Abstract
Sorafenib is a multi-kinase inhibitor that has been proven effective for the treatment of unresectable hepatocellular carcinoma (HCC). However, its precise mechanisms of action and resistance have not been well established. We have developed high-density fluorescence reverse-phase protein arrays and used them to determine the status of 180 phosphorylation sites of signaling molecules in the 120 pathways registered in the NCI-Nature curated database in 23 HCC cell lines. Among the 180 signaling nodes, we found that the level of ribosomal protein S6 phosphorylated at serine residue 235/236 (p-RPS6 S235/236) was most significantly correlated with the resistance of HCC cells to sorafenib. The high expression of p-RPS6 S235/236 was confirmed immunohistochemically in biopsy samples obtained from HCC patients who responded poorly to sorafenib. Sorafenib-resistant HCC cells showed constitutive activation of the mammalian target of rapamycin (mTOR) pathway, but whole-exon sequencing of kinase genes revealed no evident alteration in the pathway. p-RPS6 S235/236 is a potential biomarker that predicts unresponsiveness of HCC to sorafenib. The use of mTOR inhibitors may be considered for the treatment of such tumors.
Hepatocellular carcinoma (HCC)1 is the third most common cause of cancer-related death worldwide (1). Advanced HCC often cannot be managed with local treatments (surgical resection, ethanol injection, radiofrequency ablation, chemoembolization), but no systemic chemotherapy with conventional cytotoxic agents had been shown to be effective until a landmark phase III clinical trial (the Sorafenib HCC Assessment Randomized Protocol) revealed significant survival prolongation in patients treated with sorafenib (Nexavar; Bayer Healthcare Pharmaceuticals Inc. Berlin, Germany) (2). Furthermore, it has been reported that some patients show remarkable tumor shrinkage after short-term administration of sorafenib (3). Based on these results, sorafenib monotherapy has been employed as the current standard first-line treatment for unresectable HCC. However, not all HCC patients show the desired therapeutic benefits of sorafenib. The overall survival prolongation of unselected patients in the SHARP trial was limited to 2.8 months (2), and an objective tumor response was observed only in a small proportion of patients (0.6% to 2%) (2, 4). Given the relatively high cost and occasional severe adverse events (diarrhea, hand-foot skin reaction, hypertension, and others) (2, 4), there is an urgent need to identify a predictive biomarker that could exclude advanced HCC patients who are unlikely to benefit from sorafenib therapy.
Sorafenib is a multi-kinase inhibitor that blocks tumor cell proliferation and angiogenesis through the inhibition of c-RAF and b-RAF, as well as many receptor tyrosine kinases, including vascular endothelial growth factor receptors 2 and 3, platelet-derived growth factor receptor-α, Fms-related tyrosine kinase 3, RET, and c-KIT (5). In view of this broad inhibitory spectrum, the precise mechanisms underlying the anti-tumor activity remain elusive. To date, factors that have been identified as correlated with the efficacy of sorafenib include phosphorylated extracellular signal-regulated kinase 1 (p-ERK) (6), serum des-γ-carboxyprothrombin level (7), phosphorylated c-Jun protein (8), and fibroblast growth factor-3/4 gene amplification (3), but their clinical utility as predictive biomarkers has not been established.
In the present study, we developed a new technique, high-density fluorescence reverse-phase protein array (RPPA), and used it to search for a biomarker that would identify patients in whom sorafenib would be effective, employing a large library of phosphorylation-site-specific antibodies. RPPA represents an emerging technology for proteomics, and it is well suited for the profiling of phosphorylated proteins. It involves micro-format dot immunoblotting of lysates from tissues or cells (9), allowing simultaneous monitoring of the expression of a particular phosphoprotein in hundreds to thousands of samples under identical conditions in a highly quantitative manner (10). In this study we profiled the activation status of 180 key signaling nodes across a panel of 23 HCC cell lines and identified de novo activation of mTOR signaling in sorafenib-resistant HCC cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Antibodies
Cell lines used for generating the cancer cell line RPPA are listed in supplemental Table S1 and were maintained according to their suppliers' recommendations. Recombinant EGF was obtained from R&D Systems (Minneapolis, MN). A total of 180 phosphorylation-site-specific antibodies and their dilutions used for RPPA analysis are listed in supplemental Table S2. The specificity of each antibody was verified by immunoblotting or had been previously described by other investigators.
RPPA
Cells were collected by scraping and stored at −80¦°C until use. Cell lysates were prepared with RIPA buffer (Thermo Scientific, Rockford, IL) supplemented with phosphatase (Thermo Scientific) and protease (Sigma, St. Louis, MO) inhibitor cocktails. Protein concentrations of lysates were determined via the Bradford method (Bio-Rad Laboratories, Hercules, CA). The lysates were serially diluted 2-fold four times and printed in quadruplicate onto ProteoChip glass slides (Proteogen, Seoul, South Korea) using a robotic spotter (Genex Arrayer, Kaken Geneqs Inc., Chiba, Japan).
The RPPA slides were incubated overnight with primary antibodies. Following tyramide signal amplification (Dako Cytomation, Glostrup, Denmark), streptavidin Alexa Fluor 647 conjugate (Invitrogen, Carlsbad, CA) was applied to the slides (11). Fluorescence images were captured by an InnoScan 700 microarray scanner (Innopsys, Carbonne, France) and quantified using Mapix software (Innopsys). After background subtraction, values relative to γ-tubulin were subjected to quantile normalization (12) to ensure a uniform distribution of values for each slide in a set of slides. Unsupervised hierarchical clustering, using the Euclidean metric and Ward's method, was conducted with R 2.13.0. The signaling components of the mTOR and MAPK pathways were selected based on KEGG pathway maps and used for clustering analyses.
Immunoblot Analysis
Immunoblot analyses were performed using the NuPAGE Bis-Tris or Tris-Acetate electrophoresis system (Invitrogen) as described previously (13). All antibodies except for an anti-p-RSK (S380) antibody (R&D Systems) were obtained from Cell Signaling Technology (Danvers, MA). Signals were detected with the ImageQuant LAS 4010 system (GE Healthcare, Giles, UK) and quantified using the ImageQuant TL software package (GE Healthcare).
