Abstract
The scaffold protein Vac14 acts in a complex with the lipid kinase PIKfyve and its counteracting phosphatase FIG4, regulating the interconversion of phosphatidylinositol-3-phosphate to phosphatidylinositol-3,5-bisphosphate. Dysfunctional Vac14 mutants, a deficiency of one of the Vac14 complex components, or inhibition of PIKfyve enzymatic activity results in the formation of large vacuoles in cells. How these vacuoles are generated and which processes are involved are only poorly understood. Here we show that ectopic overexpression of wild-type Vac14 as well as of the PIKfyve-binding deficient Vac14 L156R mutant causes vacuoles. Vac14-dependent vacuoles and PIKfyve inhibitor-dependent vacuoles resulted in elevated levels of late endosomal, lysosomal, and autophagy-associated proteins. However, only late endosomal marker proteins were bound to the membranes of these enlarged vacuoles. In order to decipher the linkage between the Vac14 complex and regulators of the endolysosomal pathway, a protein affinity approach combined with multidimensional protein identification technology was conducted, and novel molecular links were unraveled. We found and verified the interaction of Rab9 and the Rab7 GAP TBC1D15 with Vac14. The identified Rab-related interaction partners support the theory that the regulation of vesicular transport processes and phosphatidylinositol-modifying enzymes are tightly interconnected.
Lipid kinases and phosphatases tightly regulate the interconversion and abundance of different phosphoinositide lipid derivatives (PIPs),1 which are crucial components for the identity of eukaryotic membranes (1, 2). PIPs and their modifying proteins control multiple cellular functions such as signal transduction, cytoskeletal dynamics, and membrane trafficking (1, 2). Synthesis and turnover of the low-abundant lipid phosphatidylinositol-3,5-bisphosphate is controlled by the Vac14 complex. This complex contains the scaffold protein Vac14 (ArPIKfyve), the lipid kinase PIKfyve (Fab1), and its counteracting lipid phosphatase FIG4 (SAC3) (3, 4).
The overall protein structure of Vac14 is conserved from yeast to human and consists of multiple tandem HEAT (huntingtin, elongation Factor 3, PR65/A, TOR) repeats and a rod-like helical domain that controls protein–protein interactions (5, 6). Previous studies showed that the aminoterminal part of Vac14 mediates the binding to PIKfyve. The carboxyterminal region is crucial for the interaction with FIG4 and also contains a PDZ-binding motif, which binds to nNOS (5, 7–11). Moreover, Vac14-oligomerization, which is essential for the nucleation of an active Vac14-complex, overlaps with the FIG4 interaction binding site (5, 6, 12, 13).
The lack of Vac14 or the ectopic overexpression of a PIKfyve binding-deficient Vac14 mutant in vivo results in a reduced level of phosphatidylinositol-3,5-bisphosphate and is accompanied by enhanced formation of enlarged intracellular vesicles, hereinafter called vacuoles (5, 14, 15). This was initially observed in yeast, where it leads to enlargement of the yeast vacuole, which is comparable to the mammalian lysosome (12, 16, 17). The vacuoles in mammalian cells are heterogeneous, positive for early or late endosomal structures, and involved in vesicular trafficking processes from the late endosome to the trans-Golgi network (14, 18, 19).
We now report that in addition to overexpression of the PIKfyve binding deficient mutant, overexpression of the Vac14 wild type was sufficient to induce vacuolization in human cell cultures. The Vac14-based vacuolization leads to a significant accumulation of predominantly late endosomal and autophagosomal marker proteins with only late endosomal proteins decorating the vacuolar membranes. These results almost completely phenocopy previously described effects of the inhibitor YM201636, which specifically blocks PIKfyve lipid kinase activity (20–24).
To identify proteins involved in Vac14-induced vacuolization, we used a protein affinity approach combined with multidimensional protein identification technology (MudPIT). The evaluation of the identified peptides elucidated numerous potential Vac14 interacting proteins involved in intracellular trafficking and membrane dynamics, with Rab9 and TBC1D15 being the most promising candidates. The specificity of the Vac14 interaction with Rab9 and the Rab7 regulator TBC1D15 was confirmed by co-immunoprecipitation assays. Moreover, we were able to demonstrate that Rab9 accumulates during vacuolization and localizes on the limiting membranes of vacuoles as a result of Vac14 overexpression. In summary, the identification of Rab9 and TBC1D15 as novel interaction partners provides new insights into the molecular functions of Vac14 in vesicular transport processes.
EXPERIMENTAL PROCEDURES
Constructs and Cloning
The coding sequences of full-length human Vac14 were amplified using the mCitVac14 construct described previously with primers carrying AscI and PacI extensions and inserted into pENTR-D/TOPO vectors (Invitrogen) according to the manufacturer's instructions (5). The Vac14 ingls variant L156R was generated via site-directed mutagenesis. Human Rab9, CD63, and Lamp2a cDNAs were amplified from a human immortalized podocyte library and inserted into pENTR-D/TOPO vectors (Rab9, CD63) or cloned via XhoI in pEGFP-N1 or pQCXIP mCherry-C (Lamp2a) vectors. The cDNA used to express the constitutively active form of Rab9 was generated via site-directed mutagenesis. The cDNAs were subsequently shuttled into pcDNA-DEST53 via LR-clonase reaction (Invitrogen) or subcloned into the pEGFP expression vectors (Clontech) or the V180 plasmid (Addgene plasmid 11707), which enabled the expression of N-terminal 3xFLAG-tagged fusion proteins (25, 26).
The pQCXIP mCherry-N vector was described previously (27). Plasmids to express 3xFLAG-tagged proteins in stable cell lines were generated via PCR derived from the V180 plasmids and inserted via EagI/NotI into pQCXIP. Afterward, the cDNA of Vac14 was inserted into these plasmids using NotI/PacI, and these constructs were used to generate stable 3xFLAG- and mCherry-Vac14-expressing cell lines. To obtain pQCXIP-DD-Vac14 wild type (wt), the mCherry cDNA fragment of pQCXIP-mCherry Vac14 was replaced by a cDNA encoding a mutated variant of the degradation tag (DD-tag, kindly provided by Britta Eickholt) derived from E. coli dihydrofolate reductase.
To generate doxycycline inducible stable cell lines, we replaced the GFP module of pINDUCER21 (a gift from Thomas Westbrook) with the puromycin expression cassette of pQCXIP to obtain pINDUCER21 puromycin via an overlap extension PCR technique using NdeI/SphI restriction sites (28). For the inducible expression of GFP-tagged Vac14, the EGFP cassette from pEGFP (Clontech) was amplified and subcloned via EagI/NotI to generate pENTR-EGFP Vac14. The resulting pENTR-EGFP Vac14 cassettes were shuttled into pINDUCER21 puromycin using LR-clonase. EGFP-tagged Rab5A Q79L (plasmid 28046), Rab7A Q67L (plasmid 28049) (29), and pEGFP-LC3 (plasmid 11546) (30) were obtained via Addgene. pEGFP-tagged Rab6 Q67R (31) and pEGFP 2xFYVEhrs (32) were gifts from Angelika Barnekow and Harald Stenmark, respectively. Plasmid pEGFP Vps4 E235Q was received from Geoffrey D. Holman (33), and EGFP-Tsg101 was provided by Eric Freed. The plasmid encoding myc-TBC1D15 was a kind gift from Aimee Edinger (34). All constructs were verified by DNA sequencing. Further details concerning constructs, cloning, and primers are available from U.S. and T.W.
Cell Culture and Transient Transfection
HEK293T and HeLa (SS6) cells and the Retro-X packing cell line GP2–293 (Clontech) were cultivated in standard medium (Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum or iron-supplemented calf serum (HEK293) and 1% antibiotics (Pen/Strep) as described elsewhere (35, 36). Transient transfection of HeLa cells with various expression constructs for live cell imaging and immunofluorescence analyses were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. GP2–293 (for retrovirus production; Retro-X system) and HEK293 cells (for lentivirus production; pINDUCER system) were transiently transfected using the calcium-phosphate method (35, 36).
Generation of Stable Cell Lines
To generate HEK293T and HeLa cell lines for the continuous overexpression of recombinant proteins (3xFLAG-, mCherry, DD-Vac14wt/L156R), we used a retroviral packaging system (Retro-X, Clontech). In brief, GP2–293 cells (10-cm dish) were simultaneously transfected with 5 μg of pVSV-G envelope (Clontech) and 5 μg of pQCXIP-based expression plasmids using the calcium-phosphate method.
Stable doxycycline-inducible cell lines were obtained by using the recently established pINDUCER system (28). Briefly, a 10-cm dish with 50% confluent HEK293T was simultaneously transfected with 10 μg of the pINDUCER21 puromycin GFP-Vac14 construct together with 6.5 μg of psPAX2 (Addgene plasmid 12260) and 3.5 μg of pMD2.G (Addgene plasmid 12259) (28).
