Background: PARP1 and UHRF1 participate in heterochromatin dynamics and the maintenance of DNA methylation, raising the question of whether both proteins cooperate in these events.
Results: We reveal a physical and functional poly(ADP-ribose)-mediated interaction of PARP1 with UHRF1 that helps to adjust UHRF1-regulated biological activities.
Conclusion: PARP1 is a regulator of UHRF1-controlled H4K20me3 accumulation and DNMT1 expression.
Significance: PARP1 associates and cooperates with UHRF1 to regulate heterochromatin-associated events.
Keywords: DNA Methylation, Heterochromatin, Histone Modification, Post-translational Modification (PTM), Transcription, Ubiquitination, UHRF1, Poly(ADP-ribose) Polymerase
Abstract
Poly(ADP-ribose) polymerase 1 (PARP1, also known as ARTD1) is an abundant nuclear enzyme that plays important roles in DNA repair, gene transcription, and differentiation through the modulation of chromatin structure and function. In this work we identify a physical and functional poly(ADP-ribose)-mediated interaction of PARP1 with the E3 ubiquitin ligase UHRF1 (also known as NP95, ICBP90) that influences two UHRF1-regulated cellular processes. On the one hand, we uncovered a cooperative interplay between PARP1 and UHRF1 in the accumulation of the heterochromatin repressive mark H4K20me3. The absence of PARP1 led to reduced accumulation of H4K20me3 onto pericentric heterochromatin that coincided with abnormally enhanced transcription. The loss of H4K20me3 was rescued by the additional depletion of UHRF1. In contrast, although PARP1 also seemed to facilitate the association of UHRF1 with DNMT1, its absence did not impair the loading of DNMT1 onto heterochromatin or the methylation of pericentric regions, possibly owing to a compensating interaction of DNMT1 with PCNA. On the other hand, we showed that PARP1 controls the UHRF1-mediated ubiquitination of DNMT1 to timely regulate its abundance during S and G2 phase. Together, this report identifies PARP1 as a novel modulator of two UHRF1-regulated heterochromatin-associated events: the accumulation of H4K20me3 and the clearance of DNMT1.
Introduction
Post-translational modifications of histones and CpG methylation into DNA are defined as fundamental epigenetic mechanisms that elicit specific effects on various chromatin-associated biological events including gene expression, chromatin structure, and integrity or differentiation. Among the chromatin-associated proteins that catalyze or modulate these processes, an explosion of recent findings has introduced poly(ADP-ribose) polymerase 1 (PARP1,6 also known as ARTD1) and its activity as a major actor with both structural and regulatory roles (1–3). After binding to specific DNA structures or nucleosomes, PARP1 catalyzes an NAD+-dependent polymerization of negatively charged ADP-ribose units to form a ramified polymer called poly(ADP-ribose) (PAR) onto a variety of relevant chromatin-associated targets such as histones, histone-binding proteins, chromatin modulators, and PARP1 itself. The process by which PARP1 introduces covalently bound PAR onto target proteins is known as PARylation. Like other post-translational modifications, PARylation regulates the biochemical and functional properties of the modified target. Furthermore, PAR, either protein-free or covalently linked on proteins, is capable of noncovalent binding with specific proteins owing to the presence of a PAR-binding motif (4).
Interestingly, PARP1 has been particularly studied for its implication in the structure and function of heterochromatin sometimes in redundancy with PARP2. PARP1 binds to and interacts with specific components of constitutive heterochromatin such as telomeres (5, 6), centromeres (7, 8), and pericentromeres (9–11) or silent ribosomal RNA repeats (12, 13). The PARP1+/−;PARP2−/− background displays specific female embryonic lethality associated with X chromosome instability, suggesting a role in the maintenance of facultative heterochromatin as well (14). Recent reports imply a particular contribution of PARP1 to the propagation of the repressive heterochromatin marks after the passage of the replication fork. This has been suggested first by its interaction with the SWI/SNF-like chromatin remodeler SMARCAD1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1) involved in the inheritance of the silenced pericentric heterochromatin (11) and has been next exemplified by its association with the nucleolar remodeling complex NoRC, where it serves to perpetuate silent ribosomal DNA heterochromatin (12).
It is also becoming increasingly clear that PARP1-catalyzed PARylation participates in the DNA methyltransferase-1 (DNMT1)-mediated establishment and maintenance of DNA methylation patterns (15). A series of cellular studies shows that the blockage of PARylation causes anomalous DNA hypermethylation on genomic DNA and in particular on CpG islands (16–19), whereas increased PARylation is associated with widespread DNA hypomethylation (20). Furthermore, PARP1 and DNMT1 were found in a complex also containing PAR (21). The current working model proposes that PARylated-PARP1 binds to the DNMT1 promoter, attracts DNMT1 through its PAR-binding motif, and inhibits its catalytic activity, thereby protecting the DNMT1 promoter from methylation (15, 22). In this model, PARP1 automodification is promoted by the chromatin insulator CTCF (CCCTC-binding factor) (20).
Recently, UHRF1 (ubiquitin-like, with PHD and RING finger domains 1, also known as NP95 or ICBP90) has also emerged as a central mediator in the faithful inheritance of DNA methylation in mammals. Deletion of UHRF1 in mice is embryonic lethal, and the derived embryonic stem cells display loss of DNA methylation, altered chromatin structure, and enhanced transcription of repetitive elements (23). The initial model was that UHRF1 binds to hemi-methylated DNA via its SET and RING-associated (SRA) domain and acts as a recruitment factor for DNMT1 to copy the methylation pattern onto the newly synthesized daughter strand during DNA replication (24, 25). Subsequent reports have identified an additional binding of UHRF1 to di/trimethylated histone H3 on lysine 9 (H3K9me2 and H3K9me3) involving its tandem Tudor domain (TTD) and its plant homeodomain (PHD), which may contribute to its localization to pericentric heterochromatin (26–29). A recent study highlights the significant contribution of TTD-mediated binding to H3K9me3 in DNA methylation maintenance, likely by regulating DNMT1 stability during mitosis (30). Together, these studies suggest that UHRF1 mediates cross-talk between histone modifications and DNA methylation maintenance.
Interestingly, UHRF1 also contains a Ring domain endowed with E3 ubiquitin ligase activity and has been shown to ubiquitinate itself, histone H3, and DNMT1 but with different outcomes (31, 32). Although the UHRF1-dependent ubiquitylation of H3 has been reported to act as a platform for the recruitment of DNMT1 to DNA replication sites, the ubiquitination of DNMT1 along with its deubiquitination by Usp7 (also known as HAUSP) represents a major process for the tight regulation of DNMT1 levels (33–36).
In addition to its association with DNMT1, UHRF1 interacts with many other chromatin modulators such as the de novo methyltransferases DNMT3a and DNMT3b, the histone deacetylase HDAC1, the histone methyltransferase G9a, or the histone acetyltransferase Tip60 (37–40). Most of these associations involve the SRA domain. Therefore, beyond its critical role in DNA methylation maintenance, UHRF1 functions in several other chromatin-related pathways including DNA repair, silencing of viral promoters, and replication and silencing of pericentric heterochromatin (37, 41, 42).
As both PARP1 and UHRF1 were found to act in common chromatin-related pathways and share DNMT1 as a protein partner, we anticipated a possible physical and functional cooperation between the two proteins. We identified PARP1 as a novel interacting partner of UHRF1 that modulates two of its biological properties. First, we identify a related contribution of PARP1 and UHRF1 in the maintenance of the repressive mark H4K20me3 at pericentric heterochromatin, which possibly helps to control overall transcriptional silencing. We also show that PARP1 helps to maintain the association of UHRF1 with DNMT1 although with no consequence on the loading of DNMT1 to heterochromatic sites or the DNMT1-mediated methylation of major satellite repeats. Second, we describe PARP1 as a negative regulator of the ubiquitin ligase activity of UHRF1 onto DNMT1, thereby introducing PARP1 as an additional modulator of DNMT1 abundance during S and G2 phases. This can represent an additional way to maintain DNA methylation and transcriptional silencing, more specifically during the replication of pericentric heterochromatin and onward.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
Plasmids encoding GST-fused full-length or truncated versions of human PARP1 were described elsewhere (13). Plasmids encoding Myc-tagged full-length and deleted versions of UHRF1 or GFP-DNMT1 also were described elsewhere (27, 31). The GFP-UHRF1 single domain constructs for Ubl and Ring domain expression constructs were generated by PCR using the corresponding wild-type full-length GFP-UHRF1 construct (27). The GFP-PHD, TTD, and SRA expression constructs have been described previously (27, 43). Details on individual plasmid constructs, which were verified by sequencing, are available upon request. Mouse monoclonal anti-Myc antibody (9E10: WB, 1/250; IP, 3 μg/sample) and rabbit anti-DNMT1 antibody (H-300: WB, 1/200; IF, 1/100) were from Santa Cruz Biotechnology. Rabbit polyclonal anti-GST (G7781: WB, 1/10000), the mouse monoclonal anti-actin antibody (A2066: WB, 1/500) and rabbit polyclonal anti-GAPDH antibody (G9545: WB, 1/10000) were from Sigma. The rabbit polyclonal anti-poly(ADP-ribose) antibody (4335-MC-100: WB, 1/1000) was from Trevigen. The mouse monoclonal anti-PCNA antibody (PC-10: WB, 1/2000; IP, 4 μg/sample) was from Dako-Cytomation. The mouse monoclonal anti-GFP antibody (11814460001: WB, 1/10000) was from Roche. The mouse monoclonal anti-HA.11 antibody (16B12: WB, 1/10000) was from Covance. The rabbit anti-H3K4me3 (pAB-003–050: IF, 1/200) was from Diagenode. The rabbit anti-H3K9me3 (ab8898: IF, 1/2000), mouse anti-H4K16ac (ab23352: IF, 1/100), and rabbit polyclonal anti-H4K20me3 (ab9053: IF, 1/500; WB, 1/1000) were from Abcam. The mouse monoclonal anti-H4 was from Millipore (07-108: WB, 1/10000) was from Millipore. The mouse monoclonal anti-UHRF1 (IF, 1/1000) has been described elsewhere (44). The mouse monoclonal anti-PARP1 antibody (EGT-69: WB, 1/10000) and rabbit polyclonal anti-UHRF1 antibody (WB, 1/2000; IP, 5 μl/sample; IF, 1/1000) are described in Refs. 45 and 31, respectively. The rabbit polyclonal anti-PARP1 (2869-70: IP, 15 μl/sample) was produced in-house. The Alexa-conjugated antibodies for IF (Alexa Fluor 568 goat anti-rabbit IgG, Alexa Fluor 568 goat anti-mouse IgG, and Alexa Fluor 488 goat anti-mouse IgG: IF, 1/1500) were from Molecular Probes.
Cell Culture, Synchronization, and siRNA Knockdown
COS-1 and PARP1+/+ and PARP1−/− 3T3 cells were grown in DMEM (1 g/liter d-glucose, Invitrogen) supplemented with 10% FBS (PanBiotech) and 0.1% gentamicin (Invitrogen) at 37 °C in 5% CO2.
Synchronization of 3T3 cells was performed by serum starvation (DMEM (1 g/liter), 0.1% FBS, and 0.1% gentamicin) for 48 h. After release in fresh medium, cells were collected at different time points as determined by preliminary flow cytometry experiments (T14 h for G1, T22 h for S, and T24 h for G2) for protein detection by Western blotting. To inhibit protein synthesis, cells were treated with cycloheximide (Sigma) at 20 μg/ml for 24 h (including release time) before collecting the cells.