Growth Inhibition Assay
Sorafenib, RAD001 (everolimus), and SL0101 were purchased from Toronto Research Chemicals Inc. (North York, Ontario, Canada). CI-1040 and AZD8055 were from Selleck Chemicals (Houston, TX). Stock solutions of the chemicals were prepared in dimethyl sulfoxide and stored at −20¦°C until use. Cells were seeded into 96-well cell culture plates in triplicate at a density of 3000 cells per well. On the following day, serially diluted drugs were added, and 72 h later cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Fitchburg, WI). Relative cell viability was calculated as a percentage of a control treated with 0.1% dimethyl sulfoxide after background subtraction. All experiments were repeated at least three times. The data were modeled using a four-parameter log-logistic nonlinear regression curve fit with a sigmoid dose response. These curves were drawn using R 2.13.0, and IC50 values were calculated accordingly.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections of needle biopsy samples obtained from nine HCC patients before administration of sorafenib at Wan Fang Hospital and the Taipei Medical University Hospital were immunostained with anti-p-RPS6 Ser235/236 (#2211, Cell Signaling Technology) or anti-RPS6 (#2217, Cell Signaling Technology) antibody, as described previously (13). The stained slides were evaluated by pathologists and classified according to the percentage of positively stained cells (0 = 0%, 1 = 1% to 25%, 2 = 26% to 50%, 3 = 51% to 75%, and 4 = 76% to 100%) and the intensity of staining (0, absent; 1, weak; 2, moderate; and 3, strong). A specimen was defined as positive when either the percentage of positively stained cells or the intensity of staining was 3 or higher. The use of clinical materials was approved by the respective institutional review boards.
Kinome Sequencing
Genomic DNA was extracted from 20 HCC cell lines using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), in accordance with the manufacturer's protocol. DNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific). Three micrograms of genomic DNA was used to construct libraries for sequencing. The quality of the constructed libraries was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). All the exon and 5′- and 3′-flanking sequences (200 bp) of 511 kinase genes were captured using a customized SureSelect Target Enrichment System (Agilent Technologies) according to the Illumina Paired-End Sequencing Platform Library Prep Protocol Version 1.0 (Agilent Technologies). Captured DNA fragments (∼300 bp) were sequenced using a Genome Analyzer IIx sequencer (Illumina, San Diego, CA). Base calling was performed using the Illumina Pipeline (v1.4) with default parameters. Only paired end (2 × 75 bases) sequence reads that passed the quality control were mapped to the human reference genome build hg19 (UCSC hg19) using BWA (14) with default parameters. Sequencing artifacts were eliminated using Picard MarkDuplicates. Variants were called with SAMtools (15) and annotated using Annovar ENREF 25 (16). The final set of novel variant calls was identified using the following thresholds: SNP quality ≥ 228, coverage ≥ 20 reads, frequency ≥ 10%, and not deposited in the dbSNP database (www.ncbi.nlm.nih.gov/projects/SNP/) (version 135).
Evaluation of Synergistic Drug Combinations
The synergistic interaction of drug combinations was evaluated using the Chou–Talalay median-dose effect method (17) with CompuSyn software. AZD8055 and CI-1040 were mixed at the ratio of their IC50 values (1:300). The mixed solution was 2-fold serially diluted five times and added to the cells. Combination Index values were calculated at the points causing 50%, 75%, and 90% reduction of cell viability. Combination Index values equal to 1, >1, and <1 indicate additive, antagonistic, and synergistic interactions, respectively.
RESULTS
Generation of the High-density RPPA and Phosphoprotein Profiling
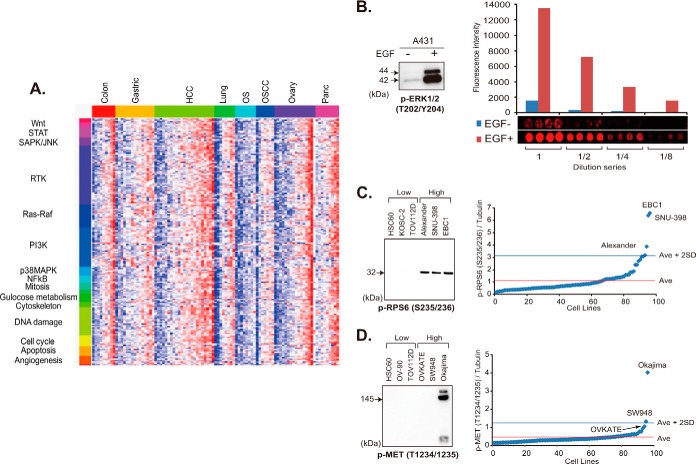
We constructed an RPPA onto which lysates of 95 cell lines derived from eight different types of cancer (listed in supplemental Table S1) cultured in the presence and absence of 10% fetal calf serum (FCS) for 17 h and A431 cells untreated or treated with 200 ng/ml EGF for 10 min were randomly plotted. Each lysate was serially diluted (1:1, 1:2, 1:4, and 1:8) and spotted in quadruplicate (16 spots per lysate). This level of high-density spotting (3072 samples per array slide) was achievable because of the highly hydrophobic surface of the array slides, which prevented diffusion of the protein samples.
By applying 180 phosphorylation-site-specific antibodies (listed in supplemental Table S2), we determined the activation status of signaling proteins (Fig. 1A). A lysate of A431 cells treated with EGF was included as a positive internal control. A431 cells carry amplification of the EGFR (EGF receptor) gene. We confirmed that a >6-fold increase in the signal intensity of ERK1/2 proteins phosphorylated at the threonine 202/tyrosine 204 residue (p-ERK1/2 T202/Y204) was detectable after treatment with EGF (Fig. 1B).
Fig. 1.