In both cases (retro- and lentiviral-based transductions), the medium of transiently transfected GP2–293 or HEK293T cells was changed after 6 to 8 h, and cells were grown for an additional 72 h. Next, the virus-containing supernatant was collected and filtered through a sterile 0.45-μm syringe-driven filter unit (Millipore, Darmstadt, Germany). Subsequently, the HEK293T or HeLa target cells (growing in six-well dishes) were infected for 24 h using one volume (up to 2 ml) of fresh DMEM medium and one volume of the virus-containing filtrate supplemented with polybrene (final concentration: 8 μg/ml). Finally, the virus-containing medium was replaced by fresh medium and cells were regenerated for 24 h. This transduction procedure was repeated once.
After the second regeneration period, cells were selected through puromycin treatment (4 μg/ml; Sigma) without sorting of monoclonal cell clones. Overexpression of ectopic Vac14 protein (encoded by the pQCXIP constructs) in the stable cell populations was verified by Western blotting and immunofluorescence analysis. The expression of GFP-Vac14 wt or Vac14 L156R (via the pINDUCER system) was induced by the addition of 125 ng/ml doxycycline (Sigma) to the culture medium. In the stable DD-Vac14 wt–expressing HeLa cells, expression of the recombinant protein was regulated by the addition of 10 μm trimethoprim (Sigma), which prevents the degradation of DD-Vac14.
Preparation of Cell Lysates
Transfected cells were scraped into co-immunoprecipitation (CoIP) buffer (20 mm Tris HCl, pH 7.4; 150 mm NaCl; 1 mm EDTA; 1% TritonX-100) containing complete protease inhibitor (Roche, Mannheim, Germany). To verify the expression of recombinant proteins, we used lysis buffer (1%Triton-X 100; 20 mm Tris-HCl, pH 7.5; 25 mm NaCl; 50 mm NaF; 15 mm Na4P2O7; and 1.5 mm EDTA). In both cases we homogenized cell lysates by passing the samples through a 26-gauge needle. After the removal of insoluble material via centrifugation (4 °C for 30 min at 10,000g), supernatants were directly used to perform the CoIP/pulldown assays or stored at −20 °C for later use.
CoIP/Pulldown Procedures
Agarose beads with covalently bound anti-FLAG tag antibodies (M2 beads, Sigma) were prepared according to the manufacturer's instructions. Cell lysates in CoIP buffer were added to the washed M2 beads and incubated for 2 h at room temperature. After being washed with TBS buffer (50 mm Tris/HCl, 150 mm NaCl, pH 7.5), 20% of the loaded beads were resolved in Laemmli sample buffer. Proteins were eluted from the M2 beads by boiling for 5 min at 96 °C, separated via SDS-PAGE, and analyzed via Western blotting and/or Coomassie Blue staining (see below). The residual amounts of the immunoprecipitation samples (80%) were stored at −80 °C until MudPIT analysis.
Western Blot Analysis
Quantitative Western blot analysis was done as described elsewhere (37, 38). Briefly, for quantitative Western blotting, assay cells were grown on 24-well plates until confluency and then lysed in 1× Laemmli (4% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromphenol blue, 0.0625 m Tris-HCl, pH 6.8). Samples were shaken at 1000 rpm for 6 h and then subjected to supersonic treatment for 15 min. After being boiled for 5 min, equal volumes of cell lysates were separated onto 8%–15% SDS-PAGE gels (Bio-Rad). Proteins were transferred to a PVDF membrane (Millipore) and incubated for 1 h at room temperature in blocking buffer (5% skim milk powder dissolved in TBS containing 0.05% Tween-20). The lysates were equalized using α-actinin 4 (Enzo, Loerrach, Germany) or GAPDH (Cell Signaling, Frankfurt, Germany) as the loading control. Primary antibodies from rabbit against Vac14 and monoclonal antibodies against the FLAG tag (M2) and Tubulin were obtained from Sigma. Antibodies against p62/SQSTM1 were acquired from BD Transduction Laboratories. Antibodies against Rab5, Rab7, and ATG5 came from Cell Signaling. Antibodies against Rab9 and myc-tag came from Abcam, Cambridge, UK. Antibodies against LC3 were obtained from nanotools, and antibodies against Lamp2 and CD63 came from the Developmental Studies Hybridoma Bank, Iowa. All primary antibodies were used in a 1:1000 dilution (except anti-FLAG M2, which was used at 1:2000) in TBS–Tween-20 and incubated at 4 °C overnight or for 1 h at room temperature. After being washed three times with TBS–Tween-20, the membrane was incubated with horseradish peroxidase–coupled secondary antibodies (Jackson Immunoresearch, via Dianova, Hamburg, Germany) diluted 1:5000 in blocking buffer for 45 min at room temperature. After three washes, the Western blot was developed using a chemiluminescence detection reagent (Roche).
Signals derived from the same immunoblot were densitometrically quantified using ImageJ and analyzed in relation to the loading control. Samples from the same gel but from noncontiguous lanes are marked by boxes in the figures. The evaluation was done using GraphPad Prism (GraphPad Software, La Jolla, CA). All data show the S.E. of at least three independent experiments and were analyzed using an unpaired Student's t test.
Measurement of Autophagic Flux and Apoptosis
The influence of autophagy on Vac14-associated vacuolization was examined in cells stably overexpressing Vac14 wt or Vac14 L156R or with cells treated with the specific PIKfyve inhibitor YM201636 (Cayman Chemicals, via Biomol, Hamburg, Germany; 800 nm, 24 h). Autophagic flux was induced by changing the standard medium into Earle's Balanced Salt solution for 3 h in the presence or absence of bafilomycin A (3 h, 100 nm up to1 μm).
Quantitative Western blots using antibodies against LC3-II and p62 normalized to the GAPDH loading control were used to measure autophagy (flux). The level of apoptosis was monitored using specific antibodies from Cell Signaling Technologies against cleaved PARP and cleaved caspase 9.
Multidimensional Protein Identification Technology
After protein preparation, proteins were resuspended in 100 μl of 50 mm ammonium bicarbonate buffer (Fluka, via Sigma, Müchen, Germany) and 20% methanol (Baker, via VWR International GmbH, Darmstadt, Germany). Next 5 μg of sequencing-grade trypsin (Promega, Mannheim, Germany) was added for an overnight proteolytic digestion at 37 °C. Subsequently, extracted peptides were diluted in 0.1% trifluoroacetic acid and 2% acetonitrile and were loaded directly onto a fused silica capillary (100-μm inner diameter; Polymicro, Phoenix, AZ) packed with reversed-phase (C18 Luna, 3 μm, 100 Å, Phenomenex, Aschaffenburg, Germany) and strong cation exchange (PolySULPHOETHYL A, 5 μm, 200 Å, PolyLC, Basel, Switzerland) chromatography material (10 to 12 cm reversed-phase/4 cm strong cation exchange/3 cm reversed-phase). MudPIT analysis was performed using a quaternary U-HPLC pump directly connected by a PEEK microcross to a Thermo LTQ XL ion trap mass spectrometer (Thermo Fisher Scientific). For optimal peptide separation, an effective flow rate of 200 (± 50) nl/min, a spray voltage of 1.8 kV, and a transfer capillary temperature of 180 °C were applied. In the eight-step MudPIT experiment, each step was represented by one instrument method file with identical settings for the HPLC gradient program. The mass spectrometer was set to measure a full MS spectrum between 400 and 2000 m/z for precursor ions followed by MS/MS scans of the most abundant precursor ions from the MS1 scan to detect fragment ions by the ion trap (39–41).
Database Generation
A non-redundant target-decoy database was generated using the human proteome annotation from UniProt (release version 2013 01 (downloaded on January 9, 2013)), resulting in a database with 70,425 target sequences. Decoy sequences were generated via tryptic peptide amino acid shuffling (length over eight amino acids) or all possible permutations (length less than eight amino acids). Arginine and lysine positions were fixed to ensure an equal length distribution of the target and decoy peptides and ensure the same number of target and decoy peptides. Immutable tryptic peptides (e.g. AAR) were not altered.
Peptide Identification
LC-MS/MS data in RAW format were converted to the mzML format (42) using msConvert (part of ProteoWizard, version 2.0.1905 (43)) and subsequently to the mascot generic format (mgf) as input for OMSSA and X! Tandem using pymzML (44) for rapid mzML file processing and the Python scripting language. Peptides were identified using the peptide-spectrum-match algorithms OMSSA (45) (version 2.19) and X! Tandem (46) (version 2011.12.01). OMSSA and X! Tandem were executed using Proteomatic (47). Most parameters were maintained as default values. The precursor ion accuracy was set at 250 ppm, and the collision-induced dissociation fragmentation ion accuracy was set at 0.5 Da. Oxidation of methionine was allowed as a variable modification, and two missed cleavage sites were allowed. Additionally, X! Tandem considers acetylation of N termini (+42.0106 Da), loss of water (−18.0106 Da), and desamidation (−17.0265 Da). Peptide spectrum matches (PSMs) were sanitized by allowing only one best hit per spectrum and search engine. PSMs were accepted only if the E-values of the best and second best hit differed at least by a factor of 100. For all PSM results obtained by OMSSA and X! Tandem, the posterior error probability and the q-value were determined using the program qvality (48, 49) to allow filtering of the PSMs. The application of posterior error probability and q-value allowed qualitative comparison of the PSMs from the different search engines.