For UHRF1 knockdown in 3T3 cells, gene-specific ON-TARGETplus SMARTpool siRNAs (pool of four sequences) for UHRF1 (L-055507-01-0010) and the control ON-TARGET nontargeting pool siRNA (D-001810-10-20) were from Dharmacon. Cells in suspension were electroporated with 50 nm siRNA pools using the Neon transfection system (Invitrogen) according to the manufacturer's instructions. A second electroporation with 50 nm siRNA pools was performed 48 h later. The transfected cells were collected 96 h after the first electroporation and processed for RNA extraction, protein extraction, or immunofluorescence staining.
Western Blot Analysis, GST Pulldown, GFP-Trap Capture, and Immunoprecipitation
For Western blotting and immunoprecipitation of endogenous proteins, cells were lysed in RIPA-like buffer (50 mm Tris, pH 8, 0.5% Triton, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 50 mm sodium fluoride, 20 mm sodium pyrophosphate, pH 7.2, 1 mm sodium orthovanodate, 1 mm Pefabloc, and one tablet of protease inhibitor complex/10 ml (Complete Mini, Roche Diagnostics)) and incubated on ice for 20 min. After centrifugation at 10,000 rpm at 4 °C for 20 min, cleared suspensions were quantified by Bradford protein assay, and 50 μg of protein was analyzed by SDS-PAGE electrophoresis and immunoblotting using the appropriate antibodies.
Histones were acid extracted from a total of 1 × 106 PARP1+/+ and PARP1−/− 3T3 cells following the Abcam histone extraction protocol. The protein content was determined using the Bradford assay, and 30 μg of protein was analyzed by SDS-PAGE electrophoresis and immunoblotting using the appropriate anti-histone antibodies.
For pulldown experiments, 1 × 106 COS-1 cells were transfected by jetPEI (Polyplus transfection) with 8 μg of total recombinant DNA. Forty-eight hours later, cells were lysed by three cycles of freeze/thaw in 50 mm Tris-HCl, pH 8, 150 mm NaCl, 0.1% Nonidet P-40, 0.5 mm Pefabloc, and one tablet of protease inhibitor complex/10 ml. Cleared lysates were incubated with glutathione-Sepharose beads (GE Healthcare) for purification of GST-tagged proteins for 2 h at 4 °C. Beads were subsequently washed twice with washing buffer (10 mm Tris-HCl, pH 8, 0.1% Nonidet P-40, 0.5 mm Pefabloc, and protease inhibitor complex) containing 500 mm NaCl and twice with washing buffer containing 150 mm NaCl. For the experiment shown in Fig. 2C, beads were washed twice with washing buffer containing 750 mm NaCl, once with washing buffer containing 500 mm NaCl, and twice with washing buffer containing 150 mm NaCl. The final pellets were resuspended in Laemmli buffer and subjected to 10% SDS-PAGE.
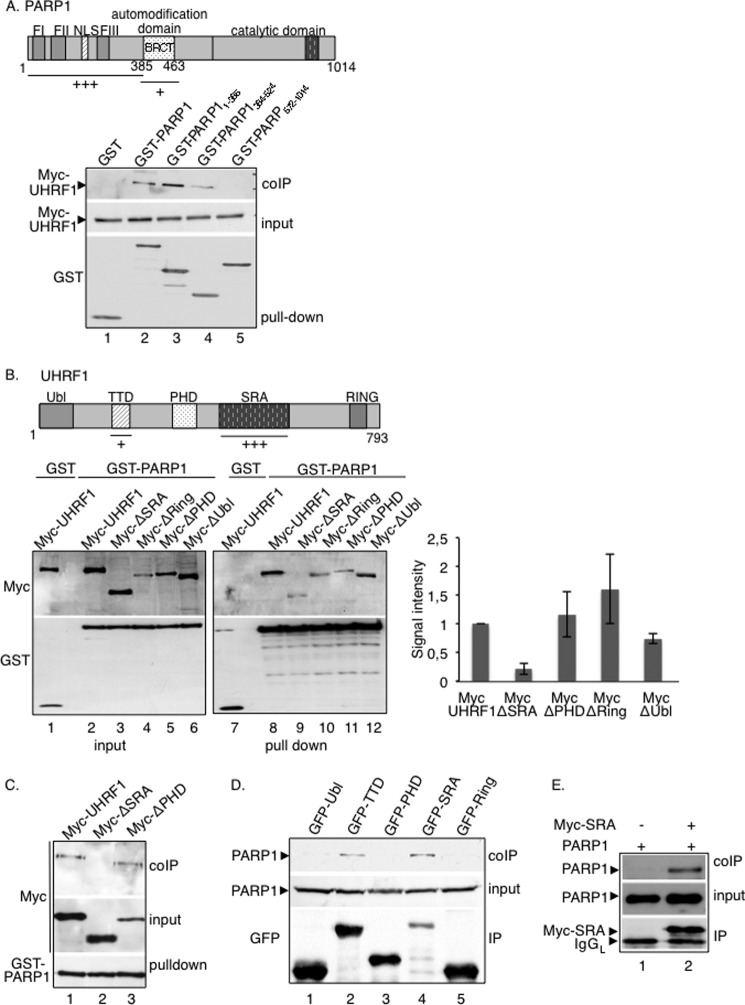
FIGURE 2.
The UHRF1-PARP1 interaction preferentially involves the DNA-binding domain of PARP1 and the SRA domain of UHRF1. A, UHRF1 interacts preferentially with the DNA-binding domain and weakly with the automodification domain of PARP1. Top, schematic representation of PARP1 indicating the interacting domains identified. Bottom, GST (lane 1), GST-PARP1 (lane 2), GST-PARP1-(1–385) expressing the DNA-binding domain (lane 3), GST-PARP1-(384–524) expressing the automodification domain (BRCT) (lane 4), and GST-PARP1-(572–1014) expressing the catalytic domain (lane 5) were expressed in COS-1 cells together with Myc-UHRF1 (lanes 1–5). Interacting proteins were analyzed by GST pulldown and Western blotting with subsequent anti-Myc and anti-GST antibodies. Input corresponds to one-tenth the amount of cell extract used for GST pulldown. B, the absence of the SRA domain impairs the interaction of UHRF1 with PARP1. Top, schematic representation of UHRF1 indicating the interacting domains identified. Bottom left, Myc-UHRF1 (lanes 1, 2, 7, and 8) or Myc-tagged deletion mutants of UHRF1 (lanes 3–6 and 9–12) were expressed in COS-1 cells together with either GST (lanes 1 and 7) or GST-PARP1 (lanes 2–6 and 8–12). Interacting proteins were analyzed as described in A (lanes 7–12). Input corresponds to 1/60th the amount of cell extract used for GST pulldown (lanes 1–6). Bottom right, the signal intensities of the coprecipitating Myc-tagged proteins relative to their expression and GST-PARP1 pulldown were measured in three independent experiments using ImageJ. The coprecipitation of Myc-UHRF1 was set to 1. Mean values ± S.D. are indicated. C, at higher stringency conditions of the washing buffer, the association of Myc-ΔSRA with GST-PARP1 is lost (lane 2), whereas the association of Myc-UHRF1 (lane 1) or Myc-ΔPHD (lane 3) with GST-PARP1 is maintained. The experiment was done as described in B except that the stringency conditions of the washing buffers were increased (beads were washed twice with washing buffer containing 750 mm NaCl, once with washing buffer containing 500 mm NaCl, and twice with washing buffer containing 150 mm NaCl). D, PARP1 interacts preferentially with the SRA domain of UHRF1 and to a lesser extent with the TTD. GFP fusion proteins expressing different domains of UHRF1 (GFP-Ubl, ubiquitin-like domain (lane 1); GFP-TTD, Tudor domain (lane 2); GFP-PHD, plant homeodomain (lane 3); GFP-SRA, SET- and Ring-associated domain (lane 4); GFP-Ring, Ring domain (lane 5)) were expressed in COS-1 cells. GFP immunoprecipitates were blotted successively with an anti-PARP1 antibody to detect coprecipitating PARP1 and an anti-GFP antibody to detect the GFP immunoprecipitates. E, in vitro interaction of PARP1 with immunopurified Myc-SRA. Purified recombinant PARP1 was incubated in a batch assay together with immunopurified Myc-SRA (lane 2) or a control anti-Myc immunoprecipitate (lane 1). Bound PARP1 was analyzed by Western blotting using, successively, anti-PARP1 and anti-Myc antibodies.
For GFP-Trap capture, 1 × 106 COS-1 cells were transfected by JetPEI with 8 μg of total recombinant DNA. Forty-eight hours later, cells were lysed in RIPA-like buffer as described above and incubated with the GFP-Trap®_A (Chromotek, Planegg-Martinsried, Germany) overnight at 4 °C for affinity purification of the GFP-tagged protein. Beads were subsequently washed twice with washing buffer (10 mm Tris-HCl, pH 8, 0.1% Nonidet P-40, 0.5 mm Pefabloc, and protease inhibitor complex) containing 500 mm NaCl and twice with washing buffer containing 150 mm NaCl. The final pellets were resuspended in Laemmli buffer and subjected to 4–20% SDS-PAGE for protein analysis or processed for in vitro PARylation as detailed below.
For the in vitro binding assays, 4 × 106 COS-1 cells were transfected by JetPEI with 8 μg of Myc-SRA. Forty-eight hours later, cells were lysed in RIPA-like buffer as describe above, incubated with the anti-Myc antibody overnight at 4 °C followed by a 2-h incubation at 4 °C with protein A/G-Sepharose (GE Healthcare). Next, beads were resuspended in 100 μl of dilution buffer (20 mm Tris-HCl pH 7,5, 0.1% Nonidet P-40, 0.5 mm Pefabloc) and incubated together with 300 ng of purified recombinant PARP1 for 20 min at 30 °C. Beads were then washed twice with dilution buffer containing 750 mm NaCl, twice with dilution buffer containing 400 mm NaCl, and twice with dilution buffer containing 150 mm NaCl. The final pellets were resuspended in Laemmli buffer and subjected to 10% SDS-PAGE.
For immunoprecipitation experiments of endogenous proteins, RIPA-like cell extracts were precleared by incubation on protein A/G-Sepharose beads for 1 h at 4 °C before incubation with the indicated antibodies overnight at 4 °C followed by a 2-h incubation at 4 °C with protein A/G-Sepharose (GE Healthcare). Beads were washed four times with 20 mm Tris-HCl, pH 7.5, 150–750 mm NaCl, 0.1% Nonidet P-40, and 0.5 mm Pefabloc, resuspended in Laemmli buffer, and analyzed by SDS-PAGE and immunoblotting. Blots were incubated with the appropriate antibodies as indicated. When indicated, 100 nm PARP inhibitor Ku-0058948 was added to the culture medium 2 h before lysis and maintained throughout the experiment.
In Vitro PARylation
For poly(ADP-ribosyl)ation of immunopurified proteins, purified PARP1 (1 μg) was incubated with immunopurified Myc-tagged UHRF1, Myc-TRF2 as a positive control, GFP, or GFP-tagged single domains of UHRF1 for 10 min at 25 °C in 480 μl of activity buffer (50 mm Tris-HCl, pH 8, 0.2 mm dithiothreitol, 4 mm MgCl2, 0.1 μg/μl BSA, and 100 nm NAD+ (for PARP1) or 1 μm NAD+ (for PARP2)) containing 2 μCi of [α-32P]NAD+ (800 Ci/mmol, PerkinElmer Life Sciences) and 900 ng of DNase I-activated calf thymus DNA. The reaction was stopped by the addition of 500 μl of cold washing buffer (50 mm Tris-HCl, pH 8, 400 mm NaCl, 0.1% Nonidet P-40, and 0.5 mm PMSF) on ice, and beads were washed five times with washing buffer and resuspended in 20 μl of Laemmli buffer. Reaction products were analyzed by gel electrophoresis on 10% SDS-PAGE and autoradiography. For PARylation of purified recombinant UHRF1 tested in ubiquitination assays, purified PARP1 (100 ng) was incubated alone or together with purified UHRF1 (100 ng) for 20 min at 25 °C in 15 μl of activity buffer (25 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 1 mm dithiothreitol, 0.1 μg/μl BSA, 5 ng/μl DNA, and 10 μm NAD+).