Phosphoprofiling of key signaling molecules by RPPA. A, phosphorylation status of 180 signaling nodes in a panel of 95 cancer cell lines cultured in the presence of 10% FCS. Red and blue colors indicate high- and low-level phosphorylation, respectively. STAT, signal transducers and activators of transcription; SAPK/JNK, stress-activated protein kinase/c-Jun NH2-terminal kinase; RTK, receptor tyrosine kinase; PI3K, phosphatidylinositol 3′-kinase; MAPK, mitogen-activated protein kinase; NFκB, nuclear factor-kappaB; OS, osteosarcoma; OSCC, oral squamous cell carcinoma. B, immunoblot (left) and RPPA (right) analyses of A431 cells cultured without (−) and with (+) EGF for 10 min with anti-p-ERK1/2 (T202/Y204) antibody. The mean fluorescence intensity in arbitrary units (top) and images (bottom) of quadruplicate RPPA spots of lysate undiluted (1) and diluted 1:2 (1/2), 1:4 (1/4), and 1:8 (1/8) -fold are shown (right). C, D, relative p-RPS6 S235/236 (C) and p-Met T1234/1235 (D) expression of 95 cell lines determined via RPPA (right). Cell lines with the three highest and three lowest levels of expression were selected and subjected to immunoblotting with the same antibody (left). Ave, average.
To further verify the data obtained via RPPA, lysates of representative cell lines were electrophoresed and blotted with the same antibodies. In the RPPA analysis, EBC1, SNU-398, and Alexander cells showed a high signal intensity for anti-p-RPS6 S235/236 antibody, exceeding the average plus 2 S.D. for 96 cell lines, whereas TOV112D, KOSC-2, and HSC60 cells showed a signal intensity below the average (Fig. 1C, right). The results we obtained from immunoblotting were consistent (Fig. 1C, left).
The glass slides that we used for construction of the RPPA were free of any autofluorescence noise. The use of fluorescent dyes and original signal enhancement significantly increased the sensitivity of signal detection. In fact, MET protein with a high level of phosphorylation (p-MET T1234/1235) in Okajima cells was detectable via immunoblotting, whereas the MET protein with a relatively low level of phosphorylation in SW948 and OVKATE cells was undetectable (Fig. 1D).
p-RPS6 S235/236 Correlates with the Sensitivity of HCC Cells to Sorafenib
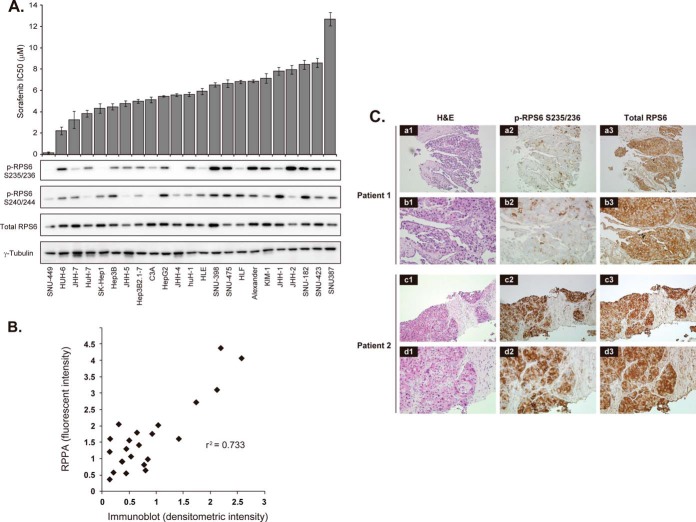
The cancer cell protein array contained 23 HCC cell lines exhibiting a wide variety of sensitivities to sorafenib (Fig. 2A, upper portion). SNU-449 was the most sensitive, with a half-maximal (50%) inhibitory concentration (IC50) of 0.172 μm. It was ∼70-fold more sensitive than the least sensitive cell line, SNU-387 (IC50 = 12.68 μm). We then compared the IC50 value of each HCC cell line with the phosphorylation level of 180 signaling nodes. Spearman's correlation coefficient analysis (supplemental Table S3) revealed that p-RPS6 S235/236 had the highest positive correlation (r = 0.58, p = 0.0044), followed by p-RPS6 at the serine 240/244 residues (p-RPS6 S240/244) (r = 0.55, p = 0.0070). 90-kDa ribosomal S6 kinase 2 (RSK2) protein phosphorylated at the serine 227 residue showed the third most significant correlation. RSK2 is one of the enzymes that phosphorylate RPS6 (18).
Fig. 2.
p-RPS6 S235/236 correlates with the sensitivity of HCC to sorafenib. A, upper graph, IC50 values for sorafenib against 23 HCC cell lines sorted from the most sensitive (left) to resistant (right) ones. Columns and error bars represent the mean and S.D. of three independent experiments, respectively. Lower panels, immunoblot analysis of pRPS6 S235/236, pRPS6 S240/244, RPS6, and γ-tubulin (loading control) expression in the 23 HCC cell lines. B, correlation between RPPA and immunoblot analyses of p-RPS6 S235/236 expression in the 23 HCC cell lines (R2 = 0.733). C, detection of p-RPS6 S235/236 in pretreatment biopsy samples. Hematoxylin and eosin (H&E) (a-d1) and immunoperoxidase staining with anti-p-RPS6 S235/236 (a-d2) and total RPS6 (a-d3) antibodies of HCC biopsy specimens obtained from a responder (patient 1 (a and b)) and a representative non-responder (patient 2 (c and d)) to sorafenib. Original magnification was ×40 (a1–3 and c1–3) and ×200 (b1–3 and d1–3).