Protein/Peptide Assignment Evaluation
All identified peptides (posterior error probability ≤ 0.001) were analyzed according to their occurrence/distribution in pulldown and control assays/samples. Peptides were assigned to the control and/or pulldown assays if they were identified in at least two independent biological replicates of these samples (n ≥ 2). Proteotypic peptides could be matched directly to their proteins, and thus the proteins could be assigned to control and/or pulldown. Proteins sharing nonproteotypic peptides were pooled together to form one protein group. Because those first groups contained only one peptide and the proteins within a group could theoretically also be members of multiple groups, an iterative process was undertaken to combine those groups that shared proteins into bigger, complete protein groups. A complete protein group contains all proteins that share peptides. These groups have been visualized using node maps, with each node representing a protein and each edge representing a nonproteotypic peptide (see Fig. 6B and supplemental information SI1). Consecutively, all peptides within a group were checked for their occurrence in the VAC14 pulldown and/or the control pulldown assays. Finally, each protein group was assigned to one of four “localization” tags. These four tags were (a) pulldown only (PD) (all peptides of this group were reproducibly identified in at least two pulldowns but nowhere else), (b) control only (C) (all peptides of this group were reproducibly identified in at least two controls but nowhere else), (c) pulldown and control (PD&C) (the peptides were identified in at least two pulldowns and two controls), and (d) ambiguous (MIXED) (peptides of this group were reproducibly identified in at least two pulldowns and/or in at least two control samples). As a result, the localization tags for groups allowed the incorporation of PD proteins into further analyses that are not definable by proteotypic peptides. Thus all proteins that could be assigned to PD based on their proteotypic peptides and all proteins that were in PD-only groups (i.e. shared nonproteotypic peptides that were all PD) were defined as Vac14 interacting proteins.
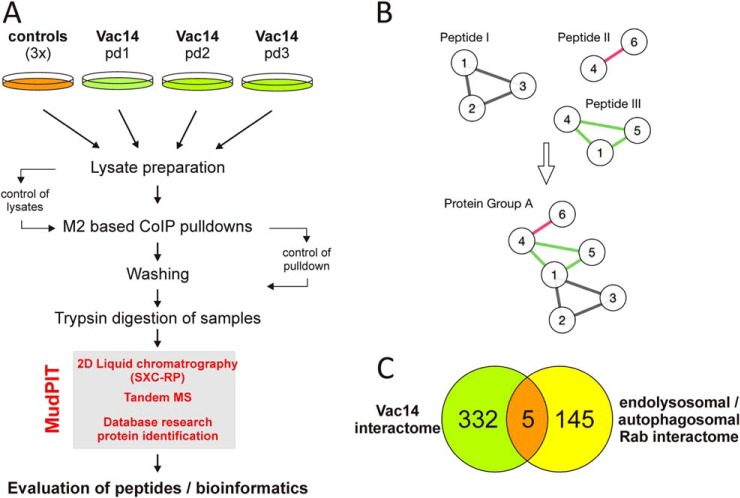
Fig. 6.
Schematic overview of the used MudPIT strategy. A, an overview of the used strategy is shown in the workflow scheme. Three independent pulldown assays in HEK293 cells expressing 3xFLAG Vac14 wt or 3xFLAG alone were used for the MudPIT assays. The CoIP/pulldown assays (pd1–pd3 and controls) were performed with beads covalently linked to anti-FLAG M2 antibodies. To verify successful coupling of FLAG-Vac14 to the M2-beads, aliquots of the CoIP/pulldown samples were controlled by SDS-PAGE and Western blot or Coomassie Blue staining (not shown). After stringency washings, protein samples were digested with trypsin and analyzed via the MudPIT technique, including two-dimensional LC and tandem MS (box). B, illustration of an evaluation example of protein groups and their localization tags. Proteins are shown as nodes, and peptides are shown as edges. Peptide I, shared by proteins 1, 2, and 3, was found in the pulldown (PD) and the control (C), and therefore the edges are colored gray. Peptide II, shared by proteins 4 and 6, was found in C only (red). Peptide III was found in PD only and was shared by proteins 1, 4, and 5 (green). Iterative merging of all protein groups sharing proteins resulted in protein group A, which showed a mixed localization tag, and thus none of those proteins could be incorporated in further analysis. However, if all edges/peptides were green in the final group, then those proteins could be incorporated into further analysis (supplemental information SI 1 and SI 2). C, analysis of the Vac14 interactome resulted in 337 proteins reviewed by the UniProtKB/Swiss-Prot databases. These were compared with the Rab family members and its interaction partners displayed in supplemental Table S1. TBC1D15, Rab9, SUN2, IDH3A, and NSF represent the intersection of both networks.
Bioinformatical Functional Analyses
Putative Vac14 interacting proteins were analyzed in detail using Cytoscape and the appropriate interactomes (50). Briefly, the interactomes of Rab GTPases 4–11, 14, 22–24, and 34 associated genes (see supplemental Table S1) were generated in Cytoscape using the APID, BIND, IntAct, MINT, and UniProt interaction databases. The UniProt entries for the resulting protein networks were exported and compared with the Vac14 interacting proteins (supplemental data SI 2).
Immunofluorescence Analysis and Live Cell Imaging
To visualize acidic compartments for live cell imaging studies, cells grown on ibidi μ-dishes (Ibidi GmbH, Martinsried, Germany) were incubated with LysoTracker green according to the manufacturer's instructions (Invitrogen). Pictures and movies were processed using ImageJ.
Quantification of Vacuoles
Cells were plated on ibidi μ-dishes and were incubated with CellTracker blue CmF2Hc (Invitrogen) at 5 mm final concentration for 16 h. The cells were washed twice with PBS to reduce background signals, and pictures were taken using a Nikon A1 microscope with a 60× oil immersion objective. The quantification and the size measurements of the vacuoles were done using NIS-elements software (Nikon, Düsseldorf, Germany). A minimum of 15 cells from two experiments were analyzed, and the CellTracker positive areas (cytoplasm) were manually encircled. The CellTracker negative structures (vacuoles) were counted and their area was determined using the three-point circle tool. Afterward, the data were exported into Microsoft Excel and evaluated using GraphPad Prism.
Measurement of Vesicle Marker Proteins
24 h post-transfection, the living cells grown on the μ-dishes were analyzed on a Zeiss Observer Z1 equipped with Apotome technology and a 63× oil immersion objective. Pictures were collected from at least two independent experiments and processed using ImageJ. Vesicular size measurements were performed manually. Therefore, single cells were magnified and only GFP- or mCherry-positive structures with a visible lumen were quantified using the straight line tool. These data were exported and normalized to the results obtained from control cells, and statistics were done in GraphPad Prism indicating the fold change in vesicular size.
RESULTS
Overexpression of Ectopic Vac14 L156R and Vac14 wt Proteins Leads to Vacuolization
In homozygote ingls mice, intracellular vacuolization is caused by the Vac14 L156R mutant, which is unable to bind the PI3 kinase PIKfyve. We mimicked this in vivo phenotype in cell culture experiments through stable overexpression of this Vac14 mutant. Surprisingly, our data revealed that high levels not only of Vac14 L156R but also of Vac14 wt led to large perinuclear localized vacuoles (Fig. 1A). To further prove that increased levels of Vac14 alone are sufficient for vacuolization, we took advantage of inducible systems that allow the conditional overexpression of ectopic Vac14. Using two different inducible expression systems, we found that high levels of Vac14 induced vacuoles independently of the epitope tags (DD, FLAG, GFP, mCherry) and cell lines used (HeLa, HEK293; supplemental Fig. S1).
Fig. 1.
Vac14 overexpression causes vacuolization. With the use of CellTracker blue in live cell imaging experiments, vacuoles were clearly distinguished from the blue labeled cytoplasm. A, both overexpression of a 3xFLAG Vac14 wt and the PIKfyve-binding deficient 3xFLAG Vac14 L156R mutant result in strong vacuolization of HeLa cells. Vacuoles are almost absent in the control cells (Ctrl) expressing the 3xFLAG tag alone. B, vacuoles caused by PIKfyve inhibitor YM201636 (24 h, 800 nm) versus untreated cells (Ctrl). C–E, quantification of the vacuoles illustrated in A and B, concerning (C) the amount of vacuoles per cell, (D) the vacuolar area as a percentage per cell, and (E) the size distribution of vacuoles caused by YM201636 treatment or Vac14 wt and Vac14 L156R overexpression, respectively. Detail shows the magnification of the indexed areas. Bar: 20 μm; differential interference contrast (DIC).
