTCAACAA-3′; GDF9/Antisense, 5′-TGGGGAGAAAGAGCTCTCCAA-3′; FSHβ/Sense, 5′-TGGAGACTCTGGCATGATTG-3′; FSHβ/Antisense, 5′-GAGTTGAGCAGCCTAACCTT-3′; LHβ/Sense, 5′-TGTCCTAGCATGGTCCGAGT-3′; LHβ/Antisense, 5′-CCCCCACAGTCAGAGCTACT-3′; CGA/Sense, 5′-TAGGAGCCCCCATCTACCAG-3′; CGA/Antisense, 5′-GCTACAGTGGCACTCCGTAT-3′; ALK5/Sense, 5′-GATCCATCACTAGATCGCCCTT-3′; ALK5/Antisense, 5′-CCGACCTTTGCCAATGCTT-3′; inhibin α/Sense, 5′-TGCCATCGAGCTGCTCTCAA-3′; inhibin α/Antisense, 5′-CACCAAAAACAGGGGCTGAA-3′; SMAD2/Sense, 5′-CTTGGCTGTCCTCATACACGAA-3′; SMAD2/Antisense, 5′-CCGAGTCTCCTGTTCCCGTA-3′; SMAD3/Sense, 5′-GAATTTGCTGCCCTCCTAGCTC-3′; SMAD3/Antisense, 5′-CTCCCCAGCCTTTGACGAAG-3′; rps11/Sense, 5′-CGTGACGAAGATGAAGATGC-3′; rps11/Antisense, 5′-GCACATTGAATCGCACAGTC-3′. Primer sequences targeting GDF9 primary transcript were: GDF9_primary/Sense, 5′-CACTTCCTGCTTTCTCTAGTTG-3′ and GDF9_primary/Antisense, 5′-CGGTCCAGGTTAAACAGC-3′. Primer sequences targeting CGA primary transcript were: CGA_primary/Sense, 5′-CTGTTAGAGTGAAAGCGAAC-3′ and CGA_primary/Antisense, 5′-GCTACAGTGGCACTCCGTAT-3′. GDF9 primary transcript was validated by sequencing at the Mount Sinai Genomics Core Facility.
Illumina Array
Two million LβT2 cells were transfected with scrambled siRNA for 48 h. RNA samples were snap-frozen in dry ice and sent to the Keck Genomic Facility at Yale University for whole genome expression profiling analysis, using MouseWG-6 v2.0 Expression BeadChip (Illumina, San Diego, CA). There were three independent replicates. The microarray data were deposited in Gene Expression Omnibus (GEO; GSE52631).
RNA-Seq Assays
RNA-Seq assays and data analysis were carried out as described previously (20). Briefly, LβT2 cells were transfected with scrambled siRNA for 48 h. There were four replicates, and total RNA (2.5 μg) from each replicate was sequenced at the Mount Sinai Genomics Core Facility using an Illumina platform (Illumina) and a HiSeq 2000 sequencing system (100-nucleotide length, single read type, multiplexing three samples per lane). The RNA-Seq data were deposited in GEO (GSE42120).
Western Blot Analysis
Western blot analysis was described previously (5).
Statistical Analysis
Statistical calculations were performed using the GraphPad Prism statistical software package version 5 (GraphPad Inc., San Diego, CA). Data were analyzed for normality followed by calculation of ANOVA. Error bars in all figures represent S.E.
RESULTS
GnRH Suppresses GDF9 Gene Expression in a Pulse Frequency-sensitive Manner
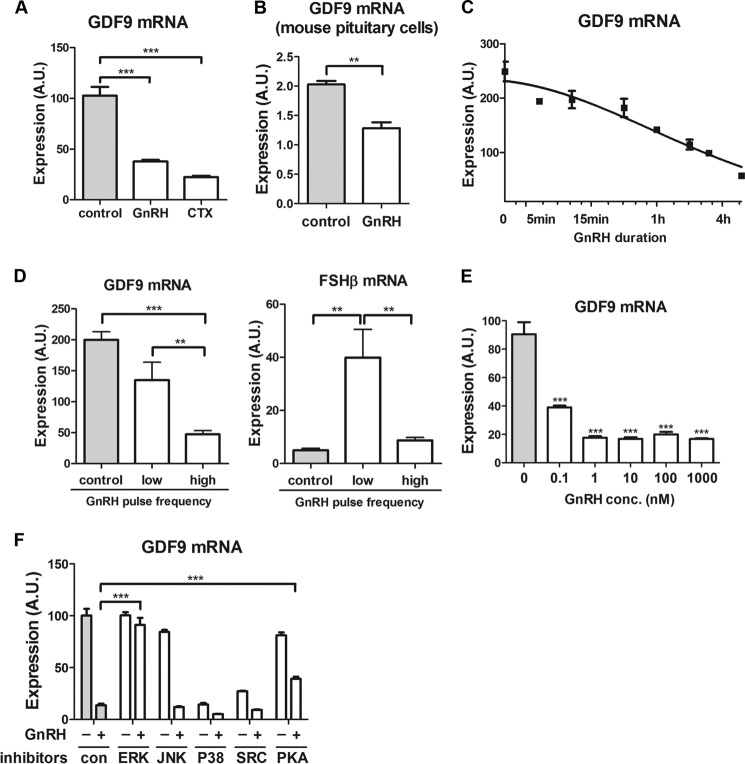
Because activation of Gαs by GnRH or CTX suppresses FSHβ gene expression (5), we studied their effects on GDF9. Either GnRH or CTX exposure greatly reduced the expression of GDF9 mRNA in LβT2 gonadotrope cells (63 and 78%, respectively, compared with untreated cells at 10 h; Fig. 1A). Similarly, GnRH exposure resulted in a significant reduction in GDF9 mRNA in mouse primary pituitary cells (Fig. 1B). In LβT2 cells, GDF9 expression decreased monotonically with increasing duration of GnRH exposure (Fig. 1C).
FIGURE 1.
GDF9 gene responsiveness to GnRH stimulation in gonadotrope cells. A, LβT2 cells were serum-starved overnight and stimulated with 5 μg/ml CTX, 1 nm GnRH, or vehicle (control) for 10 h. One-way ANOVA was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B, primary pituitary cells from adult mice were stimulated with 5 nm GnRH or vehicle for 12 h. C, LβT2 cells were stimulated with 1 nm GnRH or vehicle for the indicated times and harvested 6 h after exposure to GnRH. The data are shown on a log2 scale. D, LβT2 cells were stimulated with 2 nm GnRH for 8 h at either low pulse frequency (2-h interpulse intervals) or high pulse frequency (30-min interpulse intervals). E, LβT2 cells were stimulated with increasing concentrations of GnRH ranging from 0.1 nm to 1 μm for 5 h. F, LβT2 cells were pretreated with various pharmacological inhibitors (JNK, p38, Src, PKA, and ERK inhibitors at 10 μm final concentration) for 30 min, and cotreated with 1 nm GnRH for 6 h. GDF9 and FSHβ mRNA expression levels were determined by qPCR. One-way ANOVA with Bonferroni post-test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. The data shown are mean ± S.E. (error bars) of triplicate samples from one experiment and are representative of three independent experiments. A.U., arbitrary units.