Immunofluorescence Studies
Immunofluorescence was performed essentially as described previously (13). Briefly, cells grown on glass coverslips were washed twice with PBS and fixed for 15 min with PBS, 3.7% formaldehyde, followed by three washes in blocking buffer (PBS, 0.1% Triton X-100, and 0.1% milk). Cells were then incubated overnight at 4 °C with the primary antibodies diluted in blocking buffer. After three washes with PBS, 0.1% Triton X-100, and 0.1% milk, cells were incubated for 3 h at room temperature with the appropriate conjugated secondary antibodies diluted in blocking buffer. DNA was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI, 25 ng/ml in PBS). Slides were mounted using Mowiol 4-88 (Hoechst), and immunofluorescence microscopy was performed using a Leica microscope (Leica Microsystems, Heidelberg, Germany) and the capture software (Improvision, PerkinElmer Life Sciences).
DNA Methylation Analysis
Methylation of mouse major and minor satellite repeats was determined by pyrosequencing of bisulfite-treated, PCR-amplified, genomic DNA as described (46).
RT-PCR Analysis
Total RNA was extracted from ∼3 × 106 PARP1+/+ and PARP1−/− cells with TRIzol (Invitrogen) according to the manufacturer's instructions. The remaining DNA was digested by incubation with RNase-free DNase I (Promega) (1 unit/μg RNA), and isolated RNA was purified using the RNA Clean-up XS kit (Machery-Nagel). Reverse transcription (RT) was done on 800 ng of purified RNA using oligo(dT) primers (Sigma) and the AMV reverse transcriptase (Finnzyme). PCR reactions were performed on 5% of the RT volume using the Phusion polymerase (Finnzyme) and the following primer pairs: major satellite repeat forward, 5′- GACGACTTGAAAAATGACGAAATC-3′; major satellite repeat reverse, 5′-CATATTCCAGGTCCTTCAGTGTGC-3′; GAPDH forward, 5′- TTCTGAGTGGCAGTGATGGC-3′; and GAPDH reverse, 5′- AACAACCCCTTCATTGACCTC-3′. Transcripts were analyzed on ethidium bromide-stained agarose gels using the Typhoon instrument and quantified by ImageJ.
In Vivo Ubiquitination Assay
PARP1+/+ and PARP1−/− cells were co-transfected with 5 μg of GFP, GFP-DNMT1, or Myc-UHRF1 and 5 μg of HA-Ub using the jetPEI method. Thirty-six hours later, the cells were treated with 5 μm MG-132 (Enzo) for another 12 h and lysed with RIPA-like buffer as described above. After GFP quantification by Western blotting, equivalent amounts of GFP-DNMT1 were immunopurified using GFP-Trap®_A (Chromotek) as described above. Myc-UHRF1 was immunoprecipitated using an anti- Myc antibody as described above. Beads were washed twice with washing buffer A (20 mm Tris-HCl, pH 7.5, 0.1% Nonidet P-40, 400 mm NaCl, and 0.5 mm Pefabloc), twice with washing buffer B (20 mm Tris-HCl, pH 7.5, 0.1% Nonidet P-40, 150 mm NaCl, and 0.5 mm Pefabloc), resuspended in Laemmli buffer, and processed for Western blotting. Twenty-five % of the sample was loaded for detecting GFP-DNMT1 or Myc-UHRF1, and 75% of the sample was loaded for detecting GFP-Dnmt-1Ub or Myc-UHRF1Ub using the anti-HA antibody.
In Vitro Ubiquitination Assay
Purified recombinant UHRF1 (100 ng (31)) was first incubated alone or together with purified recombinant PARP1 (100 ng) in a PARP activity buffer with or without NAD+ as described above. After 20 min at 25 °C, PARylated or non-PARylated (PARP assay performed without NAD+) UHRF1 was subsequently incubated alone or together with purified recombinant GST-DNMT1 (300 ng, BPS Bioscience) as indicated in a standard ubiquitination reaction mixture (15 μl) containing 100 ng of human recombinant ubiquitin-activating enzyme (Boston Biochem), 300 ng of human recombinant UbcH5b (Boston Biochem), and 10 μg of Ha-tagged human recombinant ubiquitin (Boston Biochem) in 25 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 1 mm DTT, 2 mm ATP, and 100 mm NaCl. During ubiquitination, PARP activity was inhibited by adding 330 nm Ku-0059848. The reaction was incubated at 37 °C for 1 h and stopped by adding 1,25× final Laemmli buffer. Ubiquitinated proteins were analyzed by Western blot analysis; one-half of the final volume was used for HA-ubiquitin detection and one-eighth of the volume was used for poly(ADP-ribose) polymer detection.
RESULTS
PARP1 Interacts with the E3 Ubiquitin Ligase UHRF1 in a PAR-dependent Manner
Previous studies have identified PARP1 together with DNMT1 in a proteomic analysis of anti-UHRF1 immunoprecipitates, thus making PARP1 a possible candidate for modulating UHRF1-DNMT1 functional interplay (24).
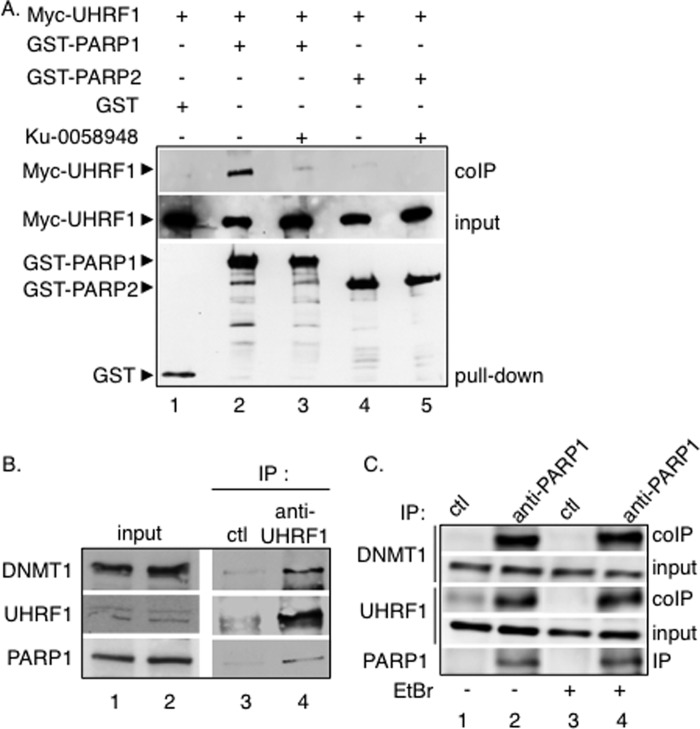
To expand on these findings, we decided to investigate the comparative association of PARP1 and PARP2 with UHRF1 (Fig. 1A). COS-1 cells were transfected with Myc-tagged UHRF1 together with GST, GST-fused PARP1, or GST-fused PARP2. After glutathione-Sepharose beads were trapped, copurified Myc-UHRF1 was assessed by Western blot analysis using an anti-Myc antibody. To test the role of PARylation in the interaction, pulldown assays were performed in the presence of the potent PARP inhibitor Ku-0058948. As shown in Fig. 1A, UHRF1 was clearly coprecipitated with GST-PARP1 (lane 2) but not efficiently with GST-PARP2 (lanes 4–5) and not with GST alone (lane 1), defining a preferential association of UHRF1 with PARP1 compared with PARP2. Furthermore, a significantly weaker copurification of UHRF1 with GST-PARP1 was observed in the presence of Ku-0058948 (Fig. 1A, lane 3), revealing a critical role of PARylation in their association. The UHRF1-PARP1 interaction and the association with DNMT1 was next verified by coimmunoprecipitation experiments with the endogenous proteins (Fig. 1, B and C). Using an anti-UHRF1 antibody, we efficiently coimmunoprecipitated both PARP1 and DNMT1 from 3T3 mouse extracts (Fig. 1B, lane 4), whereas no coprecipitation was detected using a control antibody (Fig. 1B, lane 3). In a reciprocal experiment (Fig. 1C), when 3T3 cell extracts were immunoprecipitated with an anti-PARP1 antibody, significant fractions of UHRF1 and DNMT1 were detected in the PARP1 immunoprecipitate (lane 2) but not in the control immunoprecipitate (lane 1). To further prevent any coprecipitation of either of the proteins through DNA, we performed the coprecipitation experiments in the presence of ethidium bromide, which intercalates into DNA and thereby competes for interacting proteins (Fig. 1C, lanes 3 and 4). The addition of ethidium bromide did not abolish the DNMT1-UHRF1-PARP1 interaction, suggesting that DNA was not involved (Fig. 1C, compare lanes 4 and 2). Together these results describe the existence of a protein complex containing PARP1, UHRF1, and DNMT1 in mammalian cells.
FIGURE 1.
Selective PAR-dependent association of UHRF1 with PARP1 in mammalian cells. A, selective coprecipitation of Myc-UHRF1 with GST-PARP1. GST (lane 1), GST-PARP1 (lanes 2 and 3), and GST-PARP2 (lanes 4 and 5) were expressed in COS-1 cells together with Myc-tagged UHRF1 (lanes 1–5). Interacting proteins were analyzed by GST pulldown and Western blotting with subsequent anti-Myc and anti-GST antibodies. Input corresponds to 1/60th the amount of cell extract used for GST pulldown. In lanes 3 and 5, the PARP inhibitor Ku-0058948 was added throughout the experiment. B, coimmunoprecipitation of PARP1 and DNMT1 with UHRF1 in mouse 3T3 cells. Wild-type mouse cell extracts were immunoprecipitated with an anti-UHRF1 antibody (lane 4) or a control antibody (Ctl, lane 3) and analyzed by Western blotting. Input (lanes 1 and 2) corresponds to 1/30th the amount of cell extract used for immunoprecipitation. C, coimmunoprecipitation of UHRF1 and DNMT1 with PARP1 in mouse 3T3 cells. Wild-type mouse cell extracts were immunoprecipitated with an anti-PARP1 antibody (lanes 2 and 4) or a control antibody (lanes 1 and 3) and analyzed by Western blotting. To prevent any coprecipitation of either partner through DNA, EtBr (10 μg/ml) was added throughout the immunoprecipitation when indicated (lanes 3 and 4). Input corresponds to 1/30th the amount of cell extract used for immunoprecipitation. The association of PARP1 with UHRF1 is not mediated by DNA.
To further characterize the UHRF1-PARP1 association, we aimed to identify the region of PARP1 to which UHRF1 binds (Fig. 2A). GST fusion proteins expressing truncated versions of PARP1 (amino acids 1–385 (DNA-binding domain), amino acids 384–524 (automodification domain), and amino acids 572–1014 (catalytic domain)) were tested for their interaction with Myc-UHRF1. Myc-UHRF1 coprecipitated efficiently with full-length PARP1 and its DNA-binding domain (Fig. 2A, lanes 2 and 3) and less efficiently with the automodification domain, defined as the site of autoPARylation (lane 4). No coprecipitation was detected with GST alone or the PARP catalytic domain (Fig. 2A, lanes 1 and 5). As noted above, the association of Myc-UHRF1 with the DNA-binding domain of PARP1 was not impaired by the presence of ethidium bromide (not shown).