Consistent with the RPPA data, intense signals for p-RPS6 S235/236 were detected in the sorafenib-resistant cell lines via immunoblotting (Fig. 2A, lower portion). The quantified immunoblot data correlated well with those of RPPA (r2 = 0.733), thus confirming the precision of the RPPA (Fig. 2B). p-RPS6 S235/236 and p-RPS6 S240/244 exhibited different phosphorylation patterns among several cell lines (e.g. HLF, KIM1, JHH-1, and JHH-2) (Fig. 2A, lower portion), suggesting that phosphorylation of S235/236 and S240/244 residues may be mediated by distinct regulatory processes.
p-RPS6 S235/236 Is a Potential Predictor of Response to Sorafenib
We next evaluated whether high levels of p-RPS6 S235/236 were indicative of HCC resistance to sorafenib in clinical samples (supplemental Table S4). Expression of p-RPS6 S235/236 was examined in biopsy specimens collected from nine HCC patients prior to sorafenib treatment (400 mg twice a day). Eight patients showed intense staining for p-RPS6 (Fig. 2C). Four patients (Cases 4, 6, 7, and 9) with p-RPS6-positive tumors discontinued sorafenib treatment because of disease progression within 2.3 months. Four patients (Cases 2, 3, 5, and 8) died as a result of disease progression after starting sorafenib treatment. In contrast, the remaining patient (Case 1), whose tumor was negative for p-RPS6, received sorafenib for 24 months and survived for 27 months. In this particular patient, tumor regression was confirmed by computed tomography scans performed three months after sorafenib administration and remained stable for another three months. In addition, the α-fetoprotein level dropped from 1621 to 314 ng/ml and remained low for 10 months. These results provide preliminary evidence that that high expression of p-RPS6 S235/236 might be useful for predicting which patients are unlikely to respond to sorafenib. As biopsy is not performed routinely before sorafenib treatment, we were unable to further validate the clinical significance of p-RPS6 S235/236 by examining additional cases.
mTOR Pathway Activation in Sorafenib-resistant Cells
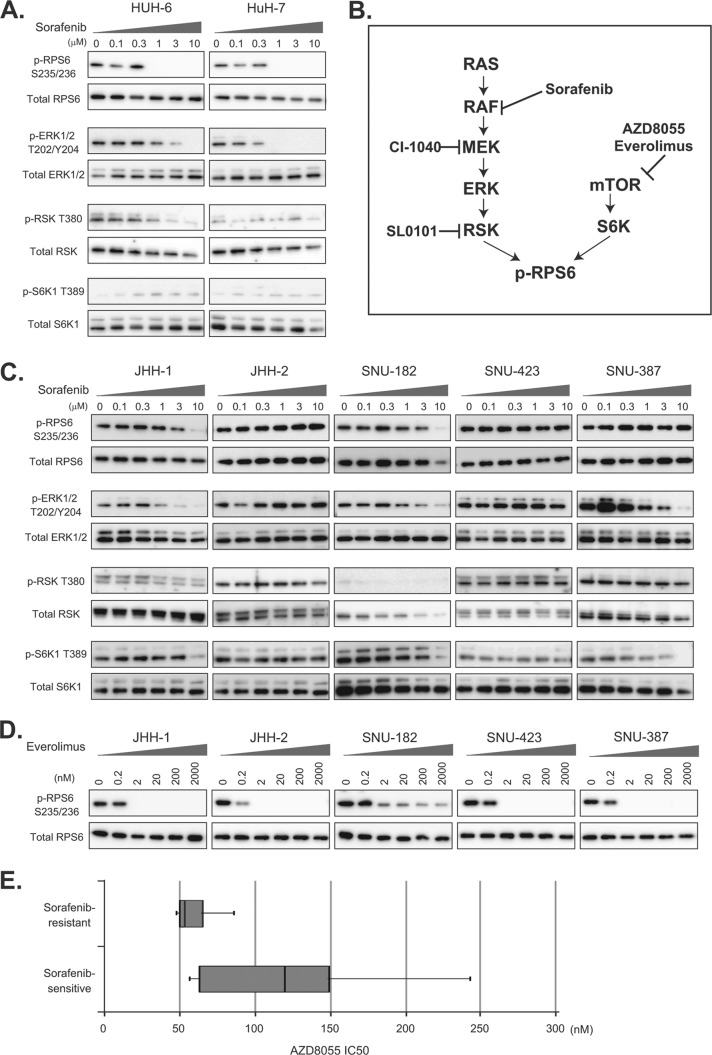
Given the association between p-RPS6 and sensitivity of HCC cell lines to sorafenib, we assessed the effects of sorafenib on p-RPS6 in representative sorafenib-sensitive and -resistant cell lines. The sorafenib-sensitive HUH-6 and HuH-7 cell lines demonstrated substantial dose-dependent decreases in p-RPS6 levels following treatment with sorafenib (Fig. 3A). Sorafenib also diminished the phosphorylation of downstream molecules in the MAPK pathway, ERK and RSK, in a dose-dependent manner (Fig. 3A), implying that the reduction of p-RPS6 in sorafenib-sensitive cells was likely attributable to blockade of the MAPK pathway (Fig. 3B).
Fig. 3.
Alternative mTOR signal activation in sorafenib-resistant HCC cells. A, C, representative sorafenib-sensitive (HUH-6 and HuH-7) and -resistant (JHH-1, JHH-2, SNU-182, SNU423, and SNU-387) HCC cells were treated with the indicated concentrations of sorafenib for 3 h, and the expression of p-RPS6 S235/236, total RPS6, p-ERK1/2 T202/Y204, total ERK, p-RSK T380, total RSK, p-S6K T389, and total S6K was determined via immunoblotting. B, schematic representation of the mTOR and MAPK pathways and their inhibitors. D, representative sorafenib-resistant (JHH-1, JHH-2, SNU-182, SNU423, and SNU-387) HCC cells were treated with the indicated concentrations of everolimus for 3 h, and the expression of p-RPS6 S235/236 and total RPS6 was determined via immunoblotting. E, distribution of IC50 values of representative sorafenib-sensitive (SNU-449, HUH-6, JHH-7, HuH-7, and SK-Hep1) and -resistant (JHH-1, JHH-2, SNU-182, SNU-423, and SNU-387) HCC cells to AZ8055. Boxes indicate 25th to 75th percentiles.
In contrast, the phosphorylation of RPS6 S235/236 in sorafenib-resistant cell lines, especially JHH-2, SNU-423, and SNU-387 cells, was insensitive to the same sorafenib treatment (Fig. 3C). The level of p-RPS6 in JHH-1 and SNU-182 cells decreased to some extent after sorafenib treatment, but a high concentration (10 μm) of sorafenib was necessary in order to suppress the phosphorylation of RPS6 completely (Fig. 3C), reflecting that the regulation of p-RPS6 in sorafenib-resistant cell lines is different from that in sensitive cell lines. RPS6 is also known to be phosphorylated by 70-kDa ribosomal S6 kinases (S6K) downstream of mTOR (Fig. 3B) (19, 20). We therefore speculated that the mTOR pathway might be alternatively activated in sorafenib-resistant cell lines. In fact, we found that sorafenib-resistant cells had a high level of p-S6K1 (Fig. 3C), whereas p-S6K1 was barely detectable in sorafenib-sensitive cells (Fig. 3A).