We next studied the response of GDF9 to different frequencies of GnRH receptor stimulation. High pulse GnRH frequency resulted in a marked decrease in GDF9 expression, whereas low pulse frequency did not significantly alter GDF9 mRNA levels (Fig. 1D). Notably, FSHβ gene expression was most stimulated at low GnRH pulse frequency and was not significantly regulated by high GnRH pulse frequency (Fig. 1D). Hence, both GDF9 and FSHβ mRNAs show lower levels of expression at higher pulse frequency GnRH exposure.
GnRH Regulation of GDF9 Gene Expression Is Concentration-dependent and Involves ERK and PKA
We sought to investigate further the mechanisms involved in GDF9 gene response to GnRH stimulation in LβT2 cells. Only high GnRH pulse frequency caused a decrease in GDF9 mRNA expression. The average GnRH concentration is proportional to frequency; thus, an increase in frequency increases the average GnRH concentration. Therefore, we next determined the concentration response of GDF9 suppression by GnRH. We found that GDF9 mRNA reduction by GnRH showed a marked concentration dependence, with the IC50 ∼ 0.1 nm GnRH and the maximal effect achieved at 1 nm GnRH (Fig. 1E). These results are consistent with the preferential suppression of GDF9 at high frequency GnRH stimulation resulting from the higher average GnRH concentration caused by this exposure.
To identify the signaling pathways involved in GDF9 gene regulation by GnRH, GnRH-stimulated cells were pretreated with pharmacological inhibitors of various kinases, i.e. JNK, p38, Src, PKA, and ERK. The suppressive effects of GnRH on GDF9 mRNA expression were completely abolished by ERK inhibition and partially eliminated by PKA inhibition (Fig. 1F). All other inhibitors either had no significant effects (JNK inhibitor) or they affected basal GDF9 mRNA expression (p38 and Src inhibitors). Overall, these data indicate that the frequency effect of GnRH on GDF9 mRNA expression likely reflects a concentration effect and that the GnRH inhibitory effect on GDF9 is significantly dependent on ERK and PKA pathways. The involvement of PKA in mediating GDF9 mRNA suppression by GnRH is consistent with the inhibitory effect of CTX on GDF9 expression (see Fig. 1A).
GnRH Decreases GDF9 Gene Transcription
We investigated whether GnRH regulated GDF9 mRNA via transcriptional or post-transcriptional mechanisms. To determine the influence of GnRH exposure on GDF9 transcription rate, we established an assay for GDF9 primary transcript levels using qPCR to measure sequence-confirmed exon-intron junction expression. GnRH decreased both GDF9 mRNA levels (Fig. 2A) and GDF9 primary transcript concentrations (Fig. 2B). As a control experiment, we showed that both CGA mRNA (Fig. 2A) and CGA primary transcript (Fig. 2B) were increased by GnRH stimulation. Thus, these data provide evidence that sustained GnRH exposure suppresses GDF9 gene transcription.
FIGURE 2.
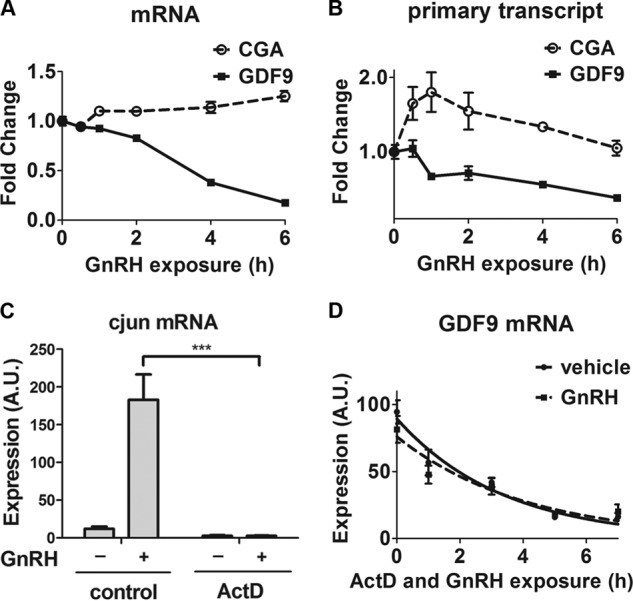
Effect of GnRH on GDF9 mRNA synthesis rate and stability. A and B, LβT2 cells were serum-starved overnight, stimulated with 1 nm GnRH, and harvested at the indicated times. One-way ANOVA was used. C, LβT2 cells were pretreated for 10 min with either 2 μg/ml actinomycin D (ActD; 1.6 μm) or vehicle (control), stimulated with 5 nm GnRH or vehicle for 45 min. D, LβT2 cells were exposed to 2 μg/ml actinomycin D (1.6 μm) in either the presence or absence of 5 nm GnRH and harvested at the indicated time points. CGA and GDF9 mRNA (A and D, respectively) and their primary transcript levels (B), as well as c-jun mRNA levels (C) were determined by qPCR. The data shown are mean ± S.E. (error bars) of triplicate samples from one experiment and are representative of three independent experiments. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001. A.U., arbitrary units.
We then assessed whether GnRH affected the stability of GDF9 transcripts. New mRNA transcription was inhibited by actinomycin D in cells stimulated with GnRH. We verified the effectiveness of actinomycin D on the induction of the immediate early gene c-jun by GnRH. As shown in Fig. 2C, actinomycin D treatment completely abolished the induction of c-jun mRNA. We measured the decay of GDF9 mRNA over time following the addition of actinomycin D. As shown in Fig. 2D, the half-life of GDF9 mRNA (1.6 ± 0.3 h) was not significantly altered by GnRH (1.4 ± 0.1 h). Thus, GnRH suppressed GDF9 transcription but did not alter the stability of GDF9 mRNA.
Both Basal and GnRH-induced FSHβ Transcript Levels Are GDF9-dependent
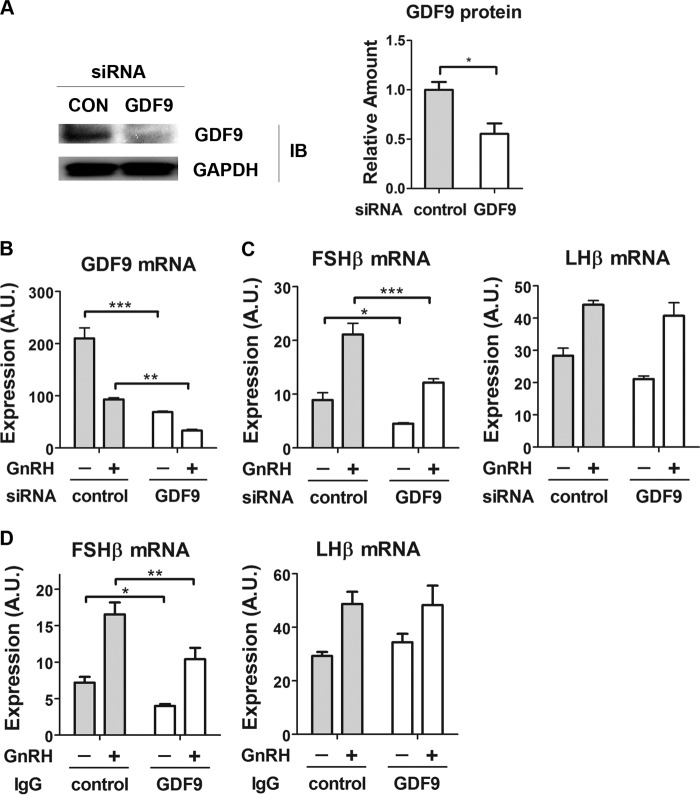
We studied the effect of GDF9 on FSHβ gene expression by siRNA-mediated knockdown. GDF9 protein levels were reduced by 50% by GDF9 knockdown (Fig. 3A). Consistent with this, GDF9 transcript levels were reduced by ∼66% compared with scrambled siRNA-transfected cells (Fig. 3B). GDF9 knockdown led to a significant attenuation of both basal and GnRH-induced FSHβ mRNA expression (49 and 43%, respectively; Fig. 3C). In contrast, LHβ gene expression was unaffected. To evaluate whether this effect was mediated by secreted GDF9, we examined the influence of an extracellular anti-GDF9 antibody on FSHβ mRNA expression. GDF9-specific antibodies significantly decreased both basal and GnRH-induced FSHβ expression (Fig. 3D), whereas LHβ expression remained unchanged. Hence, these data indicate that locally secreted GDF9 may contribute to both basal and GnRH-stimulated FSHβ mRNA expression.