In reciprocal experiments intended to identify the PARP1 interaction domain within UHRF1, Myc fusion proteins expressing various UHRF1 deletion domains were tested for their interaction with GST-PARP1 (Fig. 2B). When compared with the expression profile of each fusion protein (Fig. 2B, lanes 1–6) and the efficient binding of the full-length Myc-UHRF1 (lane 8), only the internal deletion of the SRA domain significantly reduced UHRF1 binding to GST-PARP1 (lane 9). No unspecific binding to GST was detected (Fig. 2B, lane 1). As an additional experiment, we then compared the binding of Myc-UHRF1, Myc-ΔSRA (UHRF1 deleted for the SRA domain), and Myc-ΔPHD (UHRF1 deleted for the PHD domain) to GST-PARP1 as above but under higher stringency conditions of the washing buffer (Fig. 2C). Although the association of Myc-ΔSRA with GST-PARP1 was lost (Fig. 2C, lane 2), the association of Myc-UHRF1 and Myc-ΔPHD with GST-PARP1 was maintained (lanes 1 and 3). To further ascertain the selective interaction with the SRA domain and exclude a possible misfolding of the deleted constructs, GFP fusion proteins expressing the different domains (Ubl, TTD, PHD, and SRA) were tested for their interaction with endogenous PARP1 (Fig. 2D). When compared with the expression profile of each fusion protein, we confirmed that PARP1 coprecipitated efficiently with GFP-SRA despite its weakest expression (Fig. 2D, lane 4), and we identified a coprecipitation of PARP1 with the TTD domain (lane 2). No coprecipitation of PARP1 was detected with the Ubl, PHD, or Ring domain (Fig. 2D, lanes 1, 3, and 5). To verify the preferential interaction with the SRA in vitro, Myc-SRA was expressed in COS-1 cells, immunopurified using an anti-Myc antibody, and incubated together with purified recombinant PARP1. After Myc immunoprecipitation followed by stringent washes, copurification of purified PARP1 was analyzed by Western blotting. As shown in Fig. 2E, PARP1 efficiently copurified with Myc-SRA (lane 2), whereas no PARP1 was detected in the control anti-Myc immunoprecipitate (lane 1). Taken together, these data identified a selective and PAR-dependent physical interaction between PARP1 and UHRF1 that requires the DNA-binding domain and, to a lesser extent, the BRCT domain of PARP1 and preferentially the SRA domain, but also the TTD, of UHRF1.
UHRF1 Is PARylated by PARP1 Preferentially onto Its SRA Domain and to a Lesser Extent onto Its TTD
To investigate the functional relevance of the PARP1-UHRF1 interaction, we next evaluated the ability of PARP1 to PARylate UHRF1 in vitro. To this end, Myc-tagged full-length UHRF1 and Myc-tagged TRF2 used as a positive PARylated control were expressed in COS-1 cells, purified by anti-Myc immunoprecipitation followed by stringent washes, and incubated with PARP1 or no protein in the presence of [α-32P]NAD+ and DNase-I treated calf thymus DNA (Fig. 3A). When compared with the expression profile of each fusion protein (Fig 3A, lanes 5–8), autoradiography revealed PARylation of the positive control TRF2 as described previously (lane 3 (5)) and a significant PARylation of full-length UHRF1 (lane 1). No PARylation was observed in the absence of PARP1 (Fig 3A, lanes 2 and 4). We next aimed to map the PARylated domain within UHRF1 (Fig. 3B). To this end, a PARylation assay was performed as described above using immunopurified GFP (Fig. 3B, lanes 1 and 7) or GFP fusion proteins expressing the different Ring (lanes 2 and 8), SRA (lanes 3 and 9), PHD (lanes 4 and 10) Ubl (lanes 5 and 11), and TTD (lanes 6 and 12) single domains of UHRF1. Autoradiography revealed a significant PARylation of the SRA domain (Fig. 3B, lane 3) despite its weakest expression (lane 9) and to a lesser extent the TTD (lanes 6 and 12), whereas no PARylation was detected for the other domains or GFP alone (lanes 1, 2, 4, and 5). An efficient PARylation was also detected on the immunopurified Myc-tagged SRA, whereas a reduced PARylation was detected for the Myc-tagged SRA-deleted mutant of UHRF1 (not shown). Together, these results identified the SRA domain as a preferential site of PARylation, in agreement with the recent report by Zhang et al. (47), who identified two site-specific ADP-ribosylated residues within the SRA domain of UHRF1 by boronate affinity chromatography used to isolate ADP-ribosylated peptides. To a lesser extent, the TTD is also PARylated.
FIGURE 3.
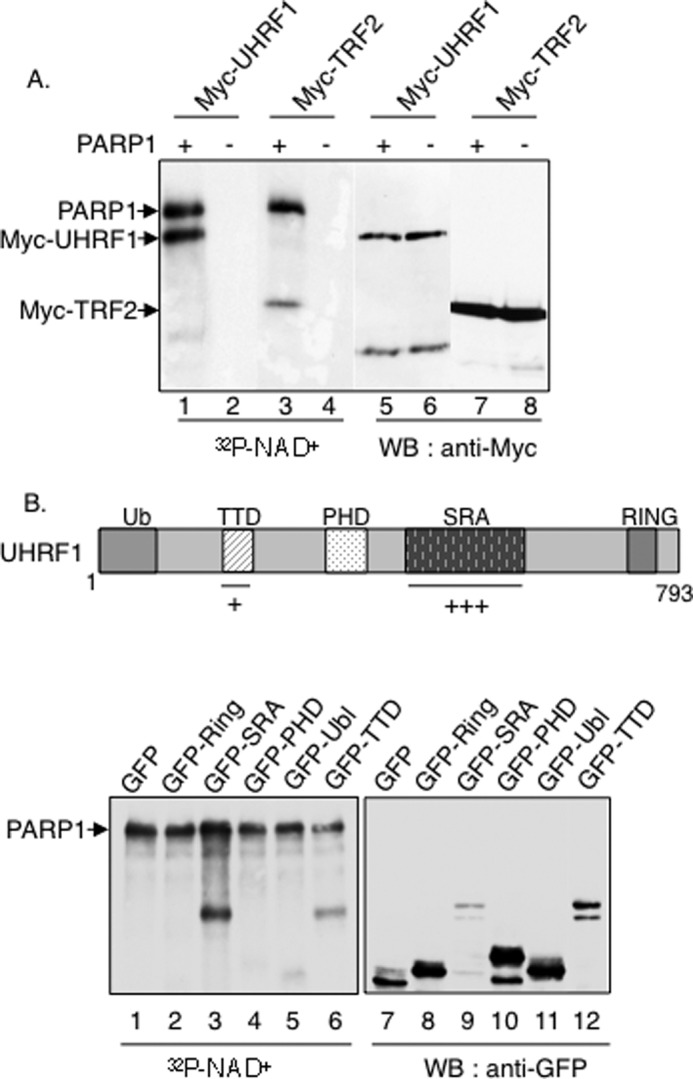
UHRF1 is PARylated preferentially onto its SRA domain but also onto its TTD. A, PARylation of full-length UHRF1 by PARP1. Myc-UHRF1 (lanes 1, 2, 5, and 6) or Myc-TRF2 (lanes 3, 4, 7, and 8) was expressed in COS-1 cells, immunopurified with an anti-Myc antibody, and incubated together with either PARP1 or without PARPs in activity buffer containing [α-32P]NAD+ and fragmented DNA. Right panel, autoradiography. Left panel, analysis of the fusion proteins with an anti-Myc antibody by Western blotting. B, upper panel, schematic representation of UHRF1 indicating the PARylated TTD and SRA domain. Lower panel, GFP fusion proteins expressing GFP alone (lanes 1 and 7) or different domains of UHRF1 (GFP-Ring (lanes 2 and 8), GFP-SRA (lanes 3 and 9), GFP-PHD (lanes 4 and 10), GFP-Ubl (lanes 5 and 11), and GFP-TTD (lanes 6 and 12)) were expressed in COS-1 cells, immunopurified by GFP trapping, and incubated together with PARP1 in the activity buffer containing [α-32]PNAD+ and fragmented DNA as in A. Left, analysis of the PARylated domains by autoradiography. The upper band represents PARylated PARP1. Right, analysis of the fusion proteins with an anti-GFP antibody by Western blotting.
PARP1 and UHRF1 Favor the Silencing of Major Satellite Repeats
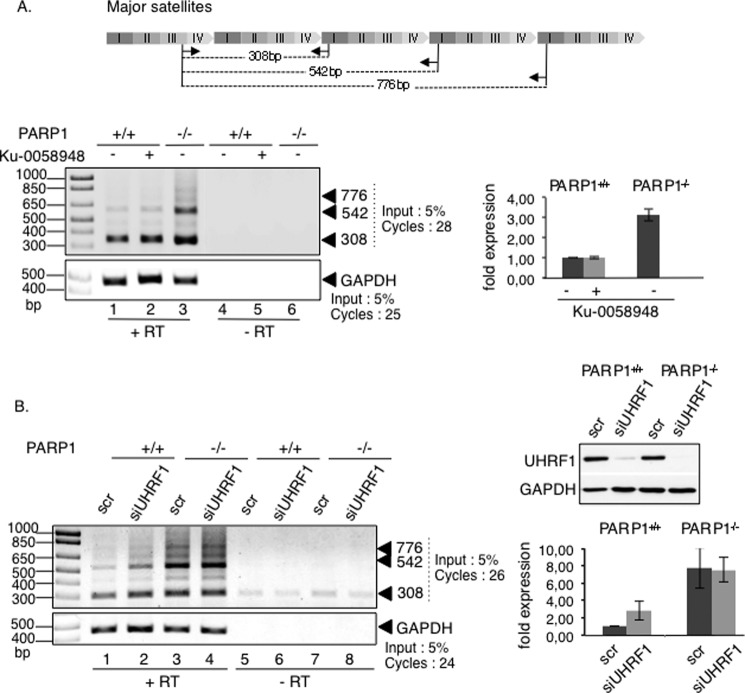
UHRF1 was described previously as contributing to the silencing of major satellites sequences, partly by targeting DNMT1 for DNA maintenance methylation during pericentric heterochromatin replication (23, 24, 42). In addition, PARP1 was found associated with the major satellite elements of pericentric heterochromatin (12). To get insights into the biological meaning of the UHRF1-PARP1 association, we first sought to determine whether PARP1 was also involved in the transcriptional silencing of major satellite repeats. To this aim, we analyzed the transcriptional activity across these elements by semiquantitative RT-PCR using RNA extracts from the PARP1+/+ and PARP1−/− cells (Fig. 4A). Our data revealed a significant increase of the satellite transcripts from pericentric regions in PARP1−/− cells compared with the PARP1+/+ cells (Fig. 4A, lane 3 versus 1). In contrast the inhibition of PARP activity in PARP1+/+ cells did not alter the overall transcription of these regions (Fig. 4A, lane 2 versus 1). To explore the contribution of UHRF1 in the transcriptional reactivation of major satellites observed in PARP1−/− cells, we examined the effect of an additional siRNA-mediated knockdown of UHRF1 (Fig. 4B). In agreement with previous data (42), the depletion of UHRF1 by siRNA in PARP1+/+ cells induced pericentric transcription, although to a significant lower extent than the absence of PARP1 (Fig. 4B, lanes 2 and 3 versus 1). In contrast, the depletion of UHRF1 in PARP1−/− cells did not significantly impair the enhanced transcriptional activity induced by the absence of PARP1, thus suggesting that both enzymes likely act in the same pathway (Fig. 4B, lane 4 versus 3). In a comparative experiment, we also analyzed the transcriptional activity of minor satellites. In agreement with the previously described association of PARP1 with centromeric regions (12), we also detected an important increase in the transcripts of centric regions, indicating a role of PARP1 in the silencing of these regions (not shown). However, the mechanism involved is likely unrelated to UHRF1, as suggested by the previously reported normal transcription of these regions in the absence of UHRF1 (42).