Inhibition of PIKfyve, which is crucial for the generation of phosphatidylinositol-3,5-bisphosphate, also results in vacuolated cells in culture (20–24). Therefore, we compared Vac14 overexpressing cells with cells treated with YM201636 (an inhibitor of PIKfyve kinase) in terms of the amount and the size distribution of the vacuoles in a live cell imaging approach (Figs. 1A and 1B). The total number of vacuoles ranged from ∼12.5 (Vac14 L156R) to 14.5 (YM201636 treatment) vacuoles per cell, with vacuoles almost absent in control cells (Fig. 1C). The vacuolar area covered nearly 36% (Vac14 L156R) or 27% (Vac14 wt) of the CellTracker surrounding area, whereas for YM201636-treated cells only 18% of the intracellular area was characterized as vacuolated (Fig. 1D). Consequently, overexpression of Vac14 L156R resulted in more than 35% large vacuoles (>30 μm2) and only 17% small vacuoles (0 to 10 μm2). Vac14 wt overexpression led to mostly medium-sized vacuoles (45%) and only 15% large vacuoles. YM201636 treatment resulted in 77% small structures and the fewest large vacuolar structures (<5%; Fig. 1E). Taken together, these observations revealed that Vac14-induced vacuoles were larger than YM201636-dependent vacuoles and represented at least 27% of the total cytoplasm in analyzed cells.
Vac14-associated Vacuoles Accumulate Marker Proteins of the Endolysosomal Trafficking Route
In order to characterize the intracytoplasmic vacuoles in detail, cells overexpressing 3xFLAG Vac14 (wt and L156R) or 3xFLAG alone (control) were analyzed via quantitative Western blotting for endogenous expression of endolysosomal proteins (Fig. 2A). Rab5 served as a marker protein for early endosomes, and Rab7, CD63, and Lamp2 represented typical proteins for late endosomal/multivesicular body and/or lysosomal structures. The quantification of this experiment revealed significantly increased levels of endogenous Rab7, CD63, and Lamp2 but not of Rab5 in Vac14 wt overexpressing cells (relative to the control). Additionally, lysates derived from the PIKfyve-binding deficient mutant (L156R) showed significantly elevated endogenous Rab5 levels, and, with the exception of CD63, an even stronger increase in late endosomal markers (Fig. 2B).
Fig. 2.
Vac14-associated vacuoles accumulate endolysosomal markers. Lysates of 3xFLAG Vac14 overexpressing HeLa cells (wt and L156R) were analyzed via quantitative Western blotting and compared with control lysates. Both Vac14 overexpressing approaches (wt and L156R) resulted in a significant increase of endogenous Rab7-, CD63-, and Lamp2-positive structures. The increase was stronger in Vac14 L156R lysates. A, sets of three independent quantifications of the Western blots were normalized to the loading control GAPDH and α-actinin 4. In all experiments, cell lines with an empty 3xFLAG plasmid served as a control. B, quantitative evaluation of at least eight samples from the proteins shown in A. ns = not significant; *p < 0.05, **p < 0.01, ***p < 0.001.
Vac14 Overexpression Causes Enlarged Endolysosomal Structures
Vac14-associated vacuoles were characterized in more detail through the transfection of fluorescent-labeled marker proteins. For this approach we used constitutively active (CA) EGFP-tagged Rab5, Rab6, and Rab7 mutants to characterize the limiting membranes of the vacuoles. As expected, both EGFP-Rab5 CA and EGFP-Rab7 CA were found on vacuoles (Figs. 3A–3C, supplemental Fig. S2A). In contrast, EGFP-Rab6 CA was present only on perinuclear structures typical for the Golgi cisternae (supplemental Fig. S2B). To exclude artificial effects of Rab CA overexpression, constructs encoding Rab5 or Rab7 wt were also transfected into the vacuolated cells. Even Rab7 wt was present on the vacuolar membranes, whereas Rab5 was found only in small dots throughout the cell, and not on vacuoles (supplemental Fig. S2C). Quantification of the vacuolar size of Rab7 showed a 6-fold (Vac14 wt) to 7-fold (Vac14 L156R) increase in the size of labeled vacuoles relative to the control. By contrast, the enlargement of Rab5-positive compartments was more moderate (1.5- and 2-fold, respectively; Fig. 3D). Together with the Western blot data (Fig. 2), these results argue for a defect in late but not in early endosomal traffic routes.
Fig. 3.
Vac14 overexpression enlarges compartments of the endolysosomal pathway. Vac14-overexpressing cells were transiently transfected with fluorescent-tagged marker proteins in order to examine the vacuole in more detail. Stable cell lines with an empty 3xFLAG plasmid (A) or expressing 3xFLAG-tagged Vac14 wt (B) and L156R mutant (C) were transiently transfected with plasmids encoding constitutively active EGFP-Rab5 and -Rab7, EGFP-2xFYVEhrs, EGFP-CD63, and Lamp2-mCherry. Vac14-associated vacuoles were predominantly positive for late endosomal marker proteins (Rab7, CD63, or Lamp2). D, quantitative analysis of EGFP/mCherry-positive compartments. The size of EGFP/mCherry protein structures (panels A–C) in Vac14-overexpressing cells (wt and L156R) is shown as the fold increase relative to the control (Ctrl; empty 3xFLAG plasmid). ns = not significant; *p < 0.05, **p < 0.01, ***p < 0.001. Size bar and detailed view: 10 μm.
Furthermore, live cell imaging approaches using LysoTracker showed low acidity of the vacuoles (supplemental Fig. S1A and supplemental movies). To analyze the late endosomal processes in more detail, we used cells overexpressing ectopic EGFP-CD63 and Lamp2a-mCherry. The limiting membrane of Vac14-induced vacuoles was also positive for fluorescent-tagged CD63 and Lamp2, which predominantly label late endosomal and/or lysosomal membranes. Quantification of these experiments is presented in Fig. 3D and reveals that Vac14-associated vacuolization (wt as well as L156R) led to significant enlargement of Lamp2-positive compartments. We also observed enlarged CD63-positive compartments (positive tendency for Vac14 wt and significant 5-fold increase for L156R). Even EGFP 2xFYVEhrs, which labels membranes positive for PI3P lipids, was found on Vac14-dependent vacuoles (Figs. 3B and 3C). Both overexpression of Vac14 wt and the PIKfyve-binding deficient L156R mutant resulted in enlargements of late endosomal compartments. The evaluation of the size of stained structures showed bigger vacuoles in the L156R mutant than in the wild type, with the exception of PI3P labeling with EGFP-2xFYVEhrs (Fig. 3D).
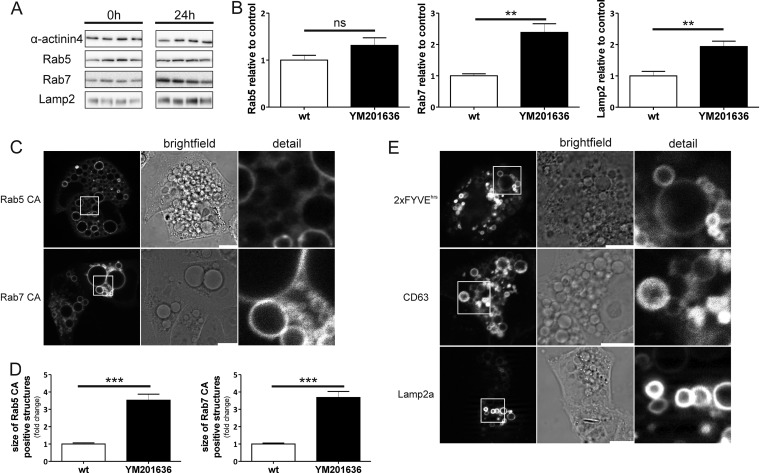
As shown, an inhibition of PIKfyve leads to a phenotype similar to that seen with the overexpression of Vac14, but these vacuoles are not as large as the Vac14-induced ones. In the next step, we compared the protein decoration of YM201636-induced vacuoles to that of Vac14-dependent vacuoles and found similar changes in protein expression. Whereas Rab5 was not significantly elevated, the late endosomal markers Rab7 and Lamp2 showed significantly increased levels (Figs. 4A and 4B). As expected, YM201636-induced vacuoles also accumulated early and late endosomal and lysosomal marker proteins similar to those caused by Vac14 overexpression (Figs. 4C and 4E). Interestingly, the increase in Rab5 CA–positive structures was as high as that for the late endosomal marker Rab7 CA, indicating a block that affects early and late endosomal structures, different from Vac14-induced vacuoles (Fig. 4D, Fig. 3D). Thus, our data, as well as other studies using YM201636, indicate that vacuolization results in a blockade or delay in endolysosomal maturation processes (20, 23).