FIGURE 3.
Effect of GDF9 inactivation on gonadotropin subunit gene expression. A, knockdown efficiency of GDF9 siRNA at the protein level. Left, 2 days after transfection, LβT2 whole cell lysates were subjected to a quantitative Western blot analysis using GDF9-specific antibodies. Right, Western blot densitometry was quantified from three independent experiments and plotted as mean ± S.E. (error bars). Two-tailed Student's t test was used; n = 3. *, p < 0.05. B, knockdown efficiency of GDF9 siRNA at the mRNA level. B and C, cells were transfected with either scrambled (control) or GDF9 siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with 1 nm GnRH for 2 h, followed by 4 h without GnRH, or with vehicle. D, cells were serum-starved overnight and treated with either 2 μg of control antibody (IgG) or 2 μg of GDF9 IgG for 8 h. After the first 2 h of antibody treatment, cells were exposed to vehicle or to 1 nm GnRH for 2 h, followed by 4 h without GnRH. GDF9, FSHβ, and LHβ mRNA expression levels were determined by qPCR. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. The data shown are representative of three independent experiments. A.U., arbitrary units.
GDF9 Stimulates FSHβ Expression in Immortalized Gonadotrope Cells and in Primary Pituitary Cultures
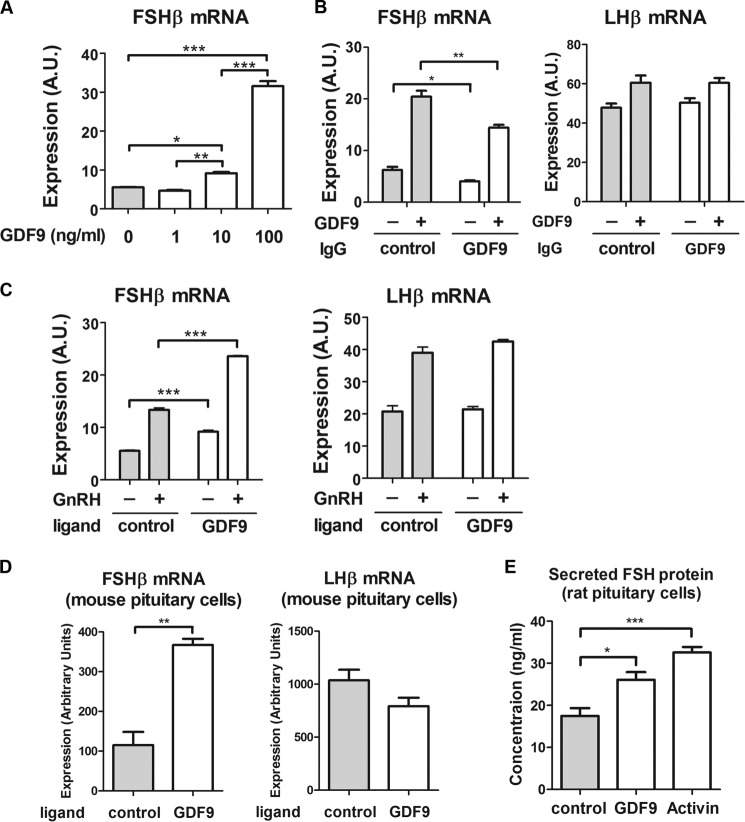
We studied whether exogenous GDF9 protein altered FSHβ mRNA expression. In LβT2 cells, we observed a significant concentration-dependent induction of FSHβ mRNA by GDF9 protein (Fig. 4A). FSHβ mRNA induction by exogenous GDF9 was significantly reduced in the presence of GDF9-specific antibodies (Fig. 4B), thus confirming the role of extracellular GDF9. In contrast, LHβ mRNA was unaffected by GDF9 exposure or by the presence of GDF9 antibody. Additionally, GDF9 increased basal FSHβ expression and enhanced GnRH-induced FSHβ expression, thus showing a synergy between GnRH and GDF9 (Fig. 4C). In contrast, GDF9 had no effect on LHβ mRNA expression. This result validated the specificity of GDF9 for regulation of the FSHβ gene.
FIGURE 4.
Effect of GDF9 treatment on gonadotropin subunit expression. A, LβT2 cells were serum-starved overnight and treated with increasing concentrations of GDF9 for 6 h. B, cells were serum-starved overnight and treated with either 2 μg of control antibody (IgG) or 2 μg of GDF9 IgG for 8 h. After 2 h of antibody treatment, cells were exposed to vehicle or 100 ng/ml GDF9 for 6 h. C, cells were serum-starved overnight and stimulated with 1 nm GnRH for 2 h, followed by 4 h without GnRH, or with vehicle. Cells were cotreated with GDF9 (10 ng/ml) for 6 h. D, primary pituitary cells from adult mice were pretreated with follistatin (500 ng/ml) overnight and treated with either vehicle or GDF9 (200 ng/ml) for 6 h. E, primary pituitary cells from ovariectomized female rats were pretreated with follistatin (500 ng/ml) overnight and treated with either vehicle, GDF9 (100 ng/ml), or activin (50 ng/ml; used as a positive control) for 6 h. Conditioned medium was harvested and sent to the Ligand Assay and Analysis Core facility at the University of Virginia for protein measurement. A and E, one-way ANOVA with Bonferroni post test corrections was used. B and C, two-way ANOVA with Bonferroni post test corrections was used. D, two-tailed Student's t test was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. The data shown are representative of three independent experiments. Error bars, S.E. A.U., arbitrary units.
To establish a physiological function for GDF9, we studied the impact of exogenous GDF9 on FSHβ expression in primary pituitary cultures of both mouse and rat origins. Treatment with GDF9 stimulated FSHβ but not LHβ mRNA expression in mouse primary pituitary cells (Fig. 4D). Additionally, we analyzed the effect of GDF9 treatment on FSH protein secretion using primary pituitary cultures from ovariectomized rats. The stimulatory effect of activin on FSH secretion was used as a positive control. Both GDF9 and activin stimulated FSH protein secretion in primary pituitary cells (Fig. 4E). Altogether, our results support a physiological role for GDF9 as a positive autocrine regulator of FSHβ.