The phosphorylation of ERK in JHH-1 and SNU-182 was suppressed to some extent by sorafenib, but the low level of p-RSK and high level of p-S6K1 indicate that the main regulator of RPS6 phosphorylation was mTOR signaling rather than MAPK signaling.
Absence of Genetic Alterations in the MAPK and mTOR Pathways
RAF kinases are among the known targets of sorafenib, but sorafenib-resistant JHH2, SNU-423, and SNU-387 cells exhibited sustained activation of molecules located downstream of RAF (ERK and RSK), even in the presence of sorafenib (Fig. 3C), suggesting sorafenib-insensitive activation of the MAPK pathway.
To clarify the molecular mechanism driving the activation of the mTOR and MAPK pathways in sorafenib-resistant cells, we sequenced the entire exons of 511 kinases (listed in supplemental Table S5) in 20 HCC cell lines using a next-generation sequencer. Supplemental Table S6 lists all of the 322 genetic alterations that were not deposited in the dbSNP database. Due to the unavailability of normal counterparts, we were unable to determine whether these alterations were somatic. Eight kinds of DNA alterations were evident in the known mTOR and MAPK pathway genes (Supplemental Table S7). b-RAF V600E, found in SK-Hep1 cells, is a known driver mutation frequently observed in malignant melanoma (21). Two kinds of alterations were identified in the ATP-binding (S72A (JHH-7)) and AGC-kinase C-terminal (K335T (huH-1, SNU-475, and SNU-185)) domains of the RSK1 genes. Three kinds of alterations were found in the proline-rich domain (A420V (11 cell lines including SNU449) and V422I (HUH-6)) and catalytic (P267L (SK-Hep1, JHH-4, Kim1, and JHH-1)) domains of the S6K2 gene.
These eight alterations were validated using a conventional sequencing method (Supplemental Table S7), but no genetic alteration was specific to sorafenib-resistant cell lines. Infrequent alteration of the mTOR and MAPK pathway genes in HCC has been demonstrated by conventional sequencing analysis of surgical samples (22), and this was consistent with the present comprehensive sequencing data. The aberrant activation of the mTOR and MAPK pathways in sorafenib-resistant HCC cells was likely attributable to complex interplay between other oncogenic and anti-oncogenic pathways, or post-translational modifications.
mTOR Inhibitors Repress the Proliferation of Sorafenib-resistant Cells
The marked inhibition of p-RPS6 S235/236 following exposure to an mTOR inhibitor, everolimus, at a concentration as low as 2 nm (Fig. 3D) confirmed that activation of the mTOR pathway is responsible for the sorafenib-insensitive phosphorylation of RPS6 S235/236 in sorafenib-resistant cells. Consistently, sorafenib-insensitive cells tended to be more sensitive to another mTOR inhibitor, AZD8055, than sorafenib-sensitive cell lines (Fig. 3E), indicating that sorafenib-resistant cells are dependent for growth on constitutive activation of the mTOR pathway.
Synergy of mTOR and MAPK Inhibitors
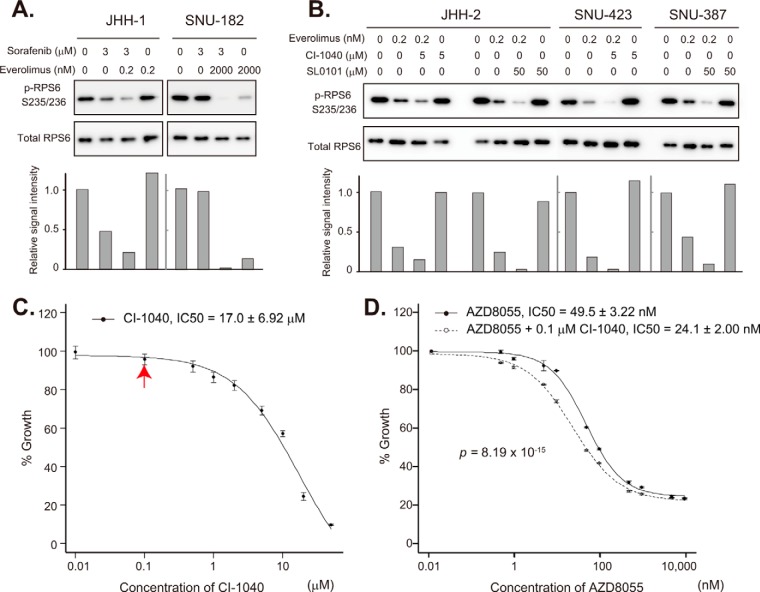
Although JHH-1 and SNU-182 cells showed resistance to sorafenib, their ERK phosphorylation was dose-dependently attenuated by sorafenib (Fig. 3C), indicating that the MAPK pathway in these cells still retained some sensitivity to the inhibition of RAF or other unknown MAPK-pathway kinases. In fact, sorafenib augmented the down-regulation of p-RPS6 S235/236 by everolimus (Fig. 4A). It is noteworthy that the low level of p-RPS6 S235/236 in SNU-182 cells sustained in the presence of 2 μm everolimus was completely abrogated by the addition of 3 μm sorafenib.
Fig. 4.