Fig. 4.
Inhibition of PIKfyve kinase and Vac14 overexpression cause similar effects. A, HeLa cells incubated with PIKfyve inhibitor YM201636 (800 nm; 24 h) showed a significant accumulation of endogenous Rab7 and Lamp2 but not of Rab5. α-actinin 4 served as a loading control. Sets of four protein samples from control cells (left) or treated cells (right) are shown. B, quantitative densitometric analysis of Western blots shown in A; quantifications of the Western blots were normalized to the loading control α-actinin 4. C, live cell imaging analysis of HeLa cells treated with YM201636 showed that the vacuoles were positive for Rab5 and Rab7. D, quantification of Rab5- and Rab7-positive structures showed significant enlargement (up to 3.5-fold) of Rab5 and Rab7 structures in YM201636-stimulated cells. E, the limiting membrane of these vacuoles was also positive for CD63, Lamp2, and PI3P lipids (EGFP 2xFYVEhrs). ns = not significant; **p < 0.01, ***p < 0.001. Size bar and detailed view: 10 μm.
Vac14-complex-associated Vacuolization Correlates with an Accumulation of Autophagy Marker Proteins but Has No Impact on the Autophagic Flux
YM201636 treatment leads to deregulated autophagy (21, 22, 24) (Fig. 5B). To test the effect of the Vac14 level on autophagy, we analyzed the intracellular levels of marker proteins LC3 and p62 (Fig. 5A). Vac14 overexpressing cells showed elevated levels of these marker proteins, arguing for a defect in autophagosomal maturation processes without changes in the endogenous apoptosis level (supplemental Figs. S4A and S5). In order to test whether the vacuoles were positive for the autophagosomal marker protein LC3, a construct to express GFP-LC3 was transiently transfected into these cells. The GFP-LC3 signal was not localized on the vacuolar structures as shown before for endolysosomal proteins (Figs. 3 and 4). Only small autophagosomal dots could be identified, which were in close proximity to the enlarged endolysosomal structures (indicated by arrows in supplemental Fig. S4D). Next, autophagic flux measurements were performed. The cells were treated with the vacuolar ATPase inhibitor bafilomycin A, which resulted in a further increase of LC3-II and p62 (supplemental Figs. 4B and 4C). Vacuolated cells, which already showed high levels of p62 and LC3-II, were less susceptible to bafilomycin-dependent effects. By contrast, starvation of cells with the standard medium changed to Earle's Balanced Salt solution led to a strong reduction of the p62 and LC3-II levels in both nonvacuolated and vacuolated cells, indicating that vacuolated cells were still able to increase their endogenous autophagic activity (see supplemental Figs. 4B and 4C). Taken together, Vac14 overexpression and YM201636 treatment led to elevated levels of LC3 and p62 without affecting the autophagic flux.
Fig. 5.
Vac14-complex-associated vacuolization causes an accumulation of autophagy marker proteins. Vacuolization due to Vac14 overexpression or YM201636 treatment was accompanied by a significant accumulation of autophagy marker proteins p62 and LC3-II. A, overexpression of Vac14 (wt and L156R) showed an increased level of p62 and LC3-II relative to the control cells (three samples each set). Densitometric evaluation of the Western blot analysis is shown on the right. B, lysates from HeLa cells treated with the PIKfyve inhibitor YM201636 for 24 h compared with nontreated cells. YM201636-derived vacuolization resulted in an accumulation of p62 and LC3-II. GAPDH served as a loading control. *p < 0.05, **p < 0.01, ***p < 0.001.
Identification of the Vac14 Interactome in Eukaryotic Cells
To further understand the processes of vacuolization, we utilized CoIP experiments with a 3xFLAG-based immunoaffinity approach in combination with MudPIT (25, 39–41). For the analysis, only peptides found in at least two of three independent Vac14 wt pulldown experiments were taken into consideration. Furthermore, a very stringent posterior error probability of 0.1% was applied to ensure high-quality identifications. Mapping of proteotypic peptides to proteins was straightforward. First, only those proteins that contained exclusively PD peptides were added to the group of Vac14 interacting proteins (supplemental information SI 1). Additionally, proteins that shared nonproteotypic peptides were merged in protein groups. Obviously, the mapping of a nonproteotypic peptide to a protein is problematic, as one cannot pinpoint which proteins have actually been pulled down during the assay. However, because all peptides shared within a PD group are all PD, one can conservatively use all proteins for further analysis. Thus, those proteins of PD groups were incorporated into the group of Vac14 interacting proteins (Fig. 6B). For further details see “Experimental Procedures.”
This approach led to the identification of 946 proteins that are known to be involved in different cellular pathways and functions (for details see supplemental information SI 1). To further reduce the number of proteins and isoforms, we mainly focused on reviewed UniProtKB/Swiss-Prot proteins, which led to 337 different proteins (supplemental information SI 2, reviewed as Vac14 interactome). A further manual-curation process led to a group of 16 interesting candidates that have been linked to endolysosomal or autophagic membrane dynamics and are summarized in Table I.
Table I. Vac14-associated proteins of the endolysosomal trafficking machinery. The table shows selected proteins identified in the CoIP/MudPIT analysis that are reported to be associated with endolysosomal homeostasis. The full list of identified proteins is given as supplemental data (supplemental information SI 1).
| UniProt identifier | Protein name | Gene I.D. | Cellular function | Reference(s) |
|---|---|---|---|---|
| P53621 | COP coat components | COPA, COPB2 | Cop coat components in concert with Arf GTPases play a crucial role in intracellular membrane budding processes | (51) |
| P35606 | α, β 2, δ, and ϵ | COPD, COPE | ||
| P48444 | ||||
| O14579 | ||||
| P40616 | Arf-like protein 1 | ARL1 | Arl1 binds to BIG1 and controls its intracellular localization | (52, 53) |
| Q9Y6D6 | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) | ARPGEP1/BIG1 | BIG controls the activation of Arf GTPases | (53) |
| BIG1 interacts with Arl1 | ||||
| P46459 | Vesicle-fusing (vesicular-fusion protein NSF) | NSF | NSF is a member of AAA+ ATPases | (54, 55) |
| Controls membrane recognition and fusion together with SNAP/SNAREs and Rab GTPases | ||||
| Q9H269 | Human vacuolar protein sorting 16 | VPS16 | Part of HOPS complex | (57) |
| Plays a role in homotypic fusion and MVB/late endosome homeostasis | ||||
| Q5VIR6 | Human vacuolar protein sorting 53 | VPS53 | Part of GARP complex that controls transport of lysosomal hydrolases to the lysosome | (56, 80) |
| Regulates the retrograde lysosome to trans-Golgi network transport | ||||
| P51151 | Rab9 | RAB9 | Rab9 localizes to late endosomal and lysosomal structures | (66, 72, 81) |
| Retrograde MVB/lysosome to trans-Golgi network trafficking | ||||
| Essential component of noncanonical autophagy | ||||
| Q14C86 | GTPase-activating protein and VPS9 domain-containing protein 1 | GAPEX5/GAPVD1 | Rab5-activating protein (Rab5 GAP); mediates insulin-stimulated PI3P production | (59, 82) |
| Is involved in trafficking of Glut4 vesicles | ||||
| Q9UH99 | SUN domain-containing protein 2 | SUN2/RAB5IP | Rab5-interacting protein that controls homotypic fusion | (60, 83, 84) |
| Nuclear-cytoplasmic connection in concert with LINC complex | ||||
| Q8TC07 | TBC1 domain family member 15 | TBC1D15 | Inactivates the Rab7 GTPase | (34, 61) |
| Provides links to the HOPS complex via VPS39 interaction | ||||
| Controls lysosome morphology | ||||
| Binding to LC3/Atg8 protein family | ||||
| Q66K14 | TBC1 domain family member 9B | TBC1D9B | Binding to LC3/Atg8 protein family | (61) |
| Q9UI12 | V-type proton ATPase catalytic subunit H | ATP6V1H | Regulatory part of cytoplasmic V1 subunit of v-ATPase | (63, 85) |
| Acidification of intracellular compartments | ||||
| Regulation of endolysosomal maturation and lysosomal activity | ||||
| P42345 | mTOR-kinase | MTOR | mTOR is part of the mTORC1 complex | (65, 86, 87) |
| Negative regulator of autophagy and re-formation of lysosomes | ||||
| Crucial for cell growth, metabolism, and cell cycle progression |
Proteins that provide direct links to the budding and fusion machinery, such as COP coat components (COPA, COPB2, COPD, and COPE), Arf-like protein 1, and its interaction partner Arf GTPase activator Brefeldin A-inhibited guanine nucleotide-exchange protein 1, were identified (51–53). Moreover, we found peptides representing the ATPase NSF, which controls membrane recognition and fusion in concert with soluble NSF attachment proteins or proteins associated with their receptors and Rab GTPases (see below and Refs. 54 and 55).