GDF9 Stimulation of FSHβ Gene Expression Involves ALK5 and Smad2/3 Signaling Proteins
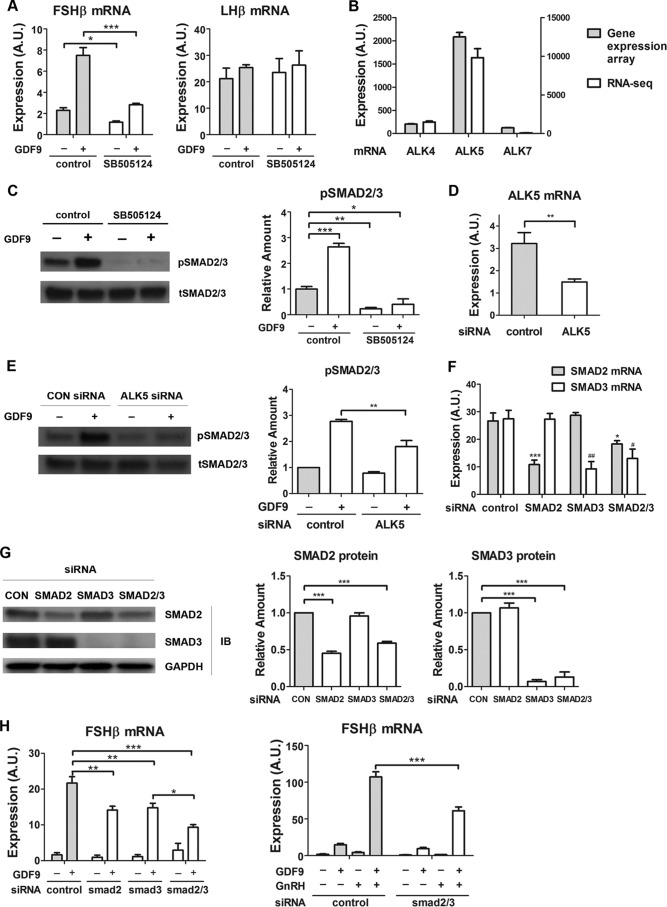
We subsequently studied the mechanism by which GDF9 regulated FSHβ mRNA. Whereas GDF9 signaling is mediated by ALK5 receptor and downstream Smad2 and Smad3 proteins in cultured ovarian granulosa cells (21), it appears to be mediated by ALK4/7 in vivo in the mouse ovary (22). We investigated the involvement of ALK and Smad2/3 in the stimulatory effect of GDF9 on FSHβ mRNA expression in LβT2 gonadotrope cells. The specific ALK4/5/7 inhibitor SB-505124 abolished GDF9 induction of FSHβ expression (Fig. 5A). In contrast, LHβ mRNA was unaffected.
FIGURE 5.
Involvement of ALK5 and Smad2/3 in GDF9 activation of FSHβ gene expression. A, LβT2 cells were serum-starved overnight, pretreated with SB-505124 (100 nm) or control (0.01% dimethyl sulfoxide) for 30 min, and stimulated with either GDF9 (100 ng/ml) or vehicle for 6 h. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B, LβT2 cells were transfected with scrambled siRNA for 48 h. RNA samples were subjected to whole genome expression profiling analysis (left y axis) and to RNA-Seq assays (right y axis), as described under “Experimental Procedures.” C, cells were serum-starved overnight, pretreated with SB-505124 (100 nm) or control (0.01% dimethyl sulfoxide) for 30 min, and stimulated with either GDF9 (100 ng/ml) or vehicle for 1 h. Left, whole cell lysates were subjected to a quantitative Western blot analysis using total Smad2/3 (tSMAD2/3)- and phospho-Smad2/3 (pSMAD2/3)-specific antibodies. Right, quantification of Western blot densitometry from three independent experiments is plotted as mean ± S.E. (error bars). One-way ANOVA was used. n = 3; *, p < 0.05. D, cells were transfected with either scrambled (control) or ALK5 siRNA. Two days after transfection, cells were harvested for qPCR measurement of ALK5 mRNA. E, cells were transfected with either scrambled (CON) or ALK5 siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with either GDF9 (100 ng/ml) or vehicle for 1 h. Left, whole cell lysates were subjected to a quantitative Western blot analysis using total Smad2/3 (tSMAD2/3)- and phospho-Smad2/3 (pSMAD2/3)-specific antibodies. Right, quantification of Western blot densitometry from three independent experiments is plotted as mean ± S.E. (error bars). Two-way ANOVA was used. n = 3; *, p < 0.05. F and G, knockdown efficiencies of Smad2, Smad3, and Smad2/3 siRNAs in LβT2 cells. LβT2 cells were transfected with either scrambled or Smad2/3 siRNA. F, 2 days after transfection, cells were harvested for qPCR measurement of Smad2 and Smad3 mRNA. One-way ANOVA with Bonferroni post test corrections was used. G, left, 2 days after transfection, whole cell lysates were subjected to a quantitative Western blot (IB) analysis using Smad2-, Smad3-, and Smad2/3-specific antibodies. Middle and right, quantification of Western blot densitometry from three independent experiments, plotted as mean ± S.E. (error bars). One-way ANOVA was used. n = 3; *, p < 0.05. H, left, LβT2 cells were transfected with either scrambled or Smad2/3 siRNA for 48 h, serum-starved overnight, and stimulated with either GDF9 (100 ng/ml) or vehicle for 6 h. Right, LβT2 cells were transfected with either scrambled or Smad2/3 siRNA for 48 h, serum-starved overnight, and stimulated with either GDF9 (100 ng/ml) for 6 h, vehicle, GnRH (1 nm) for 2 h (followed by 4 h without GnRH), or both GDF9 and GnRH. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. A.U., arbitrary units.
To determine which ALK subtype was most likely involved in this response, we interrogated LβT2 microarray datasets as well as a high sensitivity deep sequencing RNA-Seq dataset (4 samples with 30,000,000 reads/sample (20)). ALK5 mRNA showed elevated expression levels in the microarray experiment, whereas ALK4 and 7 transcript levels were at the limits of detection (Fig. 5B). The RNA-Seq data were similar, with ALK5 expression being high and very low levels of ALK4 transcript also present (Fig. 5B). GDF9 exposure was found to increase phospho-Smad2/3 protein levels by 2.7-fold, whereas total Smad2/3 protein levels remained unchanged. The stimulatory effect of GDF9 on Smad2/3 phosphorylation was blocked by ALK4/5/7 inhibitor SB-505124 (Fig. 5C). Further, siRNA-mediated inactivation of ALK5, which showed a knockdown efficiency of 55% (Fig. 5D), significantly reduced the GDF9 stimulatory effect on Smad2/3 phosphorylation (by 37%; Fig. 5E) but not significantly affecting total Smad2/3 protein levels. Altogether, these data indicate that in gonadotrope cells, GDF9 most likely activates Smad2/3 phosphorylation via ALK5.
We next tested the effect of Smad2/3 knockdown on GDF9 regulation of FSHβ gene expression. LβT2 cells were transfected with siRNA for Smad2, Smad3, or both and then exposed to GDF9. As shown in Fig. 5, F and G, Smad2 and Smad3 siRNAs were specific for their respective targets. Knockdown efficiency at the mRNA level was 60% for Smad2 and 67% for Smad3 (Fig. 5F). At the protein level, Smad2 knockdown led to a marked decrease in Smad2 (55%), and Smad3 inactivation resulted in 93% depletion of Smad3 (Fig. 5G). There was a significant attenuation of GDF9-induced FSHβ mRNA in the presence of either Smad2 or Smad3 siRNA (35 and 32%, respectively; Fig. 5H). The double knockdown suggested that the roles of Smad2 and Smad3 were additive (Fig. 5H). Finally, we examined the role of Smad2/3 in the modulation of GnRH-induced FSHβ expression by GDF9. The effect of GDF9 on GnRH-induced FSHβ gene expression was significantly attenuated by Smad knockdown (Fig. 5H). These findings indicate that in the gonadotrope, GDF9 most likely stimulates FSHβ expression through ALK5 followed by Smad2 and Smad3 activation.