FIGURE 4.
The absence of PARP1 causes derepression of major satellite transcripts. Enhanced transcriptions of major satellite repeats in PARP1−/− cells are shown. A, top, schematic representation of major satellite repeats (pericentric heterochromatin) showing the repeat distribution (I–IV) and the primers (arrows) used for PCR analysis. Lower left, representative RT-PCR analysis using total RNA extracted from PARP1+/+ (lanes 1, 2, 4, and 5) or PARP1−/− cells (lanes 3 and 6), mock-treated cells (lanes 1, 3, 4, and 6), or cells treated with Ku-0058948 (lanes 2 and 5) for 48 h. As a control, reactions were performed with mock-transcribed cDNAs (−RT, lanes 4–6). The relative amounts of input and PCR amplification cycles are indicated. Lower right, the -fold expression (histogram) represents the major satellite transcript levels (normalized against GAPDH) relative to control PARP1+/+ set to 1 for three independent experiments. Mean data ± S.D. are indicated. B, left, representative RT-PCR analysis using total RNA extracted from PARP1+/+ (lanes 1, 2, 5, and 6) or PARP1−/− cells (lanes 3, 4, 7, and 8) transfected with a scrambled siRNA (scr, lanes 1, 3, 5, and 7) or with siUHRF1 (lanes 2, 4, 6, and 8) for 96 h. As a control, reactions were performed with mock-transcribed cDNAs (−RT, lanes 5–8). Right, the -fold expression (histogram) represents the major satellite transcript levels (normalized against GAPDH) relative to control PARP1+/+ set to 1 for three independent experiments. Mean data ± S.D. are indicated.
The Absence of PARP1 Alters the Interaction of UHRF1 with DNMT1 but with No Consequences for the Methylation of Pericentric Regions
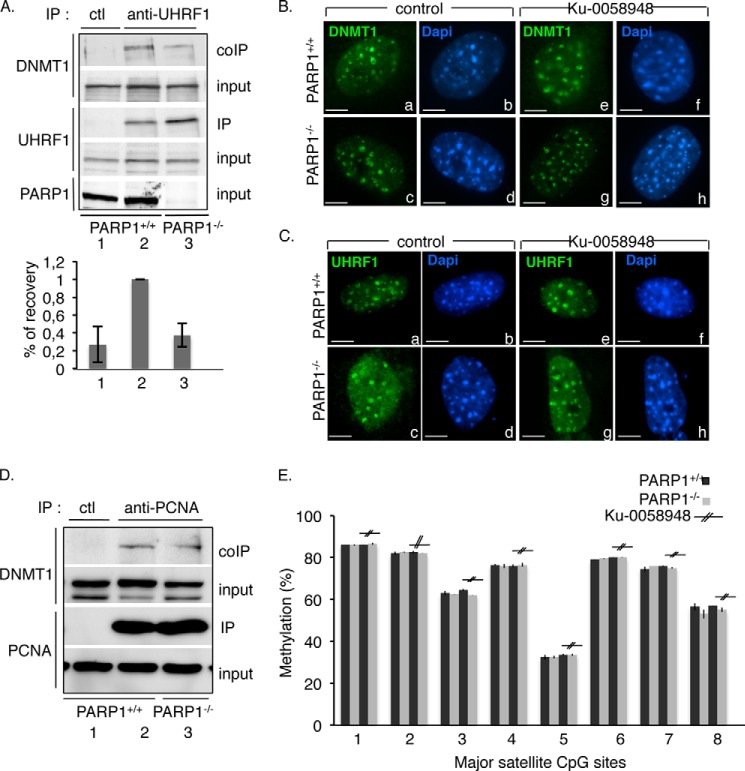
Enhanced transcriptional activity at pericentric regions could be caused by defective DNMT1-catalyzed CpG methylation. Among the UHRF1 functional domains that facilitate the loading of DNMT1 to replicating heterochromatic regions, the SRA domain mediates the recognition and preferential binding to hemi-methylated CpG sites and facilitates the interaction of UHRF1 with DNMT1 (24, 29, 30). Because we identified SRA as the preferential interaction domain (Fig. 2C) and PARylation site for PARP1 (Fig. 3B), we went on to test whether PARP1 is involved in the association of UHRF1 with DNMT1 (Fig. 5A). To address this question, whole extracts from PARP1+/+ or PARP1−/− 3T3 cells were immunoprecipitated with an anti-UHRF1 antibody and probed for the coimmunoprecipitation of DNMT1 by Western blotting. We observed a significantly reduced coimmunoprecipitation of DNMT1 with UHRF1 in PARP1−/− cells (Fig. 5A, lane 3) compared with PARP1+/+ (lane 2) cells, indicating that PARP1 is required for the efficient association of DNMT1 with UHRF1. No unspecific coprecipitation was observed with the control antibody (Fig. 5A, lane 1).
FIGURE 5.
The absence of PARP1 affects the association of UHRF1 with DNMT1 but not their targeting to replicating heterochromatin or the methylation of CpG repeats. A, the interaction of DNMT1 with UHRF1 is reduced in PARP1−/− cells. Left, equivalent amounts of total protein cell lysates from PARP1+/+ (lane 2) and PARP1−/− cells (lane 3) were immunoprecipitated using an anti-UHRF1 antibody (lanes 2 and 3) or a control antibody (ctl, lane 1) and analyzed for the coimmunoprecipitation of DNMT1 by Western blotting. Inputs correspond to 1/30th of the amount of total cell extract used for immunoprecipitation. Right, the signal intensities of the coprecipitating DNMT1 relative to DNMT1 expression and UHRF1 immunoprecipitation were measured in three independent experiments using ImageJ. The coIP in PARP1+/+ cells (lane 2) was set to 1. Mean values ± S.D. are indicated. B, the absence of PARP1 does not perturb the accumulation of DNMT1 onto pericentric heterochromatin. Shown are representative images of DNMT1 (a, c, e, and g (green)) immunostaining of the typical ring-shaped pericentric duplication bodies from PARP1+/+ (a, b, e, and f) or PARP1−/− (c, d, g, and h) cells either mock-treated (a–d) or treated with the PARP inhibitor Ku-0058948 (e–h). DNA was counterstained with DAPI (b, d, f, and h (blue)). Scale bars: 7 μm. C, the absence of PARP1 does not perturb the accumulation of UHRF1 onto pericentric heterochromatin. Shown are representative images of UHRF1 (a, c, e, and g (green)) immunostaining of the typical ring-shaped pericentric duplication bodies from PARP1+/+ (a, b, e, and f) or PARP1−/− (c, d, g, and h) cells either mock-treated (a–d) or treated with the PARP inhibitor Ku-0058948 (e–h). DNA was counterstained with DAPI (b, d, f, and h (blue)). Scale bars: 7 μm. D, the interaction of DNMT1 with PCNA is maintained in PARP1−/− cells. Equivalent amounts of total protein cell lysates from PARP1+/+ (lanes 1 and 2) and PARP1−/− cells (lane 3) were immunoprecipitated using and anti-PCNA antibody (lanes 2 and 3) or a control antibody (lane 1) and analyzed for the coimmunoprecipitation of DNMT1 by Western blotting. Input corresponds to 1/25th of the amount of total cell extract used for immunoprecipitation. E, the methylation profile of pericentric repeats is normal in PARP1−/− cells. Total DNA was isolated from PARP1+/+ or PARP1−/− cells either mock-treated or treated with Ku-0058948 for 24 h and then bisulfite-treated. Histograms show the methylation percentage at individual CpG sites as measured by pyrosequencing.
Consequently, we went on to examine by immunofluorescence analysis whether the reduced DNMT1-UHRF1 association affects the focal localization of DNMT1 to replicating heterochromatin (Fig. 5B). Because both the TTD and SRA domain of UHRF1 mediate its targeting to pericentric heterochromatin, we also verified the accumulation of UHRF1 onto these regions (Fig. 5C). Notwithstanding, we found a normal accumulation of both proteins onto the characteristic horseshoe-like replication factories in the PARP1−/− cells (Fig. 5C, compare panels c and d, with a and b) as well as after PARP inhibition (compare panels e–h with a and b) suggesting that PARP1 and PARP activity is not absolutely required for the mid-S phase-specific targeting of UHRF1 and DNMT1 to pericentric heterochromatin.
Because loading of DNMT1 onto replication foci was also shown to be promoted by PCNA (48), we explored the role of PARP1 in the association of DNMT1 with PCNA. We compared the coimmunoprecipitation of DNMT1 using an anti-PCNA antibody in PARP1+/+ versus PARP1−/− cells (Fig. 5D). We observed a similarly efficient interaction of DNMT1 with PCNA (Fig. 5D, lane 2 versus 3) in both cell lines, indicating that the association between these proteins is maintained in the absence of PARP1. No precipitation of DNMT1 was detected using the control antibody (Fig. 5D, lane 1).
Finally we verified whether the reduced DNMT1-UHRF1 association perturbs DNMT1 catalytic activity. We analyzed the methylation status of pericentric repeats by genomic bisulfite sequencing of DNA extracted from PARP1+/+ and PARP1−/− cells treated or not with the PARP inhibitor Ku-0058948 (Fig. 5E). Consistent with the normal recruitment of DNMT1 and UHRF1 onto heterochromatic foci, no apparent difference in the methylation profile of these repetitive regions was detected between both cell lines and after PARP inhibition. Similarly, the absence of PARP1 did not affect the methylation profile of minor satellites at centromeres (not shown).
Taken together, these results identified a role of PARP1 in stabilizing the interaction of UHRF1 with DNMT1. Despite the reduced interaction between DNMT1 and UHRF1 in the absence of PARP1, the recruitment of DNMT1 to heterochromatic regions and its activity are maintained owing to its efficient interaction with PCNA. Moreover, these data indicate that the derepression of the major satellites detected in the absence of PARP1 might not simply be caused by impaired DNA methylation but likely involves another defect that we aimed to identify next.