The Vac14 interactome characterization also revealed an association of 3xFLAG-Vac14 wt with VPS16 and VPS53 (vacuole protein sorting). The VPS53 protein is part of the mammalian Golgi-associated retrograde protein complex, which controls the sorting of the lysosomal protease cathepsin D to lysosomes and plays a role in the delivery of retrograde cargo into the trans-Golgi network (56). VPS16 is a component of the VPS-C complexes CORVET (class C core vacuole/endosome tethering) and HOPS (homotypic fusion and protein sorting). Studies in yeast and mammalian cells revealed that HOPS and CORVET complexes are essential for late endosome and lysosome assembly, as well as for endolysosomal trafficking, also including the maturation of autophagic compartments (57).
Given that accumulating data from other studies suggest that Rab GTPases in concert with lipids of the PIP family are crucial for membrane dynamics and subcellular membrane identity, it is interesting that we identified peptides that correspond to Rab9 and Rab regulators (58). These include Gapex-5 (GAPVD1) (59) and SUN domain-containing protein 2 (SUN2) (60), which are both GTPase activating proteins (GAPs) for Rab5, and two TBC-domain containing proteins, TBC1D9 and TBC1D15. The majority of these TBC proteins are poorly characterized, but it is assumed that many of them act as Rab GAP proteins (61, 62), like TBC1D15, which specifically acts as a GAP for Rab7 (34).
The regulatory subunit of the vacuolar ATPase (ATP6 V1H) and the serine/threonine kinase mTOR were additional promising candidates resulting from the screen. The vacuolar ATPase has an important role in the acidification of intracellular vesicles and controls the maturation of endolysosomal and autophagic compartments (63). The mTOR kinase, as part of mTOR complex 1, negatively regulates autophagy and is crucial for the re-formation of lysosomes (64, 65).
With regard to our previous results, showing that endolysosomal marker proteins, such as Rab7, significantly accumulated as a result of Vac14 wt and L156R overexpression (Figs. 2–4, supplemental Fig. S2), we compared the putative Vac14 interactome (337 hits; see supplemental information SI 1) with the complete interactomes of endolysosomal Rab GTPases (see supplemental Table S1) using Cytoscape (50). The intersections of these Cytoscape analyses with the Vac14 interactome (supplemental information SI 1) and the endosomal Rab GTPase interaction network resulted in the identification of five common proteins (TBC1D15, Rab9, SUN2, IDH3A, and NSF; Fig. 6C, supplemental information SI 2).
We focused on TBC1D15, which binds to the HOPS component VPS39 and functions as a Rab7 GAP, and Rab9, which is known to be localized to late endosomal and lysosomal membranes (34, 66). To confirm the MudPIT results, we used lysates from 3xFLAG, 3xFLAG Vac14 wt, and L156R overexpressing HEK293 cells for CoIP assays. Subsequent Western blot analysis revealed that Vac14 (wt and L156R) specifically binds to TBC1D15 and endogenous Rab9 (Fig. 7A). Moreover, lysates from Vac14-overexpressing and YM201636-treated cells showed a significant accumulation of endogenous Rab9 (Figs. 7B and 7C). Finally, immunofluorescence studies demonstrated that almost all of the limiting membranes of vacuoles caused by Vac14 overexpression or YM201636 treatment were strongly decorated with EGFP-Rab9 CA (Fig. 7D).
Fig. 7.
Vac14 interacts with the Rab7 GAP TBC1D15 and Rab9. A, stable 3xFLAG-overexpressing cell lines were transiently transfected with myc-TBC1D15. Beads that were covalently linked to the FLAG-tag specific antibody M2 were used in CoIP assays. Only lysates that expressed 3xFLAG-Vac14 wt or the Vac14 L156R mutant showed a direct association with co-expressed myc-TBC1D15 and endogenous Rab9. Cells expressing myc-TBC1D15 and the 3xFLAG alone served as negative controls and were not able to precipitate myc-TBC1D15 and endogenous Rab9. B, overexpression of 3xFLAG-Vac14 wt and Vac14 L156R in HeLa cells led to a significant accumulation of Rab9 relative to the loading control GAPDH. C, the same effect was observed in cells treated with the PIKfyve inhibitor YM201636 when compared with the loading control GAPDH (Fig. 5B). D, EGFP-Rab9 CA localized on the limiting membranes of vacuoles induced by overexpression of Vac14 wt/L156R or YM201636 treatment. *p < 0.05, ***p < 0.001. Size bar and detailed view: 10 μm.
DISCUSSION
Reduced levels of Vac14, the expression of a dominant-negative mutant of Vac14 (ingls) that is unable to bind PIKfyve lipid kinase, and the inhibition of PIKfyve activity lead to changes in the PIP homeostasis and to the formation of intracellular vacuoles (5, 14). Here we have shown for the first time that even increased levels of Vac14 are sufficient to induce vacuolization in eukaryotic cells. Vacuolization can be explained as a morphological consequence of disturbed membrane fusion of incoming vesicles with target membranes, reduced formation of intraluminal vesicles, or a defect in the budding of vesicles (67). The aim of the study was to elucidate the role of Vac14-associated vacuolization in more detail.
Vac14-induced vacuoles led to a characteristic and significant accumulation of late endosomal and lysosomal (Rab7, CD63, and Lamp2) and autophagy marker proteins (p62 and LC3). This is in line with previous studies and provides evidence that endosomal trafficking and maturation and autophagy processes could be disturbed (14, 18, 19, 68).
Live cell imaging analysis of Vac14-induced vacuoles on the other side revealed that the limiting membranes, especially those from large vacuoles, are highly positive for EGFP-Rab7, EGFP-CD63, or Lamp2-mCherry, but negative for EGFP-LC3. Thus, the alterations affect predominantly endolysosomal maturation processes as opposed to autophagy-linked events. Autophagic flux experiments proved that the vacuolated cells were still able to increase their autophagic activity during starvation. Hence, the accumulation of LC3-II could be an indirect consequence of dysfunctional endolysosomal maturation, or it could arise because of a delay in autophagic processes.
Less is known about the linkage between Vac14 and other regulators of membrane dynamics. Therefore, we investigated this in more detail via a CoIP/MudPIT approach. The subsequent bioinformatic evaluation resulted in a list of proteins involved in numerous cellular functions such as cell motility and cytoskeletal dynamics or cell growth (Table I, supplemental information SI 1).
Many of the identified proteins are involved in membrane budding or the fusion of membranes, like ATPase NSF or various COPs. In addition, proteins mediating vesicular transport processes of the endolysosomal pathway were identified, including Rab9 and regulating proteins of Rab5 (e.g. GAPEX, SUN2) and Rab7 (TBC1D15). This is of special interest because there is increasing evidence that Rab GTPases are interconnected with lipids of the PIP family in controlling the identity of intracellular membranes (58).
We also identified mTOR via the MudPIT approach, which is part of mTOR complex 1 and serves as a negative regulator of autophagic processes (65, 69). This is interesting, because it has recently been demonstrated that the mTOR complex 1 component Raptor specifically binds to phosphatidylinositol-3,5-bisphosphate lipids and is important for the proper localization of the mTOR complex 1 complex (70). This suggests that the scaffold protein Vac14 might have additional regulatory effects on mTOR activation.
To decipher the interconnection between Vac14 and regulators of the endolysosomal pathway in more detail, we focused on Rab9 and the Rab7 GAP TBC1D15 and confirmed the interaction with Vac14 via CoIP. Rab9 predominantly localizes to late endosomal and lysosomal membranes and facilitates late endosome-to-trans-Golgi-network transport, a function that was demonstrated earlier for Vac14 as well (66, 71, 72). In addition, recent studies elucidated a role of Rab9 in (noncanonical) autophagy (73, 74). The authors showed that autophago-lysosomes from trans-Golgi and late endosomal membranes are transported in a Rab9-dependent manner (73, 74). As Rab9 accumulates in vacuolated cells and almost completely localizes to the limiting membranes of these vacuoles, our data suggest a blockade of late endosome-to-trans-Golgi-network transport, as well as of Rab9-dependent autophagic processes. This could also provide an explanation for the elevated autophagy marker proteins in these vacuolated cells.