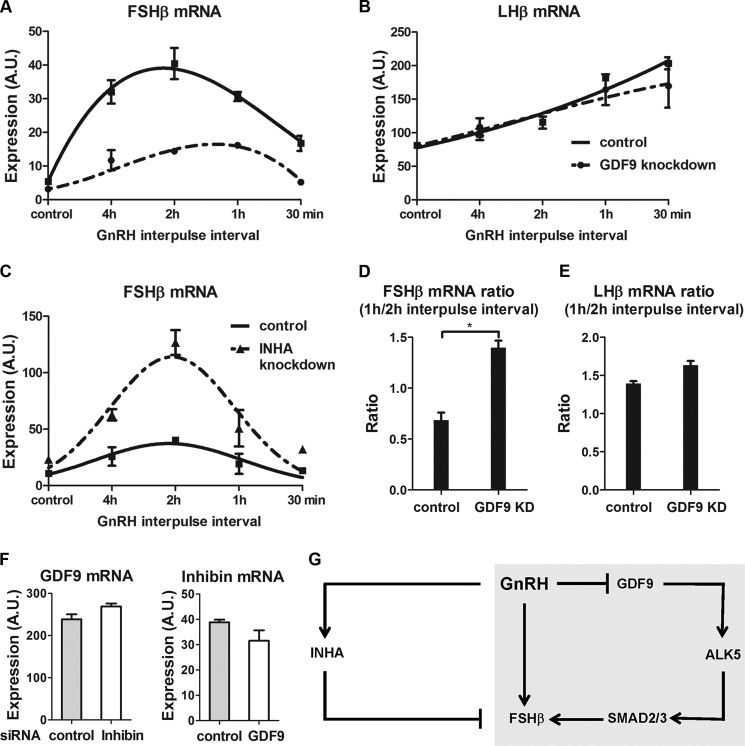
GDF9 Inactivation Impairs the GnRH Pulse Frequency Dependence of FSHβ Gene Expression
We previously identified inhibin α as a GnRH-regulated gonadotrope autocrine factor that might contribute to the frequency response characteristics of FSHβ induction (5). The present study identifies another factor, GDF9, forming an autocrine circuit regulating FSHβ. To compare the roles of GDF9 and inhibin α in the frequency tuning of the FSHβ response to GnRH, we used a custom programmable laminar flow parallel perifusion system. As expected, the maximum levels of FSHβ mRNA observed were decreased by GDF9 knockdown (Fig. 6A) and increased by inhibin α knockdown (Fig. 6C). However, the peak level of FSHβ gene expression occurred at the same frequency (2-h interpulse interval) with both control cells and inhibin α knockdown cells (Fig. 6C). The peak FSHβ induction by a 2-h GnRH interpulse interval is stable and reproducible, having been observed repeatedly in independent experiments. Strikingly, we found that GDF9 knockdown reproducibly and significantly shifted the frequency, causing the maximum levels of FSHβ mRNA to occur at a 1-h instead of a 2-h GnRH interpulse interval (Fig. 6, A and D). The effect of GDF9 was specific, as LHβ transcript levels were not significantly altered by GDF9 knockdown and were highest at a 30-min interpulse interval (Fig. 6, B and E). The possibility that GDF9 might regulate inhibin α or vice versa was examined by knocking down one and measuring mRNA expression of the other. GDF9 and inhibin α siRNAs had knockdown efficiencies of 66% (see Fig. 3B) and 90% (5), respectively. Inhibin α siRNA inactivation did not significantly alter GDF9 mRNA expression; likewise, GDF9 knockdown did not significantly affect inhibin α mRNA levels (Fig. 6F). These results indicate that whereas inhibin α and GDF9 have opposing effects on the amplitude of FSHβ expression, GDF9 is essential for tuning the GnRH frequency response characteristics of the system.
FIGURE 6.
Influence of GDF9 and inhibin α on the GnRH pulse frequency pattern of FSHβ gene expression. On day 2 after transfection, LβT2 cells were stimulated with various GnRH (2 nm) pulse frequencies for 8 h. A and B, impact of GDF9 siRNA knockdown on FSHβ (A) and LHβ (B) gene expression in GnRH-stimulated cells. C, impact of inhibin α siRNA knockdown on FSHβ gene expression in GnRH-stimulated cells. Data were normalized to the levels of expression obtained in unstimulated cells with control siRNA and either GDF9 siRNA (A and B) or inhibin α (ΙΝΗΑ) siRNA (C) in at least four independent experiments. The data shown are representative of at least three independent experiments. D and E, ratios of FSHβ (D) and LHβ (E) gene expression between 1-h and 2-h interpulse intervals of GnRH stimulation. To calculate the gene expression ratio, the average RNA expression level obtained at the 1-h pulse interval was divided by the average RNA expression level at the 2-h pulse interval. *, p < 0.05 two-tailed t test. KD, knockdown. F, LβT2 cells transfected with either control, inhibin α, or GDF9 siRNA. Two days later, mRNA levels of inhibin α and GDF9 were determined by quantitative real time-PCR. The data shown are mean ± S.E. (error bars) of triplicate samples from one experiment and are representative of three independent experiments. G, schematic of the regulation of FSHβ expression by GnRH and locally produced GDF9 and inhibin α. Exposure of the gonadotrope to fast GnRH pulses induces INHA expression but suppresses GDF9 expression. Whereas inhibin α represses FSHβ expression, GDF9 most likely stimulates FSHβ gene transcription via ALK5 and Smad2/3 protein activation. Highlighted in gray is the incoherent FFL formed by GnRH, GDF9 (and its downstream effectors), and FSHβ. An incoherent FFL provides a motif that potentially could allow FSHβ to have a preference for low frequency GnRH pulses if the inhibitory limb is better activated at higher frequency stimulation and if this effect is dominant. A.U., arbitrary units.
DISCUSSION
Our results identify GDF9 as an autocrine FSHβ-inducing factor that contributes to tuning the GnRH pulse frequency preference of the FSHβ gene. We show that GDF9 contributes to basal FSHβ expression both in immortalized gonadotrope cells and in primary pituitary cultures. GDF9 and GnRH act synergistically to induce FSHβ. The suppression of GDF9 gene transcription is dependent on the ERK and PKA pathways and occurs preferentially at high frequency, thereby leading to reduced FSHβ gene expression as the GnRH pulse frequency increases.