PARP1 and UHRF1 Cooperate to Regulate the Repressive Mark H4K20me3
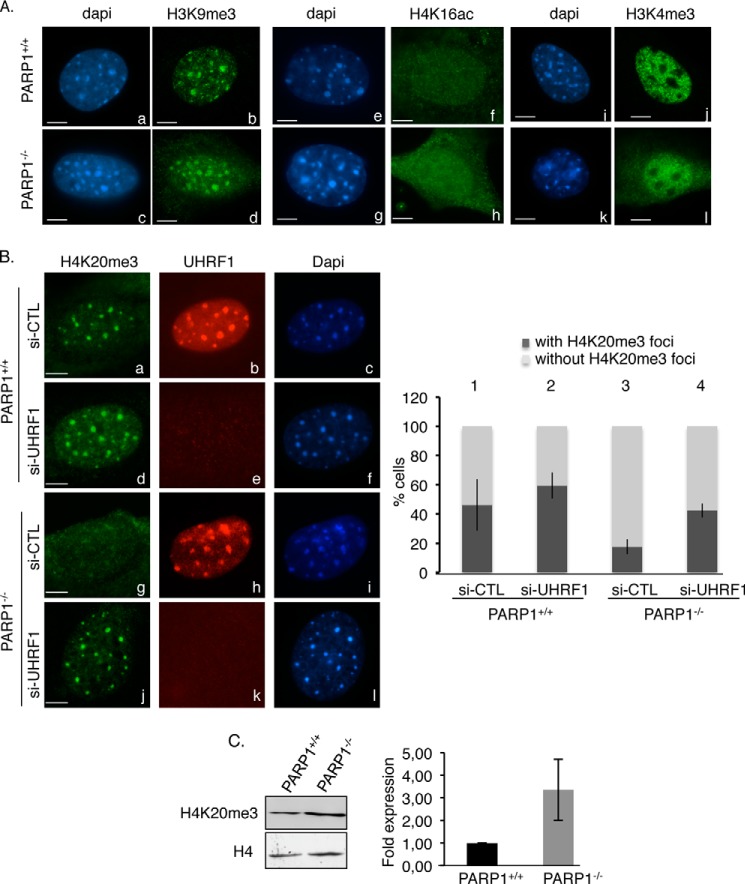
To further decipher how PARP1 regulates the silencing of pericentric regions, given the link of UHRF1 with repressive chromatin marks (24, 27, 30) we then asked whether the transcriptional activation was accompanied by modifications of the chromatin signatures of pericentric heterochromatin. We looked for H3K9 and H4K20 trimethylation, which is linked with silencing, and H4K16 acetylation or H3K4 trimethylation, which is required for gene activation (Fig. 6). The staining of H3K9m3 remained unchanged, and no detectable acetylation of H4K16 or trimethylation of H3K4 could be detected at DAPI-dense heterochromatic regions of PARP1−/− cells (Fig. 6A, compare panels d, h, and l with b, f, and j, respectively). In contrast, the absence of PARP1 led to a specific loss of the local repressive H4K20me3 mark at heterochromatic foci in most of the cells (>80%), thus indicating a less compact heterochromatin structure that could contribute to up-regulation of the heterochromatin transcripts described above (Fig. 6B, compare panel g with a and vertical bar 3 with 1). Remarkably, the additional depletion of UHRF1 rescued the localization of H4K20me3 to heterochromatin regions in the PARP1−/− cells (Fig. 6B, compare panel j with g and vertical bar 4 with 3), whereas it did not perturb H4K20me3 staining in the PARP1+/+ cells (compare panel d with a and vertical bar 2 with 1). To ascertain that the absence of H4K20me3 heterochromatic staining was not due simply to an overall reduction in H4K20me3, we evaluated the global expression of H4K20me3 by Western blot analysis of acid-extracted histones from PARP1+/+ and PARP1−/− cells (Fig. 6C). We found rather an increase in the overall expression of H4K20me3 in the PARP1−/− cells. Taken together, these findings highlight a related contribution of PARP1 and UHRF1 in the transcriptional repression of pericentric heterochromatin through a mechanism that at least partly involves a specific regulation of the repressive mark H4K20me3 at heterochromatin.
FIGURE 6.
The absence of PARP1 causes reduced staining of the repressive mark H4K20me3 at pericentric regions, which is rescued by the additional depletion of UHRF1. A, PARP1−/− cells display normal H3K9me3, H4K16ac, and H3K4me3 staining onto DAPI-dense heterochromatic regions. Shown are representative immunofluorescence images for the comparative distribution of H3K9me3 (b and d (green)), H4K16ac (f and h (green)), and H3K4me3 (j and l) in PARP1+/+ and PARP1−/− interphase cells. DNA and heterochromatic foci are counterstained with DAPI (a, c, e, g, i, and k). Scale bars: 7 μm. B, left, representative images for the loss of H4K20me3 staining at HC regions in PARP1−/− cells rescued by the additional depletion of UHRF1. Shown is immunofluorescence analysis of H4K20me3 (a, d, g, and j (green)) and UHRF1 (b, e, h, and k (red)) in PARP1+/+ and PARP1−/− cells transfected with either control siRNA (si-CTL) or si-UHRF1. DNA and heterochromatic foci are counterstained with DAPI (c, f, i, and l). Right, the histogram depicts the percentage of cells with or without H4K20me3 staining. An average of 500 cells/cell line were scored in >20 randomly selected fields. Results are averages from three independent experiments. Mean values ± S.D. are indicated. C, the overall expression of H4K20me3 is weakly increased in the PARP1−/− cells. Left, equivalent amounts of acid-extracted histones from PARP1+/+ and PARP1−/− cells were analyzed by Western blotting using an anti-H4K20me3 antibody and an anti-H4 antibody as loading control. Right, the signal intensities of H4K20me3 relative to H4 were measured in three independent experiments using ImageJ. Mean values ± S.D. are indicated.
PARP1 Restrains UHRF1-mediated Ubiquitination of DNMT1 and Modulates Its Stability during the Cell Cycle
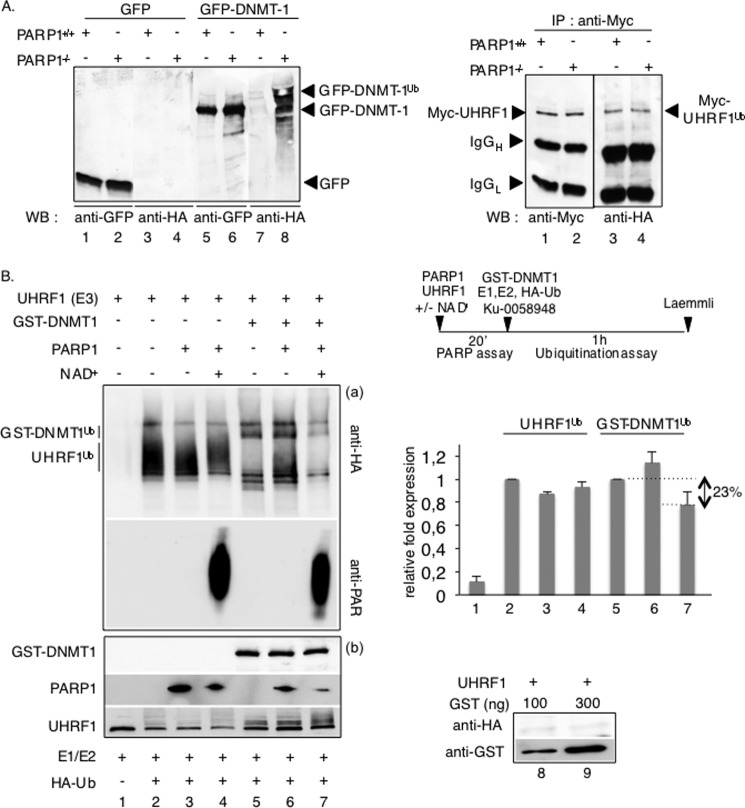
Among the various UHRF1-regulated processes, recent reports have identified an UHRF1-mediated ubiquitination of DNMT1 that coordinately regulates its stability with the completion of DNA replication (34, 35). Because PARylation has also recently been connected with ubiquitination and proteasomal degradation (4, 49, 50), we decided also to explore a possible involvement of PARP1 in this pathway. We compared the level of Myc-UHRF1-mediated ubiquitination of immunopurified GFP-DNMT1 in PARP1+/+ versus PARP1−/− 3T3 cells (Fig. 7A). Strikingly, the absence of PARP1 led to an apparent increase in the ubiquitination levels of GFP-DNMT1 (Fig. 7A, left panel, lane 8 versus 7), whereas the autoubiquitination levels of Myc-UHRF1 were weaker and remained rather unchanged in similar experimental conditions (right panel, lane 2 versus 1). No unspecific ubiquitination of GFP was detected (Fig. 7A, left panel, lanes 3 and 4). Thus, these results introduced PARP1 as a negative regulator of UHRF1-ubiquitin ligase activity onto DNMT1. To confirm and examine this hypothesis further, we performed in vitro ubiquitination assays using purified recombinant PARP1, UHRF1, and GST-DNMT1 in the presence of the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and ubiquitin (Fig. 7B). To discriminate between a structural and enzymatic role for PARP1, UHRF1 was first preincubated together with PARP1 in the absence (inactive PARP1) or presence of NAD+ (active PARP1) and subsequently tested in the ubiquitination assays (Fig. 7B, diagram). The addition of inactive PARP1 did not significantly impact the autoubiquitination of UHRF1 (Fig. 7B, lane 3 versus 2) or the ubiquitination of GST-DNMT1 (lane 6 versus 5). However, when UHRF1 was first PARylated with active PARP1 in the presence of NAD+, its ubiquitination activity onto DNMT1 was significantly reduced (Fig. 7B, lane 7 versus 5), whereas the autoubiquitination of UHRF1 was only weakly if significantly modified (lane 4 versus 2). Under similar experimental conditions, the ubiquitination of GST used as control was never observed (Fig. 7B, lanes 8 and 9). Interestingly, the addition of PARP1, when automodified previously, had no significant impact on UHRF1 activity (not shown). Altogether, these data reveal that the PARP1-catalyzed PARylation of UHRF1 inhibits its ubiquitin ligase activity essentially toward DNMT1.
FIGURE 7.
PARP1 selectively inhibits the UHRF1-driven ubiquitination of DNMT1 in vivo and in vitro. A, in vivo ubiquitination assays. The absence of PARP1 enhances the UHRF1-mediated ubiquitination of DNMT1 but not the autoubiquitination of UHRF1 in vivo. Left, PARP1+/+ and PARP1−/− cells were transfected with either GFP-DNMT1 (lanes 5–8) or GFP (lanes 1–4) together with HA-ubiquitin and treated with 5 μm MG-132 for 12 h to inhibit proteasomal degradation. GFP immunoprecipitates were blotted successively with an anti-HA antibody to detect ubiquitinated proteins (lanes 3, 4, 7, and 8) and an anti-GFP antibody (lanes 1, 2, 5, and 6) to detect immunopurified proteins. Right, PARP1+/+ (lanes 1 and 3) and PARP1−/− cells (lanes 2 and 4) were transfected with Myc-UHRF1 together with HA-ubiquitin and treated as described above. Myc immunoprecipitates were blotted successively with an anti-HA antibody to detect ubiquitinated UHRF1 (lanes 3 and 4) and an anti-Myc antibody to detect immunopurified UHRF1 (lanes 1 and 2). B, in vitro ubiquitination assays. Left, PARP1-catalyzed poly(ADP-ribosyl)ation of UHRF1 selectively inhibits its ubiquitination activity onto DNMT1. Purified UHRF1 was first preincubated alone (lanes 1, 2, and 5) or together with purified PARP1 (lanes 3, 4, 6, and 7) as indicated in the PARP activity buffer. PARP activity is induced by the addition of NAD+. The proteins were subsequently assayed for UHRF1 ubiquitination activity onto itself (lanes 1–4) or onto GST-DNMT1 (lanes 5–7). a, ubiquitinated proteins (UHRF1Ub and GST-DNMT1Ub) were detected by immunoblotting using an anti-HA antibody, and the PARP activity was verified by immunoblotting using an anti-PAR antibody. b, the purified recombinant proteins mixed in the experiment were detected by Western blotting using anti-GST, anti-PARP1, and anti-UHRF1 antibodies. The lower amount of PARP1 detected in lanes 4 and 7 is explained by its automodification, which limits its detection by the monoclonal anti-PARP1 antibody used. As a control, reactions were performed with GST (lanes 8 and 9). A representative experiment of three is shown. Upper right, a schematic diagram of the experiment is shown. Lower right, the relative -fold expression (histogram) represents the ImageJ-quantified ubiquitinated protein levels of the samples containing PARP1 relative to the samples without PARP1 (lanes 1, 3, and 4 versus 2; lanes 6 and 7 versus 5). The values represent the mean ± S.D. of three independent experiments.