Synergy of mTOR and MAPK inhibitors. A, sorafenib-resistant JHH-1 and SNU-182 cells were treated with the indicated concentrations of sorafenib and everolimus, and the expression of p-RPS6 S235/236 and total RPS6 was examined via immunoblotting (top). The bottom panel indicates intensity relative to control blots (no treatment). B, sorafenib-resistant JHH2, SNU-423 and SNU-387 cells were treated with the indicated concentrations of CI-1040, SL0101, and everolimus, and the expression of p-RPS6 S235/236 and RPS6 was examined via immunoblotting (top). The bottom panel indicates blot intensities relative to control blots (no drug treatment). C, sorafenib-resistant SNU-423 cells were treated with the indicated concentrations of CI-1040, and relative cell viability was determined 72 h later. Note that CI-1040 had no significant inhibitory effect on cell growth at 0.1 μm (indicated by a red arrow). D, sorafenib-resistant SNU-423 cells were treated with the indicated concentrations of AZD8055 in the presence (open circles) or absence (solid circles) of 0.1 μm CI-1040, and relative cell viability was determined 72 h later.
The phosphorylation of RPS6 S235/236 in JHH-2, SNU-423, and SNU-387 cells was insensitive to sorafenib (Fig. 3C). However, the active MAPK pathway in these cell lines seems to lie downstream of RAF kinases. An MEK inhibitor, CI-1040 (Fig. 3B), enhanced the attenuation of p-RPS6 S235/236 by everolimus in JHH-2 and SNU-423 cells (Fig. 4B), and an RSK inhibitor, SL0101, enhanced the attenuation of p-RPS6 S235/236 by everolimus in JHH-2 and SNU-387 cells (Fig. 4B).
Marked inhibition of p-RPS6 S235/236 by combined blockade of the MAPK pathway downstream of RAF and the mTOR pathway prompted us to examine the effect of this drug combination on HCC cell growth. CI-1040 had no inhibitory effect on the growth of SNU-423 cells at a concentration of 0.1 μm (Fig. 4C), but it was able to enhance the growth-inhibitory effect of AZD8055 (Fig. 4D). Synergy between AZD8055 and CI-1040 was confirmed via Chou–Talalay median dose effect analysis (17) (supplemental Table S8). The combination index values at 50%, 75%, and 90% growth inhibition were 0.783, 0.804, and 0.827, respectively (values of <1 are defined as representative of synergistic effects). These results suggest that HCC patients refractory to sorafenib with a high level of p-RPS6 S235/236 might be treatable with an mTOR inhibitor in combination with drugs that block the MAPK signaling pathway.
To further provide a rational basis for synergistic targeting of the mTOR and MAPK pathways in HCC, we performed unsupervised hierarchical cluster analysis of 23 HCC cell lines based on their phosphorylation status of signaling components in the mTOR and MAPK pathways listed in supplemental Table S9. Clustering analysis stratified the cell lines into two major groups, A and B (supplemental Fig. S1). In comparison with group A, group B showed higher levels of phosphorylated MAPK signaling components including p-PDGF receptor-β(Thr751), p-Raf-A(Ser299), and phosphorylated signaling modules of the JNK and p38 MAPK pathways (supplemental Fig. S1, C1). Among them, the levels of p-p53 at Ser392, Ser37, and Ser6 differed substantially between groups A (low) and B (high) (supplemental Fig. S1). In addition, cell lines clustered into group A tended to have simultaneous phosphorylation of the mTOR signaling components C2, RPS6(Ser235/236), RPS6(Ser240/244), and eIF4G(Ser1108) (supplemental Fig. S1). With some notable exceptions, sorafenib-insensitive cell lines (high IC50 values for sorafenib) and sorafenib-sensitive cell lines (low IC50 values for sorafenib) were clustered into group A and group B, respectively. Although sorafenib-insensitive SNU-387, JHH-1, and KIM-1 cells were classified into the sorafenib-sensitive group B, their phosphorylation levels of mTOR signaling components C2 were higher than those in the other cell lines in group B, suggesting that activation of mTOR signaling might be responsible for the resistance to sorafenib in these cell lines. Some of the sorafenib-insensitive cell lines (e.g. Alexander, JHH-2, and SNU-475 cells) partitioned into subtype Ab (supplemental Fig. S1) were characterized by prominent activation of mTOR signaling components C2 and MAPK signaling components C1. Together, these findings imply that there is a certain population of HCC cells showing up-regulation of both mTOR and MAPK signaling. Such an HCC subtype might respond better to combination treatment with mTOR and MAPK inhibitors. When we compared the phosphorylation status of the mTOR and MAPK signaling nodes in 95 cell lines by means of unsupervised hierarchical clustering, HCC cell lines were significantly clustered together (p = 0.011 by Fisher's exact test) in group A (supplemental Fig. S2), characterized by high levels of phosphorylation of the signaling components C1 and C2, in comparison to cell lines derived from seven other cancer types (supplemental Table S10). The signaling components C1 included previously reported targets of sorafenib such as b-RAF and PDGF receptor-β, as well as the upstream modules of the mTOR pathway (e.g. Akt and PDK). The components C2 comprised RPS6(Ser235/236), RPS6(Ser240/244), eIF4G(Ser1108), and the signaling modules of the JNK and p38 MAPK pathways. These observations may reflect the fact that the activation of MAPK signaling by itself, or in combination with mTOR signaling, is a unique feature of HCC.
It is noteworthy that some sorafenib-sensitive and -insensitive cell lines were clustered together into subgroup Aa (supplemental Fig. S1). Although this subgroup was characterized by relatively low levels of both mTOR and MAPK signaling activation, the most sorafenib-sensitive cell line, SNU-449, was classified into this subgroup. It is therefore plausible that some other signaling pathway, in addition to the mTOR and MAPK pathways, may be involved in defining the marked sensitivity of SNU-449 cells to sorafenib.