Studies of signaling networks, mostly performed in bacteria and yeast, suggest that responses result from the behavior of modular signaling motifs. A previous mathematical study of possible gonadotrope signaling motifs identified topologies referred to as incoherent feed-forward loops (FFL) as candidates for mediating FSHβ frequency decoding (13, 23, 24). In an incoherent FFL, the overall effects of two regulation paths to the target gene have opposing effects on the level of gene expression. Notably, GDF9 contributes to the formation of an incoherent FFL (see Fig. 6G), where GnRH activates FSHβ, but also inhibits FSHβ through its suppression of GDF9, a stimulatory factor for FSHβ expression. This structure is classified as a type 4 incoherent FFL (23). To our knowledge, GDF9 forms the first type 4 incoherent FFL identified in mammalian cells. An incoherent FFL provides a motif that could allow FSHβ to have a preference for low frequency GnRH pulses if the inhibitory limb is more robustly activated at higher frequency stimulation and if this effect dominates. Thus, at lower frequencies the positive limb would dominate, but as the inhibitory limb becomes more active at higher frequency the overall FSHβ response would be suppressed. The frequency sensing capacity of the GDF9 motif is supported by the effects of GDF9 knockdown on the GnRH pulse frequency response (see Fig. 6).
We have previously identified another gonadotrope autocrine factor, inhibin α, that also creates an incoherent FFL (Fig. 6 and Ref. 5). The inhibin α and GDF9 FFL motifs differ slightly in their design in terms of the location of the inhibitory effect (type 1 versus type 4 (23)). However, the overall functional behaviors of the two FFL topologies are similar. Nevertheless, we found that the role of the GDF9 incoherent FFL in contributing to the low frequency preference of the FSHβ gene is more important (see Fig. 6). Inhibin α knockdown altered the levels of FSHβ, but not the GnRH pulse frequency response curve. We speculate that the differing effects of inhibin α and GDF9 knockdown do not result from differences in the two motif structures and are more likely due to the weaker regulatory effect of GnRH on inhibin α than on GDF9 (Ref. 5 and Fig. 6). The lack of effect of inhibin α on frequency response is consistent with previous in vivo studies and models (25, 26). Our results do not exclude a frequency tuning effect of inhibin α, especially in concert with other regulatory factors such as GDF9.
Other potential mechanisms may contribute to GnRH frequency-dependent regulation of the FSHβ gene, including induction of distinct transcription factors by frequency-sensitive signaling pathways, post-translational modifications of these transcription factors, or epigenetic events like chromatin remodeling (27). As a member of the G protein-coupled receptor family (28), the GnRH receptor can activate both Gαq/11 (29–31) and Gαs (32, 33) in response to a GnRH stimulus. The Gαq pathway was reported to be subject to desensitization in response to pulse stimulation, whereas the Gαs-cAMP-PKA-dependent pathway was more sensitive to high GnRH pulse frequency (33). Conversely, recent work has revealed that at low pulse frequency the PKA-dependent pathway mediates GnRH activation of the cAMP response element-binding protein (CREB), which results in stimulation of FSHβ transcription (11). cAMP early repressor is thought to abrogate the preferential stimulation of FSHβ gene expression at high GnRH pulse frequency by reducing CREB occupation of the rat FSHβ promoter (6). In contrast, differential expression of AP1 factors and corepressors SKIL and TGIF1 was proposed to mediate GnRH pulse sensitivity of FSHβ expression (9). In another study, at low pulse frequency, GnRH induced c-Fos phosphorylation, which extended c-Fos half-life and augmented its transcriptional activity, thereby resulting in higher FSHβ induction (10). Additional studies have proposed that differences in ERK phosphorylation pattern in response to distinct GnRH pulse regimes are related to different expression patterns of MAPK phosphatase MKP1; this phenomenon may participate in the differential control of gonadotropin subunit expression. Kanasaki et al. initially observed that slow GnRH pulses triggered a more rapid and sustained phosphorylation of ERK1/2 than fast pulses (8). Subsequently, the feedback activity of MKP1, which inactivates MAPK via dephosphorylation, was found to modulate GnRH-induced ERK activation and gonadotropin response to GnRH (34, 35). Finally, MKP1 expression was shown to be predominant under high GnRH frequency compared with low GnRH frequency (36). The relationship and relative importance of these various loci which have been implicated in frequency decoding remain to be determined. Our data suggest that GDF9 serves a major role in GnRH pulse frequency decoding.
Besides the studies performed using LβT2 cells, specific induction of FSHβ mRNA and of FSH protein secretion by GDF9 exposure were shown using primary pituitary cell cultures. Additionally, we demonstrated that GDF9 mRNA is expressed in primary pituitary cells and that it is decreased by GnRH exposure. Further investigation will be required to ascertain the role of GDF9 in FSHβ regulation in vivo. Previous studies showed that GDF9 treatment promotes the growth of human ovarian follicles in vitro (37) and of rat ovarian follicles in vivo (38). However, according to a recent in vitro study, human GDF9:BMP15 heterodimers but not GDF9 homodimers are biopotent regulators of ovarian granulosa cell function; in contrast, both GDF9 homodimers and GDF9:BMP15 heterodimers are bioactive in mouse (39). Thus, there might be species-specific differences in the role of GDF9. Moreover, GDF9-deficient mice do not have fully developed follicles and are infertile, indicating that GDF9 is crucial for folliculogenesis and female fertility (40). According to that knock-out model, ovarian GDF9 has a major influence on both LH and FSH serum levels, which does not preclude a distinct autocrine/paracrine role of GDF9 in the pituitary. Overall, whereas the role of GDF9 in the ovary is clearly established, our work and the previous detection of GDF9 expression in rat primary pituitary cultures and LβT2 gonadotropes (15) and in the human (41) (GEO accession GDS3834 (42)), (GEO accession GDS1096 (43)), and bovine pituitary (44) support the view that GDF9 has an important functional role in the pituitary.
A striking aspect of our results is that the signaling network controlling a nuclear gene, FSHβ, by activation of a cell surface receptor, the GnRH receptor, involves regulatory loops that are extracellular. Of these extracellular loops, GDF9 contributes to both amplitude and frequency tuning and inhibin α predominantly to amplitude of the response. Together with previous reports of local gonadotrope regulatory factors, the view that cell signaling is distributed both intracellularly and extracellularly begins to emerge. These findings provide important new avenues for studying gene control mechanisms in the gonadotrope and potentially in other cell types. Additional local regulators of FSH, such as activin, follistatin, pituitary adenylate cyclase-activating polypeptide (PACAP), and bone morphogenetic proteins (BMPs), may play a part in the differential regulation of gonadotropin subunit expression (for review, see Ref. 4). For instance, follistatin is locally produced by the gonadotropes (45) and reduces FSHβ gene expression via the inactivation of activin (46). Because follistatin gene expression is increased by high frequency GnRH pulses, it may be linked to the repression of FSHβ transcription at that pulse frequency (47–49). PACAP potentiates GnRH action on gonadotropin release locally in the pituitary (50) and modulates gonadotropin subunit expression in gonadotrope-derived cell lines (51–53). GnRH induces PACAP expression in the gonadotropes, and the increase in PACAP transcripts is significantly more prominent at a lower pulse frequency (54). Evidence suggests that PACAP may be involved in GnRH pulse frequency-dependent stimulation of FSHβ expression in LβT2 cells (55). These extracellular factors constitute a complex regulatory network, which our results suggest need to be considered as an integral extension of intracellular signal processing in controlling gonadotrope responses.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46943.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession number GSE52631).
- GnRH
- gonadotropin-releasing hormone
- ALK
- activin receptor-like kinase
- ANOVA
- analysis of variance
- BMP
- bone morphogenetic protein
- CGA
- common α-glycoprotein subunit
- CREB
- cAMP response element-binding protein
- CTX
- cholera toxin
- FFL
- feed-forward loop
- GDF9
- growth differentiation factor 9
- LH
- luteinizing hormone
- MKP
- mitogen-activated protein kinase phosphatase
- PACAP
- pituitary adenylate cyclase-activating polypeptide
- qPCR
- quantitative real-time PCR.