The UHRF1 triggered ubiquitination of DNMT1 was described previously as targeting DNMT1 for proteasomal degradation, thereby regulating its protein stability (35). Therefore, we followed the protein expression profile of DNMT1 in PARP1+/+ and PARP1−/− 3T3 cells throughout the cell cycle after release from serum starvation (Fig. 8). To carefully address DNMT1 protein stability as opposed to its steady state levels, cells were treated with the protein synthesis inhibitor cycloheximide. We observed a similar abundance of DNMT1 in whole cell extracts from nonsynchronized and G1-synchronized PARP1+/+ and PARP1−/− cells (Fig. 8, lanes 1–4). In contrast, we detected a reduced level of DNMT1 in the S and G2 PARP1−/− cells compared with the PARP1+/+ cells (Fig. 8, compare lane 6 with 5 and 8 with 7). Therefore, consistent with the ubiquitination data, these results suggest that PARP1 likely helps to maintain DNMT1 protein stability throughout the progression of the cell cycle.
FIGURE 8.
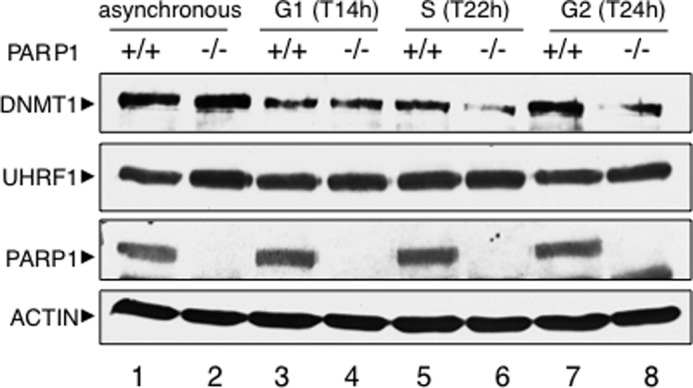
DNMT1 abundance is reduced in PARP1−/− cells. Protein expression from nonsynchronized (lanes 1 and 2) and synchronized (lanes 3–8) PARP1+/+ and PARP1−/− cells was analyzed by SDS-PAGE and Western blotting with the appropriate antibodies. To evaluate protein stability, cells were treated with cycloheximide before lysis. Progression of serum-starved cells released into fresh medium through the cell cycle was monitored by flow cytometry analysis (not shown). The time points of release as determined by FACS are indicated in parentheses.
DISCUSSION
In this study we identified a PAR-dependent physical and functional interaction of PARP1 with UHRF1 involving DNMT1 in which PARP1 helps to modulate two different UHRF1-regulated processes: the accumulation of the repressive mark H4K20me3 on one hand and the abundance of DNMT1 on the other hand. Both events possibly contribute to the silencing of pericentric heterochromatin.
We revealed enhanced transcriptional activity in PARP1−/− cells in both the centric and pericentric regions, thus implying a key role of PARP1 in their silencing. Accordingly, recent chromatin immunoprecipitation experiments have identified an association of PARP1 with minor and major satellites in NIH3T3 cells (12). Furthermore, PARP1 has been described as a key element in the repression of rRNA transcription and inheritance of silent rDNA chromatin (12). UHRF1 was shown previously to exert a selective transcriptional control on the DNA satellites of pericentric heterochromatin but not centromeric heterochromatin (42). That the additional depletion of UHRF1 in PARP1−/− cells has no additive impact on the transcription of pericentric repeats implies that both proteins likely act in the same molecular process. In addition, based on the physical and functional interaction of UHRF1 and PARP1, it is tempting to speculate on a selective coordinated interplay between both enzymes for the silencing of major satellite sequences. What would be the molecular mechanism involved?
UHRF1 has been shown to bind hemi-methylated CpG dinucleotides specifically and to associate with DNMT1 through the same SRA domain. In addition, UHRF1 is described as binding methylated H3K9 through its TTD (26, 29). Together these activities cooperate for the loading of DNMT1 to pericentric heterochromatic sites to guarantee the maintenance of DNA methylation (24, 29). The PARylation of both the SRA domain and the TTD of UHRF1 reported here and the previously described functional interaction between PARP1 and DNMT1 (15) prompted us to investigate a possible role of PARP1 in one or the other of these properties. The conserved accumulation of UHRF1 onto the DAPI-dense replicating heterochromatic regions of PARP1−/− cells or after PARP inhibition suggests that PARP1 might not be absolutely required for the binding of UHRF1 to hemi-methylated CpG sites. In contrast, we found a reduced interaction of UHRF1 with DNMT1 in the PARP1−/− cells, thus rather indicating a contribution of PARP1 to the formation or the stability of the UHRF1-DNMT1 complex. In line with this hypothesis, DNMT1 contains a PAR-binding domain and was shown to interact noncovalently with PAR (21). These observations led to the hypothesis that the PARylation of UHRF1 can serve to construct a robust interaction network among PARP1, UHRF1, and DNMT1 and to favor the association among the members of this complex by noncovalent interactions. In addition, we also found a PARP1-catalyzed poly(ADP-ribosyl)ation of DNMT1 that can contribute to its interaction with UHRF1 (not shown). Notwithstanding, the reduced UHRF1-DNMT1 association observed in the absence of PARP1 did not significantly perturb the loading or the catalytic activity of DNMT1 as revealed by its normal enrichment at heterochromatic regions of PARP1−/−- or PARP-inhibited cells and by the wild type-like methylation status detected at the major satellite repeats. Based on our findings, we propose that this is at least partly explained by a compensating activity of PCNA in promoting the localization of DNMT1 onto replication foci as supported by the PCNA-DNMT1 interaction that is maintained in the absence of PARP1. Alternatively, a compensating DNA methylation activity by DNMT3 proteins, recruited via the Suv39h-mediated H3K9me3 (51), a modification not impaired in PARP1−/− cells, is an appealing hypothesis. In any case, the overall derepression of the major satellites detected in the absence of PARP1 in the asynchronous cells cannot simply be caused by impaired DNA methylation, suggesting the involvement of another mechanism that we aimed to identify.
UHRF1 has also been reported to have key functions in sensing and controlling chromatin features other than just the CpG methylation status of pericentric heterochromatin. Therefore, a conceivable hypothesis is that PARP1 participates in the UHRF1-mediated chromatin modifications required for its silencing. UHRF1, through its SRA domain, recruits HDAC1, which deacetylates histone H4, thereby controlling the transcription of major satellites (38). However, in contrast to findings in UHRF1-depleted cells, we did not detect hyperacetylation of H4K16 at heterochromatic sites in the PARP1−/− cells, indicating that PARP1 might not be required for this mechanism. The TTD and SRA domain of UHRF1 have also been suggested to mediate the binding activity and specificity of UHRF1 onto H3K9me3 sites and regulate the organization of this epigenetic mark onto pericentric heterochromatin (26, 29, 30, 32). Furthermore, a perturbed H3K9me3 profile as reported in Suv39h−/− cells, is associated with enhanced transcription of major satellite repeats owing to defective recruitment of DNMT3b (51, 52). Again, the normal staining of H3K9me3 observed in the PARP1−/− cells suggests that PARP1 might not be involved in its accumulation onto heterochromatic regions. In contrast, our findings reveal a selective cross-talk between PARP1 and UHRF1 in the regulation of the repressive mark H4K20me3 that characterizes the correct condensation and thereby the silencing of pericentric heterochromatin. The results suggest that UHRF1 negatively regulates the accumulation of H4K20me3 onto HC foci and that PARP1 serves to control this activity. Future work will be required to further dissect the molecular link between both proteins and their activities and to identify the targets among the proteins that orchestrate the sequential methylation of H4K20 at pericentric regions, including SET8/PR-Set7, SUV4-H20.1, and SUV4-H20.2, or those that catalyze demethylation, such as PHF2 and PHF8 (53). That the depletion of UHRF1 alone is not sufficient to decrease the enhanced transcription of the major satellites observed in the PARP1−/− cells and restore their repression might simply be explained by a broader role of PARP1 at pericentric heterochromatin. In support of this view, PARP1 was found to interact with the SWI/SNF-like factor SMARCAD1, involved in the maintenance of epigenetic marks throughout pericentric heterochromatin replication (11), to cooperate with the histone deacetylase SirT1 for the maintenance of pericentric heterochromatin integrity (9) and to mediate the inheritance of silent rDNA chromatin (12). In addition, the repressive mark H4K20me3 is probably not the only requirement for the transcriptional silencing of pericentric heterochromatin. Alternatively, given the emerging function of H4K20 methylation in genome maintenance, whether derepression of satellite repeats might be partially elicited by perturbed pericentric and centromeric heterochromatin integrity as reported for BRCA-deficient cells (54) cannot be excluded. In support of this idea, both UHRF1-deficient and PARP1−/− cell lines were found to display higher genome instability and defects in chromosome segregation (14, 55).
Given the recently uncovered role of UHRF1 in the clearance of DNMT1 through its ubiquitination and proteasomal degradation (33–35), our results imply an additional possible contribution of PARP1 and its activity to this process. Much recent evidence exists to support a broad role of PARPs in ubiquitin-mediated protein degradation. A first example is the association of PARP1 with the E3 ubiquitin ligase CHFR (checkpoint with Forkhead and Ring finger domains), which was shown to induce the polyubiquitination of PARP1 and its degradation in response to mitotic stress (56). PARP1 and PARylation have also been implicated in the ubiquitination of repair proteins catalyzed by the E3 ligase RNF146/Iduna in response to genotoxic stress, and Tankyrase (PARP5a)-catalyzed PARylation has been associated with the RNF146/Iduna-triggered ubiquitination of targets from the Wnt/β-catenin signaling pathway (49, 50). In our study we define PARP1 as a negative regulator of the E3 ubiquitin ligase activity of UHRF1 onto DNMT1. In vivo this is exemplified by an enhanced ubiquitination of DNMT1 in PARP1−/− cells, whereas the autoubiquitination of UHRF1 remains rather unaffected. This regulation may preserve DNMT1 from early proteasomal degradation and consequently facilitate its accumulation throughout the cell cycle. In support of this assumption, we detected decreased DNMT1 stability in the S and G2 phases of the cell cycle in PARP1−/− cells. Recent reports have identified an additional role of Usp7 (HAUSP) in promoting DNMT1 stability both by deubiquitinating DNMT1 and by inhibiting UHRF1 activity (34, 35). Whether PARP1 also controls the deubiquitinating activity of Usp7 onto DNMT1 represents a possibility, although we have observed that the interaction of Usp7 with UHRF1 and DNMT1 is not perturbed by the absence of PARP1 (not shown).
Based on our results, we propose PARP1 as a novel player in the UHRF1-mediated fine-tuned regulation of DNMT1 abundance throughout the cell cycle. Does this cellular process also contribute to the repression of major satellites in addition to the accumulation of H4K20me3? The possibility exists. The repression of mouse pericentric HC is tightly coupled to the cell cycle. Although two burst of transcription have been detected in late G1 to early S phase just before the replication of chromocenters and during mitosis to generate transcripts that drive the reassembly of heterochromatin, major satellites are silenced in a large part of G1 and when cells are engaged in replicating chromocenters from mid-late S phase to G2 (57). It is conceivable that the enhanced UHRF1-catalyzed degradation of DNMT1 that we detected in the PARP1−/− cells from S to G2 perturbs DNA methylation at pericentric regions and participates together with the loss of H4K20me3 in the abnormal derepression of satellite repeats, specifically during the replication of these regions and onward. As a consequence, reduced pericentric heterochromatin silencing can be associated with the delay of mid-to-late S phase replication that we detected in the PARP1−/− cells (not shown). Similarly, UHRF1 and BRCA1 have been proposed to control the silencing of major satellites repeats and the replication of pericentric heterochromatin (42, 54, 58).