DISCUSSION
Derangements in the phosphorylation of signaling molecules are hallmarks of cancers, and are often considered as targets of molecular therapies. By profiling the phosphorylation status of multiple signaling components, it is possible to derive important clues for understanding the pathogenesis and classification of cancers. In this study, a high level of p-RPS6 S235/236 was detected in sorafenib-resistant HCC cells. Consistent with this in vitro observation, such high expression of p-RPS6 S235/236 was detected in pretreatment biopsy specimens from HCC patients who had shown early radiographically evident disease progression after starting sorafenib therapy. The number of patient samples analyzed in this study was small and insufficient for providing conclusive evidence, but the present findings warrant future clinical studies to evaluate the significance of p-RPS6 S235/236 as a predictor of response to sorafenib. In order to ensure accurate validation of the utility of p-RPS6 S235/236 as a predictor in future studies, standardized guidelines of immunohistochemistry for detecting p-RPS6 (Ser235/236) need to be developed, including tissue preparation, fixation, staining methods, scoring system, and the definition of a “positive“ result.
p-RPS6 has been used as a molecular surrogate for mTOR activation. Villanueva et al. (22) assessed 314 surgical specimens of HCC immunohistochemically using an anti-p-RPS6 S240/244 antibody. They detected p-RPS6 S240/244 in half of the cases examined, and positive staining was correlated with HCC recurrence (22). Although antibodies against p-RPS6 S235/236 and p-RPS6 S240/244 have been used equivalently in many studies to evaluate mTOR activation (22, 23), the phosphorylation of these serine residues was found to be differentially regulated (Fig. 2A). An earlier study demonstrated persistent phosphorylation of RPS6 S235/236 in cells derived from S6K1−/−/S6K2−/− double-knockout mice, and it was concluded that this paradoxical phosphorylation was caused by MAPK signaling. A later study revealed that RSK (MAPK pathway) predominantly phosphorylated the serine 235 and 236 residues of RPS6, whereas S6K (mTOR pathway) broadly phosphorylated the serine 235, 236, 240, 244, and 247 residues. Therefore, use of an antibody against p-RPS6 S240/244 would seem more appropriate for specific detection of the mTOR pathway activation status (20). In the present study, however, we found that phosphorylation of the serine 235 and 236 residues of RPS6 reflected cross-talk between the mTOR and MAPK pathways (Fig. 3C) and served as a predictive biomarker of sorafenib sensitivity. Although RAF kinases (MAPK pathway) are one of the main molecular types targeted by sorafenib, intervention of active mTOR signaling in the MAPK pathway seems to be one of the molecular mechanisms responsible for the resistance of HCC to sorafenib.
It is therefore conceivable that HCC patients with tumors having high levels of p-RPS6 S235/236 could benefit from inhibition of mTOR signaling. We found that mTOR inhibitors showed greater antitumor activity against sorafenib-resisntant HCC cells (Fig. 3E). A recent phase I/II study of everolimus given daily as a single agent in patients with advanced HCC showed that the drug was well tolerated and exerted preliminary antitumor activity in some patients (24). A phase III EVOLVE-1 randomized trial is now ongoing to evaluate the efficacy of everolimus in HCC patients whose disease progressed during or after sorafenib treatment or who were intolerant to sorafenib (25). This clinical trial is designed to reveal the efficacy of mTOR pathway inhibition for control of sorafenib-resistant HCC and is expected to clarify the significance of our present findings.
Clustering analysis of RPPA data revealed that 6 out of 23 HCC cell lines (Alexander, JHH-2, SNU-475, Huh-7, KIM-1, and JHH-1) had prominent activation of both the MAPK and mTOR pathways, indicating a possible subset of HCC patients who might benefit from a combination of MAPK and mTOR inhibitors (supplemental Fig. S1). We also found synergy between MAPK and mTOR pathway inhibitors in sorafenib-resistant cell lines (Fig. 4). However, there is a need for caution before this can be applied clinically. Activation of MAPK signaling occurred at various levels of RAF/MEK/ERK/RSK in sorafenib-resistant cells (Fig. 3C). In addition, clustering analysis showed activation of two other major MAPK pathways, the Jun N-terminal kinase (JNK) and p38 MAPK pathways, in more than half the HCC cell lines (supplemental Fig. S1). Cross-talk among three major MAPK pathways (RAF/MEK/ERK, JNK, and p38MAPK) has been reported previously (26). Together, these findings suggest that careful assessment is vital when selecting an appropriate MAPK inhibitor for each individual HCC patient. Despite extensive sequencing of kinase genes, we were unable to identify any alterations in the pathway that might be responsible, indicating the need to expedite pharmacoproteomics for therapy personalization.
The present study highlighted the potential power of the RPPA platform for pathway profiling. We have provided proof-of-principle support for the utility of the highly sensitive, high-throughput RPPA platform by identifying a practical biomarker with potential clinical applicability. In this study, we used only well-characterized antibodies with high specificity. The Human Antibody Initiative is an ongoing project to raise at least one monospecific antibody against all >20,000 proteins encoded by the human genome (27). It is anticipated that the completion of this project will greatly accelerate the capability of RPPA. The majority of current molecular targeting drugs are designed to target a particular signaling pathway (28). Precise determination of signaling pathways that are activated in individual patients seems to be essential for obtaining maximum benefit from any given treatment. RPPA requires only a minuscule specimen quantity (e.g. less than 1 ng of protein per array) and is applicable even to small biopsy samples. The potential clinical utility of RPPA for decision-making and monitoring of cancer therapeutics is thus enormous.
Supplementary Material
Footnotes
Author contributions: M.M. and T.Y. designed research; M.M., W.C., A.M., and Y.N. performed research; M.M. and K.K. contributed new reagents or analytic tools; M.M., W.C., K.K., T.S., and C.C. analyzed data; M.M. and T.Y. wrote the paper; M.M. developed the methodology; M.O. and K.H. contributed to the conception of the study; T.Y. supervised research.
* This work was supported by the National Cancer Center Research and Development Fund (23-A-38 and 23-A-11), the Program for Promotion of Fundamental Studies in Health Sciences conducted by the National Institute of Biomedical Innovation of Japan (10-07, 10-44, and 10-45), and the Research on Biological Markers for New Drug Development conducted by the Ministry of Health, Labor and Welfare of Japan.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HCC
- hepatocellular carcinoma
- ERK
- extracellular signal-regulated kinase
- IC50
- half-maximal (50%) inhibitory concentration
- p-RPS6 S235/236
- ribosomal protein S6 phosphorylated at the serine 235/236 residue
- MAPK
- mitogen-activated protein kinase
- RPPA
- reverse-phase protein array
- mTOR
- mammalian target of rapamycin
- RSK
- 90-kDa ribosomal protein S6 kinase
- S6K
- 70-kDa ribosomal protein S6 kinase.