Apart from Rab9, the Rab7 regulator TBC1D15 is an interesting Vac14 interacting protein. Edinger and colleagues elegantly confirmed the function of TBC1D15 as a Rab7-specific GAP and showed that this protein, in concert with VPS39, regulates lysosomal morphology (34). VPS39, like VPS16—also identified in our MudPIT analysis—is a component of the HOPS complex (57). This complex is essential for the homotypic fusion of late endosomal structures and is probably linked to the acidification of endolysosomal compartments via vacuolar ATPase function (75). Supporting studies in yeast showed an association of the HOPS complex components Vps39 and Vps41 with Ypt7p, which is the yeast orthologue of Rab7 (57, 76). In yeast the HOPS complex regulates the fusion and fission status of Ypt7-positive membranes. These processes are controlled by Gyp7p, which acts as a GAP for Ypt7p, similar to mammalian TBC1D15 for Rab7 (77). Interestingly, TBC1D15 is linked to members of the LC3 protein family, which is another indicator of an association between autophagosomal and late endosomal trafficking processes (78).
In addition to the labeling with late endosomal proteins, the Vac14-induced vacuoles were strongly decorated by the PI3P marker EGFP 2xFYVEhrs. This is in line with data from Catimel et al., who investigated the PI3P interactome in a colon cancer cell line and identified TBC1D15 as a PIP-binding protein, arguing for a link between Rab7 controlling factors like TBC1D15 and PI3P-positive membranes (79). Thus, TBC1D15 might serve as an interconnection between PIP homeostasis and Rab7-related recruitment of effector proteins onto late endosomal compartments.
In Vac14 overexpressing cells, Vac14 probably sequesters the Rab7 GAP TBC1D15 in the cytosol, resulting in a reduced inactivation of Rab7 on late endosomal membranes. This in turn might also block Rab9-dependent retrograde transport events from the multivesicular body/lysosome back to the trans-Golgi network, providing an explanation for vacuolization and the observed accumulation of late endosomal and lysosomal marker proteins.
Together our findings serve as evidence that Vac14 is crucial for Rab7-dependent endolysosomal maturation processes and Rab9-dependent transport of lysosomal membranes to the trans-Golgi network. However, further cell biological studies are necessary in order to decipher the precise role of diverse components of the Vac14 associated network.
Supplementary Material
Acknowledgments
We thank Katja Brinkmann for excellent technical assistance and Ivona Djuric for critical reading of the manuscript. Additionally, we are grateful to Angelika Barnekow (Muenster, Germany), Aimee Edinger (Irvine, CA), Britta Eickholt (Berlin, Germany), Eric O. Freed (Frederick, MD), Geoffrey D. Holman (Bristol, UK), Tony Pawson (Toronto, Ontario, Canada), Harald Stenmark (Oslo, Norway), Didier Trono (Lausanne, Switzerland), Thomas Westbrook (Houston, TX), and Lois Weisman (Ann Arbor, MI) for providing plasmids.
Footnotes
Author contributions: U.S., B.V., H.P., and T.W. designed research; U.S., B.V., D.A.B., T.L., D.G., B.F., R.K., and T.W. performed research; U.S., J.K., B.F., R.K., J.B., C.F., and D.A.W. contributed new reagents or analytic tools; U.S., B.V., B.F., R.K., J.B., C.F., D.A.W., H.P., and T.W. analyzed data; U.S., B.V., H.P., and T.W. wrote the paper.
* The work was supported by grants from Fritz-Thyssen-Stiftung (FTS Az.10.11.1.224) and from the German Research Foundation to T.W. and H.P. (DFG PA483/16-1), D.W. (DFG WO1214/4-1), and C.F. (DFG FU780/2).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PIP
- phosphoinositide lipid derivative
- CA
- constitutively active
- CoIP
- co-immunoprecipitation
- COP
- coatomer protein
- EGFP
- enhanced green fluorescent protein
- GAP
- GTPase activating protein
- HOPS
- homotypic fusion and vacuole protein sorting
- MS
- mass spectrometry
- MudPIT
- multidimensional protein identification technology
- NSF
- N-ethylmaleimide-sensitive factor
- PI3P
- phosphatidylinositol-3-phosphate
- PD
- pulldown only
- PSM
- peptide spectrum match
- SUN2
- SUN domain-containing protein 2
- TBC
- Tre2/bub2/cdc16
- wt
- wild type.
REFERENCES
- 1. Di P. G., De C. P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 2. Mayinger P. (2012) Phosphoinositides and vesicular membrane traffic. Biochim. Biophys. Acta 1821, 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCartney A. J., Zhang Y., Weisman L. S. (2014) Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. Bioessays 36, 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shisheva A. (2012) PIKfyve and its lipid products in health and in sickness. Curr. Top. Microbiol. Immunol. 362, 127–162 [DOI] [PubMed] [Google Scholar]
- 5. Jin N., Chow C. Y., Liu L., Zolov S. N., Bronson R., Davisson M., Petersen J. L., Zhang Y., Park S., Duex J. E., Goldowitz D., Meisler M. H., Weisman L. S. (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 27, 3221–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Botelho R. J., Efe J. A., Teis D., Emr S. D. (2008) Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig 4 phosphoinositide phosphatase. Mol. Biol. Cell. 19, 4273–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ikonomov O. C., Sbrissa D., Fligger J., Delvecchio K., Shisheva A. (2010) ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot-Marie-Tooth 4J disorder. J. Biol. Chem. 285, 26760–26764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ikonomov O. C., Sbrissa D., Fenner H., Shisheva A. (2009) PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J. Biol. Chem. 284, 35794–35806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sbrissa D., Ikonomov O. C., Fenner H., Shisheva A. (2008) ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J. Mol. Biol. 384, 766–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sbrissa D., Ikonomov O. C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., Shisheva A. (2007) Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 282, 23878–23891 [DOI] [PubMed] [Google Scholar]
- 11. Lemaire J. F., McPherson P. S. (2006) Binding of Vac14 to neuronal nitric oxide synthase: characterisation of a new internal PDZ-recognition motif. FEBS Lett. 580, 6948–6954 [DOI] [PubMed] [Google Scholar]
- 12. Dove S. K., McEwen R. K., Mayes A., Hughes D. C., Beggs J. D., Michell R. H. (2002) Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12, 885–893 [DOI] [PubMed] [Google Scholar]
- 13. Alghamdi T. A., Ho C. Y., Mrakovic A., Taylor D., Mao D., Botelho R. J. (2013) Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J. Biol. Chem. 288, 9363–9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., Zolov S. N., Chow C. Y., Slutsky S. G., Richardson S. C., Piper R. C., Yang B., Nau J. J., Westrick R. J., Morrison S. J., Meisler M. H., Weisman L. S. (2007) Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. U.S.A. 104, 17518–17523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zolov S. N., Bridges D., Zhang Y., Lee W. W., Riehle E., Verma R., Lenk G. M., Converso-Baran K., Weide T., Albin R. L., Saltiel A. R., Meisler M. H., Russell M. W., Weisman L. S. (2012) In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. U.S.A. 109, 17472–17477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonangelino C. J., Nau J. J., Duex J. E., Brinkman M., Wurmser A. E., Gary J. D., Emr S. D., Weisman L. S. (2002) Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156, 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rudge S. A., Anderson D. M., Emr S. D. (2004) Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig 4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell. 15, 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sbrissa D., Ikonomov O. C., Strakova J., Dondapati R., Mlak K., Deeb R., Silver R., Shisheva A. (2004) A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol. Cell. Biol. 24, 10437–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., McCartney A. J., Zolov S. N., Ferguson C. J., Meisler M. H., Sutton M. A., Weisman L. S. (2012) Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P(2) and PI(5)P. EMBO J. 31, 3442–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jefferies H. B., Cooke F. T., Jat P., Boucheron C., Koizumi T., Hayakawa M., Kaizawa H., Ohishi T., Workman P., Waterfield M. D., Parker P. J. (2008) A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 9, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaugel A., Bakula D., Hoffmann A., Proikas-Cezanne T. (2012) Defining regulatory and phosphoinositide-binding sites in the human WIPI-1 beta-propeller responsible for autophagosomal membrane localization downstream of mTORC1 inhibition. J. Mol. Signal. 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mauthe M., Yu W., Krut O., Kronke M., Gotz F., Robenek H., Proikas-Cezanne T. (2012) WIPI-1 positive autophagosome-like vesicles entrap pathogenic Staphylococcus aureus for lysosomal degradation. Int. J. Cell Biol. 2012, 179207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de L. J., Polson H., Feldman M., Shokat K., Tooze S. A., Urbe S., Clague M. J. (2009) PIKfyve regulation of endosome-linked pathways. Traffic 10, 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin S., Harper C. B., May L. M., Coulson E. J., Meunier F. A., Osborne S. L. (2013) Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One 8, e60152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Colwill K., Wells C. D., Elder K., Goudreault M., Hersi K., Kulkarni S., Hardy W. R., Pawson T., Morin G. B. (2006) Modification of the Creator recombination system for proteomics applications—improved expression by addition of splice sites. BMC Biotechnol. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., Pawson T. (2006) A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 27. Schluter M. A., Pfarr C. S., Pieczynski J., Whiteman E. L., Hurd T. W., Fan S., Liu C. J., Margolis B. (2009) Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell 20, 4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A., Schiff R., Rosen J. M., Westbrook T. F., Elledge S. J. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun Q., Westphal W., Wong K. N., Tan I., Zhong Q. (2010) Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. U.S.A. 107, 19338–19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson W. T., Giddings T. H., Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., Kirkegaard K. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matanis T., Akhmanova A., Wulf P., Del N. E., Weide T., Stepanova T., Galjart N., Grosveld F., Goud B., De Zeeuw C. I., Barnekow A., Hoogenraad C. C. (2002) Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4, 986–992 [DOI] [PubMed] [Google Scholar]
