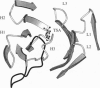Abstract
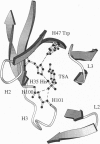
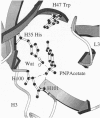
The x-ray structure of the complex of a catalytic antibody Fab fragment with a phosphonate transition-state analog has been determined. The antibody (CNJ206) catalyzes the hydrolysis of p-nitrophenyl esters with significant rate enhancement and substrate specificity. Comparison of this structure with that of the uncomplexed Fab fragment suggests hapten-induced conformational changes: the shape of the combining site changes from a shallow groove in the uncomplexed Fab to a deep pocket where the hapten is buried. Three hydrogen-bond donors appear to stabilize the charged phosphonate group of the hapten: two NH groups of the heavy (H) chain complementarity-determining region 3 (H3 CDR) polypeptide chain and the side-chain of histidine-H35 in the H chain (His-H35) in the H1 CDR. The combining site shows striking structural similarities to that of antibody 17E8, which also has esterase activity. Both catalytic antibody ("abzyme") structures suggest that oxyanion stabilization plays a significant role in their rate acceleration. Additional catalytic groups that improve efficiency are not necessarily induced by the eliciting hapten; these groups may occur because of the variability in the combining sites of different monoclonal antibodies that bind to the same hapten.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Evnin L. B., Vásquez J. R., Craik C. S. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6659–6663. doi: 10.1073/pnas.87.17.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinelli-Pimpaneau B., Gigant B., Bizebard T., Navaza J., Saludjian P., Zemel R., Tawfik D. S., Eshhar Z., Green B. S., Knossow M. Crystal structure of a catalytic antibody Fab with esterase-like activity. Structure. 1994 Mar 15;2(3):175–183. doi: 10.1016/s0969-2126(00)00019-8. [DOI] [PubMed] [Google Scholar]
- Huse W. D., Sastry L., Iverson S. A., Kang A. S., Alting-Mees M., Burton D. R., Benkovic S. J., Lerner R. A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989 Dec 8;246(4935):1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Benkovic S. J., Schultz P. G. At the crossroads of chemistry and immunology: catalytic antibodies. Science. 1991 May 3;252(5006):659–667. doi: 10.1126/science.2024118. [DOI] [PubMed] [Google Scholar]
- Roberts V. A., Stewart J., Benkovic S. J., Getzoff E. D. Catalytic antibody model and mutagenesis implicate arginine in transition-state stabilization. J Mol Biol. 1994 Jan 21;235(3):1098–1116. doi: 10.1006/jmbi.1994.1060. [DOI] [PubMed] [Google Scholar]
- Tawfik D. S., Green B. S., Chap R., Sela M., Eshhar Z. catELISA: a facile general route to catalytic antibodies. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):373–377. doi: 10.1073/pnas.90.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik D. S., Zemel R. R., Arad-Yellin R., Green B. S., Eshhar Z. Simple method for selecting catalytic monoclonal antibodies that exhibit turnover and specificity. Biochemistry. 1990 Oct 23;29(42):9916–9921. doi: 10.1021/bi00494a023. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Cunningham B. C., Graycar T. P., Estell D. A. Recruitment of substrate-specificity properties from one enzyme into a related one by protein engineering. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Stanfield R. L. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994 Dec;4(6):857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Zemel R., Schindler D. G., Tawfik D. S., Eshhar Z., Green B. S. Differences in the biochemical properties of esterolytic antibodies correlate with structural diversity. Mol Immunol. 1994 Feb;31(2):127–137. doi: 10.1016/0161-5890(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Zhou G. W., Guo J., Huang W., Fletterick R. J., Scanlan T. S. Crystal structure of a catalytic antibody with a serine protease active site. Science. 1994 Aug 19;265(5175):1059–1064. doi: 10.1126/science.8066444. [DOI] [PubMed] [Google Scholar]