REFERENCES
- 1. Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., Parkin D. M. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 [DOI] [PubMed] [Google Scholar]
- 2. Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Haussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. (2008) Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 [DOI] [PubMed] [Google Scholar]
- 3. Arao T., Ueshima K., Matsumoto K., Nagai T., Kimura H., Hagiwara S., Sakurai T., Haji S., Kanazawa A., Hidaka H., Iso Y., Kubota K., Shimada M., Utsunomiya T., Hirooka M., Hiasa Y., Toyoki Y., Hakamada K., Yasui K., Kumada T., Toyoda H., Sato S., Hisai H., Kuzuya T., Tsuchiya K., Izumi N., Arii S., Nishio K., Kudo M. (2013) FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 57, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 4. Cheng A. L., Kang Y. K., Chen Z., Tsao C. J., Qin S., Kim J. S., Luo R., Feng J., Ye S., Yang T. S., Xu J., Sun Y., Liang H., Liu J., Wang J., Tak W. Y., Pan H., Burock K., Zou J., Voliotis D., Guan Z. (2009) Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 10, 25–34 [DOI] [PubMed] [Google Scholar]
- 5. Wilhelm S. M., Adnane L., Newell P., Villanueva A., Llovet J. M., Lynch M. (2008) Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol. Cancer Ther. 7, 3129–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abou-Alfa G. K., Schwartz L., Ricci S., Amadori D., Santoro A., Figer A., De Greve J., Douillard J. Y., Lathia C., Schwartz B., Taylor I., Moscovici M., Saltz L. B. (2006) Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J. Clin. Oncol. 24, 4293–4300 [DOI] [PubMed] [Google Scholar]
- 7. Ueshima K., Kudo M., Takita M., Nagai T., Tatsumi C., Ueda T., Kitai S., Ishikawa E., Yada N., Inoue T., Hagiwara S., Minami Y., Chung H., Sakurai T. (2011) Des-gamma-carboxyprothrombin may be a promising biomarker to determine the therapeutic efficacy of sorafenib for hepatocellular carcinoma. Dig. Dis. 29, 321–325 [DOI] [PubMed] [Google Scholar]
- 8. Hagiwara S., Kudo M., Nagai T., Inoue T., Ueshima K., Nishida N., Watanabe T., Sakurai T. (2012) Activation of JNK and high expression level of CD133 predict a poor response to sorafenib in hepatocellular carcinoma. Br. J. Cancer 106, 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishizuka S., Charboneau L., Young L., Major S., Reinhold W. C., Waltham M., Kouros-Mehr H., Bussey K. J., Lee J. K., Espina V., Munson P. J., Petricoin E., 3rd, Liotta L. A., Weinstein J. N. (2003) Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl. Acad. Sci. U.S.A. 100, 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Byers L. A., Wang J., Nilsson M. B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y. H., Diao L., Masrorpour F., Shen L., Liu W., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B. S., Kalhor N., Wistuba I. I., Girard L., Lippman S. M., Mills G. B., Coombes K. R., Weinstein J. N., Minna J. D., Heymach J. V. (2012) Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2, 798–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spurrier B., Ramalingam S., Nishizuka S. (2008) Reverse-phase protein lysate microarrays for cell signaling analysis. Nat. Protoc. 3, 1796–1808 [DOI] [PubMed] [Google Scholar]
- 12. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 13. Masuda M., Maruyama T., Ohta T., Ito A., Hayashi T., Tsukasaki K., Kamihira S., Yamaoka S., Hoshino H., Yoshida T., Watanabe T., Stanbridge E. J., Murakami Y. (2010) CADM1 interacts with Tiam1 and promotes invasive phenotype of human T-cell leukemia virus type I-transformed cells and adult T-cell leukemia cells. J. Biol. Chem. 285, 15511–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and Genome Project Data Processing, S. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K., Li M. Y., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou T. C. (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 [DOI] [PubMed] [Google Scholar]
- 18. Anjum R., Blenis J. (2008) The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 19. Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. (2004) S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24, 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., Blenis J. (2007) RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ascierto P. A., Kirkwood J. M., Grob J. J., Simeone E., Grimaldi A. M., Maio M., Palmieri G., Testori A., Marincola F. M., Mozzillo N. (2012) The role of BRAF V600 mutation in melanoma. J. Transl. Med. 10, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Villanueva A., Chiang D. Y., Newell P., Peix J., Thung S., Alsinet C., Tovar V., Roayaie S., Minguez B., Sole M., Battiston C., Van Laarhoven S., Fiel M. I., Di Feo A., Hoshida Y., Yea S., Toffanin S., Ramos A., Martignetti J. A., Mazzaferro V., Bruix J., Waxman S., Schwartz M., Meyerson M., Friedman S. L., Llovet J. M. (2008) Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 135, 1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou L., Huang Y., Li J., Wang Z. (2010) The mTOR pathway is associated with the poor prognosis of human hepatocellular carcinoma. Med. Oncol. 27, 255–261 [DOI] [PubMed] [Google Scholar]
- 24. Zhu A. X., Abrams T. A., Miksad R., Blaszkowsky L. S., Meyerhardt J. A., Zheng H., Muzikansky A., Clark J. W., Kwak E. L., Schrag D., Jors K. R., Fuchs C. S., Iafrate A. J., Borger D. R., Ryan D. P. (2011) Phase 1/2 study of everolimus in advanced hepatocellular carcinoma. Cancer 117, 5094–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kudo M. (2011) Signaling pathway and molecular-targeted therapy for hepatocellular carcinoma. Dig. Dis. 29, 289–302 [DOI] [PubMed] [Google Scholar]
- 26. Wagner E. F., Nebreda A. R. (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 27. Uhlen M., Ponten F. (2005) Antibody-based proteomics for human tissue profiling. Mol. Cell. Proteomics 4, 384–393 [DOI] [PubMed] [Google Scholar]
- 28. Wulfkuhle J. D., Edmiston K. H., Liotta L. A., Petricoin E. F., 3rd (2006) Technology insight: pharmacoproteomics for cancer—promises of patient-tailored medicine using protein microarrays. Nat. Clin. Pract. Oncol. 3, 256–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.