REFERENCES
- 1. Marshall J. C., Kelch R. P. (1986) Gonadotropin-releasing hormone: role of pulsatile secretion in the regulation of reproduction. N. Engl. J. Med. 315, 1459–1468 [DOI] [PubMed] [Google Scholar]
- 2. Bédécarrats G. Y., Kaiser U. B. (2003) Differential regulation of gonadotropin subunit gene promoter activity by pulsatile gonadotropin-releasing hormone (GnRH) in perifused LβT2 cells: role of GnRH receptor concentration. Endocrinology 144, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 3. Marshall J. C., Eagleson C. A., McCartney C. R. (2001) Hypothalamic dysfunction. Mol. Cell. Endocrinol. 183, 29–32 [DOI] [PubMed] [Google Scholar]
- 4. Bernard D. J., Fortin J., Wang Y., Lamba P. (2010) Mechanisms of FSH synthesis: what we know, what we don't, and why you should care. Fertil. Steril. 93, 2465–2485 [DOI] [PubMed] [Google Scholar]
- 5. Choi S. G., Jia J., Pfeffer R. L., Sealfon S. C. (2012) G proteins and autocrine signaling differentially regulate gonadotropin subunit expression in pituitary gonadotrope. J. Biol. Chem. 287, 21550–21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciccone N. A., Xu S., Lacza C. T., Carroll R. S., Kaiser U. B. (2010) Frequency-dependent regulation of follicle-stimulating hormone β by pulsatile gonadotropin-releasing hormone is mediated by functional antagonism of bZIP transcription factors. Mol. Cell. Biol. 30, 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaiser U. B., Sabbagh E., Katzenellenbogen R. A., Conn P. M., Chin W. W. (1995) A mechanism for the differential regulation of gonadotropin subunit gene expression by gonadotropin-releasing hormone. Proc. Natl. Acad. Sci. U.S.A. 92, 12280–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanasaki H., Bedecarrats G. Y., Kam K. Y., Xu S., Kaiser U. B. (2005) Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LβT2 cells. Endocrinology 146, 5503–5513 [DOI] [PubMed] [Google Scholar]
- 9. Mistry D. S., Tsutsumi R., Fernandez M., Sharma S., Cardenas S. A., Lawson M. A., Webster N. J. (2011) Gonadotropin-releasing hormone pulse sensitivity of follicle-stimulating hormone-β gene is mediated by differential expression of positive regulatory activator protein 1 factors and corepressors SKIL and TGIF1. Mol. Endocrinol. 25, 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reddy G. R., Xie C., Lindaman L. L., Coss D. (2013) GnRH Increases c-Fos half-life contributing to higher FSHβ induction. Mol. Endocrinol. 27, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson I. R., Ciccone N. A., Xu S., Zaytseva S., Carroll R. S., Kaiser U. B. (2013) GnRH pulse frequency-dependent stimulation of FSHβ transcription is mediated via activation of PKA and CREB. Mol. Endocrinol. 27, 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kashtan N., Itzkovitz S., Milo R., Alon U. (2004) Topological generalizations of network motifs. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70, 031909 [DOI] [PubMed] [Google Scholar]
- 13. Krakauer D. C., Page K. M., Sealfon S. (2002) Module dynamics of the GnRH signal transduction network. J. Theor. Biol. 218, 457–470 [PubMed] [Google Scholar]
- 14. Yeger-Lotem E., Sattath S., Kashtan N., Itzkovitz S., Milo R., Pinter R. Y., Alon U., Margalit H. (2004) Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 101, 5934–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y., Nicholls P. K., Stanton P. G., Harrison C. A., Sarraj M., Gilchrist R. B., Findlay J. K., Farnworth P. G. (2009) Extra-ovarian expression and activity of growth differentiation factor 9. J. Endocrinol. 202, 419–430 [DOI] [PubMed] [Google Scholar]
- 16. Nuñez L., Villalobos C., Senovilla L., García-Sancho J. (2003) Multifunctional cells of mouse anterior pituitary reveal a striking sexual dimorphism. J. Physiol. 549, 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGillivray S. M., Thackray V. G., Coss D., Mellon P. L. (2007) Activin and glucocorticoids synergistically activate follicle-stimulating hormone β-subunit gene expression in the immortalized LβT2 gonadotrope cell line. Endocrinology 148, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turgeon J. L., Shyamala G., Waring D. W. (2001) PR localization and anterior pituitary cell populations in vitro in ovariectomized wild-type and PR-knockout mice. Endocrinology 142, 4479–4485 [DOI] [PubMed] [Google Scholar]
- 19. Turgeon J. L., Waring D. W. (1990) Rapid augmentation by progesterone of agonist-stimulated luteinizing hormone secretion by cultured pituitary cells. Endocrinology 127, 773–780 [DOI] [PubMed] [Google Scholar]
- 20. Wang Q., Chikina M., Zaslavsky E., Pincas H., Sealfon S. C. (2013) β-Catenin regulates GnRH-induced FSHβ gene expression. Mol. Endocrinol. 27, 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazerbourg S., Klein C., Roh J., Kaivo-Oja N., Mottershead D. G., Korchynskyi O., Ritvos O., Hsueh A. J. (2004) Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol. Endocrinol. 18, 653–665 [DOI] [PubMed] [Google Scholar]
- 22. Li Q., Agno J. E., Edson M. A., Nagaraja A. K., Nagashima T., Matzuk M. M. (2011) Transforming growth factor β receptor type 1 is essential for female reproductive tract integrity and function. PLoS Genet. 7, e1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangan S., Alon U. (2003) Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U.S.A. 100, 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva-Rocha R., de Lorenzo V. (2011) A composite feed-forward loop I4-FFL involving IHF and Crc stabilizes expression of the XylR regulator of Pseudomonas putida mt-2 from growth phase perturbations. Mol. Biosyst. 7, 2982–2990 [DOI] [PubMed] [Google Scholar]
- 25. Bertram R., Li Y. X. (2008) A mathematical model for the actions of activin, inhibin, and follistatin on pituitary gonadotrophs. Bull. Math. Biol. 70, 2211–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma T. P., Nett T. M., Karsch F. J., Phillips D. J., Lee J. S., Herkimer C., Padmanabhan V. (2012) Neuroendocrine control of FSH secretion. IV. Hypothalamic control of pituitary FSH-regulatory proteins and their relationship to changes in FSH synthesis and secretion. Biol. Reprod. 86, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferris H. A., Shupnik M. A. (2006) Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GNRH1. Biol. Reprod. 74, 993–998 [DOI] [PubMed] [Google Scholar]
- 28. Sealfon S. C., Weinstein H., Millar R. P. (1997) Molecular mechanisms of ligand interaction with the gonadotropin-releasing hormone receptor. Endocr. Rev. 18, 180–205 [DOI] [PubMed] [Google Scholar]
- 29. Grosse R., Schmid A., Schöneberg T., Herrlich A., Muhn P., Schultz G., Gudermann T. (2000) Gonadotropin-releasing hormone receptor initiates multiple signaling pathways by exclusively coupling to Gq/11 proteins. J. Biol. Chem. 275, 9193–9200 [DOI] [PubMed] [Google Scholar]