In summary, our findings define PARP1 as part of a protein complex containing UHRF1 and DNMT1 in which PARP1 regulates two UHRF1-associated biological activities. Accumulating reports in the literature show that the functional role of the UHRF1-DNMT1 association is dual. (i) On one hand, it helps to recruit DNMT1 to replicating heterochromatic regions for DNA methylation maintenance; (ii) on the other, it mediates the ubiquitination of DNMT1 to regulate DNMT1 stability. Similarly, our data suggest that the biological outcome of the PARP1-UHRF1 association is also dual. (i) Both proteins cooperate in the maintenance of the repressive mark H4K20me3 at pericentric heterochromatin to favor transcriptional silencing, and (ii) PARP1 seems to negatively regulate the UHRF1-catalyzed ubiquitination of DNMT1 to maintain the abundance of DNMT1 from S to G2 phase. This can be an additional way to control transcriptional repression, specifically during the replication of pericentric heterochromatin and onward.
Acknowledgment
We thank S. Link (Ludwig Maximilians University) for technical help with the GFP-UHRF1 plasmid constructs.
This work was supported by the Association pour le Recherche Contre le Cancer (ARC), the CNRS, and the Université de Strasbourg. This work has also been published within the LABEX ANR-10-LABX-0034_Medalis and received financial support from the French government via the Agence Nationale de la Recherche under the Programme d'Investissement d'Avenir.
- PARP
- poly(ADP-ribose) polymerase
- PAR
- poly(ADP-ribose)
- DNMT
- DNA methyltransferase
- SRA
- SET and RING-associated
- TTD
- tandem Tudor domain
- PHD
- plant homeodomain
- WB
- Western blot
- IP
- immunoprecipitation
- IF
- immunofluorescence
- RIPA
- radioimmune precipitation assay
- Ubl
- ubiquitin-like
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Krishnakumar R., Kraus W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Quénet D., El Ramy R., Schreiber V., Dantzer F. (2009) The role of poly(ADP-ribosyl)ation in epigenetic events. Int. J. Biochem. Cell Biol. 41, 60–65 [DOI] [PubMed] [Google Scholar]
- 3. Kraus W. L., Hottiger M. O. (2013) PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 34, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 4. Kalisch T., Amé J.C., Dantzer F., Schreiber V. (2012) New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 37, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dantzer F., Giraud-Panis M. J., Jaco I., Amé J. C., Schultz I., Blasco M., Koering C. E., Gilson E., Ménissier-de Murcia J., de Murcia G., Schreiber V. (2004) Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell. Biol. 24, 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomez M., Wu J., Schreiber V., Dunlap J., Dantzer F., Wang Y., Liu Y. (2006) PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell 17, 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saxena A., Saffery R., Wong L. H., Kalitsis P., Choo K. H. (2002) Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 277, 26921–26926 [DOI] [PubMed] [Google Scholar]
- 8. Saxena A., Wong L. H., Kalitsis P., Earle E., Shaffer L. G., Choo K. H. (2002) Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum. Mol. Genet. 11, 2319–2329 [DOI] [PubMed] [Google Scholar]
- 9. El Ramy R., Magroun N., Messadecq N., Gauthier L. R., Boussin F. D., Kolthur-Seetharam U., Schreiber V., McBurney M. W., Sassone-Corsi P., Dantzer F. (2009) Functional interplay between Parp-1 and SirT1 in genome integrity and chromatin-based processes. Cell. Mol. Life Sci. 66, 3219–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quénet D., Gasser V., Fouillen L., Cammas F., Sanglier-Cianferani S., Losson R., Dantzer F. (2008) The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1α. FASEB J. 22, 3853–3865 [DOI] [PubMed] [Google Scholar]
- 11. Rowbotham S. P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W. R., Webster J., Oxley D., Green C. M., Varga-Weisz P., Mermoud J. E.(2011) Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 42, 285–296 [DOI] [PubMed] [Google Scholar]
- 12. Guetg C., Scheifele F., Rosenthal F., Hottiger M. O., Santoro R. (2012) Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 45, 790–800 [DOI] [PubMed] [Google Scholar]
- 13. Meder V.S., Boeglin M., de Murcia G., Schreiber V. (2005) PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 118, 211–222 [DOI] [PubMed] [Google Scholar]
- 14. Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J. C., Dierich A., LeMeur M., Sabatier L., Chambon P., de Murcia G. (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caiafa P., Guastafierro T., Zampieri M. (2009) Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 23, 672–678 [DOI] [PubMed] [Google Scholar]
- 16. de Capoa A., Febbo F. R., Giovannelli F., Niveleau A., Zardo G., Marenzi S., Caiafa P. (1999) Reduced levels of poly(ADP-ribosyl)ation result in chromatin compaction and hypermethylation as shown by cell-by-cell computer-assisted quantitative analysis. FASEB J. 13, 89–93 [DOI] [PubMed] [Google Scholar]
- 17. Zardo G., D'Erme M., Reale A., Strom R., Perilli M., Caiafa P. (1997) Does poly(ADP-ribosyl)ation regulate the DNA methylation pattern? Biochemistry 36, 7937–7943 [DOI] [PubMed] [Google Scholar]
- 18. Zardo G., Caiafa P. (1998) The unmethylated state of CpG islands in mouse fibroblasts depends on the poly(ADP-ribosyl)ation process. J. Biol. Chem. 273, 16517–16520 [DOI] [PubMed] [Google Scholar]
- 19. Zardo G., Reale A., Passananti C., Pradhan S., Buontempo S., De Matteis G., Adams R. L., Caiafa P. (2002) Inhibition of poly(ADP-ribosyl)ation induces DNA hypermethylation: a possible molecular mechanism. FASEB J. 16, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 20. Guastafierro T., Cecchinelli B., Zampieri M., Reale A., Riggio G., Sthandier O., Zupi G., Calabrese L., Caiafa P. (2008) CTCF activates PARP-1 affecting DNA methylation machinery. J. Biol. Chem. 283, 21873–21880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reale A., Matteis G. D., Galleazzi G., Zampieri M., Caiafa P. (2005) Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene 24, 13–19 [DOI] [PubMed] [Google Scholar]
- 22. Zampieri M., Passananti C., Calabrese R., Perilli M., Corbi N., De Cave F., Guastafierro T., Bacalini M.G., Reale A., Amicosante G., Calabrese L., Zlatanova J., Caiafa P.(2009) Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One 4, e4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 24. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 25. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 26. Nady N., Lemak A., Walker J. R., Avvakumov G. V., Kareta M. S., Achour M., Xue S., Duan S., Allali-Hassani A., Zuo X., Wang Y. X, Bronner C., Chédin F., Arrowsmith C. H., Dhe-Paganon S. (2011) Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 286, 24300–24311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rottach A., Frauer C., Pichler G., Bonapace I. M., Spada F., Leonhardt H. (2010) The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 38, 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng J., Yang Y., Fang J., Xiao J., Zhu T., Chen F., Wang P., Li Z., Yang H., Xu Y. (2013) Structural insight into coordinated recognition of H3K9me3 by the plant homeodomain (PHD) and tandem tudor domain (TTD) of the ubiquitin-like, containing PHD and RING finger domains, 1 (UHRF1). J. Biol. Chem. 288, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J. (2013) UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563. [DOI] [PubMed] [Google Scholar]
- 30. Rothbart S. B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A. I., Barsyte-Lovejoy D., Martinez J. Y., Bedford M. T., Fuchs S. M., Arrowsmith C. H., Strahl B. D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P. P., Bonapace I. M. (2004) Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 24, 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karagianni P., Amazit L., Qin J., Wong J. (2008) ICBP90, a novel methyl K9 H3-binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 28, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felle M., Joppien S., Németh A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Längst G. (2011) The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 39, 8355–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin W., Leonhardt H., Spada F. (2011) Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 [DOI] [PubMed] [Google Scholar]
- 35. Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., Kao H. Y., Xu Y., Willis J., Markowitz S. D., Sedwick D., Ewing R. M., Wang Z. (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., Koseki H., Nakanishi M. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253 [DOI] [PubMed] [Google Scholar]
- 37. Meilinger D., Fellinger K., Bultmann S., Rothbauer U., Bonapace I. M., Klinkert W. E., Spada F., Leonhardt H. (2009) Np95 interacts with de novo DNA methyltransferases, Dnmt3a, and Dnmt3b and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 10, 1259–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Unoki M., Nishidate T., Nakamura Y. (2004) ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 23, 7601–7610 [DOI] [PubMed] [Google Scholar]
- 39. Achour M., Fuhrmann G., Alhosin M., Rondé P., Chataigneau T., Mousli M., Schini-Kerth V. B., Bronner C. (2009) UHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activity. Biochem. Biophys. Res. Commun. 390, 523–528 [DOI] [PubMed] [Google Scholar]
- 40. Kim J. K., Estève P. O., Jacobsen S. E., Pradhan S. (2009) UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 37, 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mistry H., Tamblyn L., Butt H., Sisgoreo D., Gracias A., Larin M., Gopalakrishnan K., Hande M. P., McPherson J. P. (2010) UHRF1 is a genome caretaker that facilitates the DNA damage response to γ-irradiation. Genome Integr. 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Papait R., Pistore C., Negri D., Pecoraro D., Cantarini L., Bonapace I. M. (2007) Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell 18, 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pichler G., Wolf P., Schmidt C. S., Meilinger D., Schneider K., Frauer C., Fellinger K., Rottach A., Leonhardt H. (2011) Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 112, 2585–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hopfner R., Mousli M., Jeltsch J. M., Voulgaris A., Lutz Y., Marin C., Bellocq J. P., Oudet P., Bronner C. (2000) ICBP90, a novel human CCAAT-binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 60, 121–128 [PubMed] [Google Scholar]
- 45. Boehler C., Gauthier L. R., Mortusewicz O., Biard D. S., Saliou J. M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F., Dantzer F. (2011) Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 108, 2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frauer C., Rottach A., Meilinger D., Bultmann S., Fellinger K., Hasenöder S., Wang M., Qin W., Söding J., Spada F., Leonhardt H. (2011) Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One 6, e16627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Wang J., Ding M., Yu Y. (2013) Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 10, 981–984 [DOI] [PubMed] [Google Scholar]
- 48. Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. (1997) Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 [DOI] [PubMed] [Google Scholar]
- 49. Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A., Myer V. E., Finan P. M., Porter J. A., Huang S. M., Cong F. (2011) RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13, 623–629 [DOI] [PubMed] [Google Scholar]
- 50. Kang H. C., Lee Y. I., Shin J. H., Andrabi S. A., Chi Z., Gagné J. P., Lee Y., Ko H. S., Lee B. D., Poirier G. G., Dawson V. L., Dawson T. M. (2011) Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 108, 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 52. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 53. Jørgensen S., Schotta G., Sørensen C. S. (2013) Histone H4 Lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41, 2797–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu Q., Pao G. M., Huynh A. M., Suh H., Tonnu N., Nederlof P. M., Gage F. H., Verma I. M. (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muto M., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K. (2002) Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 277, 34549–34555 [DOI] [PubMed] [Google Scholar]
- 56. Kashima L., Idogawa M., Mita H., Shitashige M., Yamada T., Ogi K., Suzuki H., Toyota M., Ariga H., Sasaki Y., Tokino T. (2012) CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J. Biol. Chem. 287, 12975–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu J., Gilbert D. M. (2007) Proliferation-dependent and cell cycle-regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 179, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pageau G. J., Lawrence J. B. (2006) BRCA1 foci in normal S-phase nuclei are linked to interphase centromeres and replication of pericentric heterochromatin. J. Cell Biol. 175, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]