- 32. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whitley P., Reaves B. J., Hashimoto M., Riley A. M., Potter B. V., Holman G. D. (2003) Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J. Biol. Chem. 278, 38786–38795 [DOI] [PubMed] [Google Scholar]
- 34. Peralta E. R., Martin B. C., Edinger A. L. (2010) Differential effects of TBC1D15 and mammalian Vps39 on Rab7 activation state, lysosomal morphology, and growth factor dependence. J. Biol. Chem. 285, 16814–16821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duning K., Rosenbusch D., Schluter M. A., Tian Y., Kunzelmann K., Meyer N., Schulze U., Markoff A., Pavenstadt H., Weide T. (2010) Polycystin-2 activity is controlled by transcriptional coactivator with PDZ binding motif and PALS1-associated tight junction protein. J. Biol. Chem. 285, 33584–33588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duning K., Schurek E. M., Schluter M., Bayer M., Reinhardt H. C., Schwab A., Schaefer L., Benzing T., Schermer B., Saleem M. A., Huber T. B., Bachmann S., Kremerskothen J., Weide T., Pavenstadt H. (2008) KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 19, 1891–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. George B., Vollenbroker B., Saleem M. A., Huber T. B., Pavenstadt H., Weide T. (2011) GSK3beta inactivation in podocytes results in decreased phosphorylation of p70S6K accompanied by cytoskeletal rearrangements and inhibited motility. Am. J. Physiol. Renal Physiol. 300, F1152–F1162 [DOI] [PubMed] [Google Scholar]
- 38. Vollenbroker B., George B., Wolfgart M., Saleem M. A., Pavenstadt H., Weide T. (2009) mTOR regulates expression of slit diaphragm proteins and cytoskeleton structure in podocytes. Am. J. Physiol. Renal Physiol. 296, F418–F426 [DOI] [PubMed] [Google Scholar]
- 39. Franzel B., Wolters D. A. (2011) Advanced MudPIT as a next step toward high proteome coverage. Proteomics 11, 3651–3656 [DOI] [PubMed] [Google Scholar]
- 40. Washburn M. P., Wolters D., Yates J. R., III (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 41. Wolters D. A., Washburn M. P., Yates J. R., III (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 42. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bald T., Barth J., Niehues A., Specht M., Hippler M., Fufezan C. (2012) pymzML—Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28, 1052–1053 [DOI] [PubMed] [Google Scholar]
- 45. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 46. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 47. Specht M., Kuhlgert S., Fufezan C., Hippler M. (2011) Proteomics to go: Proteomatic enables the user-friendly creation of versatile MS/MS data evaluation workflows. Bioinformatics 27, 1183–1184 [DOI] [PubMed] [Google Scholar]
- 48. Kall L., Storey J. D., Noble W. S. (2008) Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 24, i42-i48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kall L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res. 7, 40–44 [DOI] [PubMed] [Google Scholar]
- 50. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Popoff V., Adolf F., Brugger B., Wieland F. (2011) COPI budding within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3, a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Donaldson J. G., Jackson C. L. (2011) ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christis C., Munro S. (2012) The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J. Cell Biol. 196, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao C., Smith E. C., Whiteheart S. W. (2012) Requirements for the catalytic cycle of the N-ethylmaleimide-Sensitive Factor (NSF). Biochim. Biophys. Acta 1823, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 56. Perez-Victoria F. J., Mardones G. A., Bonifacino J. S. (2008) Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol. Biol. Cell. 19, 2350–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Solinger J. A., Spang A. (2013) Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 280, 2743–2757 [DOI] [PubMed] [Google Scholar]
- 58. Jean S., Kiger A. A. (2012) Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 13, 463–470 [DOI] [PubMed] [Google Scholar]
- 59. Lodhi I. J., Chiang S. H., Chang L., Vollenweider D., Watson R. T., Inoue M., Pessin J. E., Saltiel A. R. (2007) Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 5, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffenberg S., Liu X., Nikolova L., Hall H. S., Dai W., Baughn R. E., Dickey B. F., Barbieri M. A., Aballay A., Stahl P. D., Knoll B. J. (2000) A novel membrane-anchored Rab5 interacting protein required for homotypic endosome fusion. J. Biol. Chem. 275, 24661–24669 [DOI] [PubMed] [Google Scholar]
- 61. Popovic D., Akutsu M., Novak I., Harper J. W., Behrends C., Dikic I. (2012) Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 64. Laplante M., Sabatini D. M. (2012) mTOR signaling. Cold Spring Harb. Perspect. Biol. 4, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., Hailey D. W., Oorschot V., Klumperman J., Baehrecke E. H., Lenardo M. J. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. (1993) Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wada Y. (2013) Vacuoles in mammals: a subcellular structure indispensable for early embryogenesis. Bioarchitecture 3, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ferguson C. J., Lenk G. M., Meisler M. H. (2009) Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18, 4868–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bridges D., Ma J. T., Park S., Inoki K., Weisman L. S., Saltiel A. R. (2012) Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 23, 2955–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ikonomov O. C., Sbrissa D., Mlak K., Deeb R., Fligger J., Soans A., Finley R. L., Jr., Shisheva A. (2003) Active PIKfyve associates with and promotes the membrane attachment of the late endosome-to-trans-Golgi network transport factor Rab9 effector p40. J. Biol. Chem. 278, 50863–50871 [DOI] [PubMed] [Google Scholar]
- 72. Barbero P., Bittova L., Pfeffer S. R. (2002) Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 156, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 [DOI] [PubMed] [Google Scholar]
- 74. Shimizu S., Arakawa S., Nishida Y. (2010) Autophagy takes an alternative pathway. Autophagy 6, 290–291 [DOI] [PubMed] [Google Scholar]
- 75. Takeda K., Cabrera M., Rohde J., Bausch D., Jensen O. N., Ungermann C. (2008) The vacuolar V1/V0-ATPase is involved in the release of the HOPS subunit Vps41 from vacuoles, vacuole fragmentation and fusion. FEBS Lett. 582, 1558–1563 [DOI] [PubMed] [Google Scholar]
- 76. Brocker C., Kuhlee A., Gatsogiannis C., Balderhaar H. J., Honscher C., Engelbrecht-Vandre S., Ungermann C., Raunser S. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. U.S.A. 109, 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brett C. L., Plemel R. L., Lobingier B. T., Vignali M., Fields S., Merz A. J. (2008) Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cell Biol. 182, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Catimel B., Kapp E., Yin M. X., Gregory M., Wong L. S., Condron M., Church N., Kershaw N., Holmes A. B., Burgess A. W. (2013) The PI(3)P interactome from a colon cancer cell. J. Proteomics 82, 35–51 [DOI] [PubMed] [Google Scholar]
- 80. Perez-Victoria F. J., Bonifacino J. S. (2009) Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol. Cell. Biol. 29, 5251–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yan H., Li W. L., Xu J. J., Zhu S. Q., Long X., Che J. P. (2013) D2 dopamine receptor antagonist raclopride induces non-canonical autophagy in cardiac myocytes. J. Cell. Biochem. 114, 103–110 [DOI] [PubMed] [Google Scholar]
- 82. Lodhi I. J., Bridges D., Chiang S. H., Zhang Y., Cheng A., Geletka L. M., Weisman L. S., Saltiel A. R. (2008) Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol. Biol. Cell 19, 2718–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Haque F., Mazzeo D., Patel J. T., Smallwood D. T., Ellis J. A., Shanahan C. M., Shackleton S. (2010) Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 285, 3487–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W., Chen H., Jin C., Wei J., Zhang W., Yang Y., Liu Q., Zhou Y., Zhang C., Wu Z., Xu W., Zhang Y., Liu T., Yu D., Zhang Y., Chen L., Zhu D., Zhong X., Kang L., Gan X., Yu X., Ma Q., Yan J., Zhou L., Liu Z., Zhu Y., Zhou T., He F., Yang X. (2011) Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 7, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hinton A., Bond S., Forgac M. (2009) V-ATPase functions in normal and disease processes. Pflugers Arch. 457, 589–598 [DOI] [PubMed] [Google Scholar]
- 86. Settembre C., Fraldi A., Medina D. L., Ballabio A. (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.