- 30. Hsieh K. P., Martin T. F. (1992) Thyrotropin-releasing hormone and gonadotropin-releasing hormone receptors activate phospholipase C by coupling to the guanosine triphosphate-binding proteins Gq and G11. Mol. Endocrinol. 6, 1673–1681 [DOI] [PubMed] [Google Scholar]
- 31. Naor Z., Azrad A., Limor R., Zakut H., Lotan M. (1986) Gonadotropin-releasing hormone activates a rapid Ca2+-independent phosphodiester hydrolysis of polyphosphoinositides in pituitary gonadotrophs. J. Biol. Chem. 261, 12506–12512 [PubMed] [Google Scholar]
- 32. Liu F., Usui I., Evans L. G., Austin D. A., Mellon P. L., Olefsky J. M., Webster N. J. (2002) Involvement of both Gq/11 and Gs proteins in gonadotropin-releasing hormone receptor-mediated signaling in LβT2 cells. J. Biol. Chem. 277, 32099–32108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsutsumi R., Mistry D., Webster N. J. (2010) Signaling responses to pulsatile gonadotropin-releasing hormone in LβT2 gonadotrope cells. J. Biol. Chem. 285, 20262–20272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lim S., Pnueli L., Tan J. H., Naor Z., Rajagopal G., Melamed P. (2009) Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS One 4, e7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen K. A., Intriago R. E., Upadhyay H. C., Santos S. J., Webster N. J., Lawson M. A. (2010) Modulation of gonadotropin-releasing hormone-induced extracellular signal-regulated kinase activation by dual-specificity protein phosphatase 1 in LβT2 gonadotropes. Endocrinology 151, 4882–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Purwana I. N., Kanasaki H., Mijiddorj T., Oride A., Miyazaki K. (2011) Induction of dual-specificity phosphatase 1 (DUSP1) by pulsatile gonadotropin-releasing hormone stimulation: role for gonadotropin subunit expression in mouse pituitary LβT2 cells. Biol. Reprod. 84, 996–1004 [DOI] [PubMed] [Google Scholar]
- 37. Hreinsson J. G., Scott J. E., Rasmussen C., Swahn M. L., Hsueh A. J., Hovatta O. (2002) Growth differentiation factor-9 promotes the growth, development, and survival of human ovarian follicles in organ culture. J. Clin. Endocrinol. Metab. 87, 316–321 [DOI] [PubMed] [Google Scholar]
- 38. Vitt U. A., McGee E. A., Hayashi M., Hsueh A. J. (2000) In vivo treatment with GDF-9 stimulates primordial and primary follicle progression and theca cell marker CYP17 in ovaries of immature rats. Endocrinology 141, 3814–3820 [DOI] [PubMed] [Google Scholar]
- 39. Peng J., Li Q., Wigglesworth K., Rangarajan A., Kattamuri C., Peterson R. T., Eppig J. J., Thompson T. B., Matzuk M. M. (2013) Growth differentiation factor 9: bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc. Natl. Acad. Sci. U.S.A. 110, E776–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dong J., Albertini D. F., Nishimori K., Kumar T. R., Lu N., Matzuk M. M. (1996) Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 383, 531–535 [DOI] [PubMed] [Google Scholar]
- 41. Fitzpatrick S. L., Sindoni D. M., Shughrue P. J., Lane M. V., Merchenthaler I. J., Frail D. E. (1998) Expression of growth differentiation factor-9 messenger ribonucleic acid in ovarian and nonovarian rodent and human tissues. Endocrinology 139, 2571–2578 [DOI] [PubMed] [Google Scholar]
- 42. She X., Rohl C. A., Castle J. C., Kulkarni A. V., Johnson J. M., Chen R. (2009) Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics 10, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S. M., Aburatani H. (2005) Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 86, 127–141 [DOI] [PubMed] [Google Scholar]
- 44. Hosoe M., Kaneyama K., Ushizawa K., Hayashi K. G., Takahashi T. (2011) Quantitative analysis of bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) gene expression in calf and adult bovine ovaries. Reprod. Biol. Endocrinol. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaiser U. B., Lee B. L., Carroll R. S., Unabia G., Chin W. W., Childs G. V. (1992) Follistatin gene expression in the pituitary: localization in gonadotropes and folliculostellate cells in diestrous rats. Endocrinology 130, 3048–3056 [DOI] [PubMed] [Google Scholar]
- 46. Shimonaka M., Inouye S., Shimasaki S., Ling N. (1991) Follistatin binds to both activin and inhibin through the common subunit. Endocrinology 128, 3313–3315 [DOI] [PubMed] [Google Scholar]
- 47. Besecke L. M., Guendner M. J., Schneyer A. L., Bauer-Dantoin A. C., Jameson J. L., Weiss J. (1996) Gonadotropin-releasing hormone regulates follicle-stimulating hormone-β gene expression through an activin/follistatin autocrine or paracrine loop. Endocrinology 137, 3667–3673 [DOI] [PubMed] [Google Scholar]
- 48. Kanasaki H., Mutiara S., Oride A., Purwana I. N., Miyazaki K. (2009) Pulse frequency-dependent gonadotropin gene expression by adenylate cyclase-activating polypeptide 1 in perifused mouse pituitary gonadotroph LβT2 cells. Biol. Reprod. 81, 465–472 [DOI] [PubMed] [Google Scholar]
- 49. Winters S. J., Ghooray D., Fujii Y., Moore J. P., Jr., Nevitt J. R., Kakar S. S. (2007) Transcriptional regulation of follistatin expression by GnRH in mouse gonadotroph cell lines: evidence for a role for cAMP signaling. Mol. Cell. Endocrinol. 271, 45–54 [DOI] [PubMed] [Google Scholar]
- 50. Culler M. D., Paschall C. S. (1991) Pituitary adenylate cyclase-activating polypeptide (PACAP) potentiates the gonadotropin-releasing activity of luteinizing hormone-releasing hormone. Endocrinology 129, 2260–2262 [DOI] [PubMed] [Google Scholar]
- 51. Burrin J. M., Aylwin S. J., Holdstock J. G., Sahye U. (1998) Mechanism of action of pituitary adenylate cyclase-activating polypeptide on human glycoprotein hormone α-subunit transcription in αT3-1 gonadotropes. Endocrinology 139, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 52. Counis R., Laverrière J. N., Garrel-Lazayres G., Cohen-Tannoudji J., Larivière S., Bleux C., Magre S. (2007) What is the role of PACAP in gonadotrope function? Peptides 28, 1797–1804 [DOI] [PubMed] [Google Scholar]
- 53. Ferris H. A., Walsh H. E., Stevens J., Fallest P. C., Shupnik M. A. (2007) Luteinizing hormone β promoter stimulation by adenylyl cyclase and cooperation with gonadotropin-releasing hormone 1 in transgenic mice and LβT2 cells. Biol. Reprod. 77, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 54. Grafer C. M., Thomas R., Lambrakos L., Montoya I., White S., Halvorson L. M. (2009) GnRH stimulates expression of PACAP in the pituitary gonadotropes via both the PKA and PKC signaling systems. Mol. Endocrinol. 23, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanasaki H., Purwana I. N., Miyazaki K. (2013) Possible role of PACAP and its PAC1 receptor in the differential regulation of pituitary LHβ- and FSHβ-subunit gene expression by pulsatile GnRH stimulation. Biol. Reprod. 88, 35. [DOI] [PubMed] [Google Scholar]