Abstract
The tumor microenvironment plays key roles in cancer biology, but its impact on the regulation of signaling pathway activity in cancer cells has not been systemically investigated. We designed an analytical strategy that allows differential analysis of signaling between cancer and stromal cells present in tumor xenografts. We used this approach to investigate how in vivo growth conditions and PI3K inhibitors regulate pathway activities in both cancer and stromal cell populations. We found that, despite inducing more modest changes in protein expression, in vivo growing conditions extensively rewired protein kinase networks in cancer cells. As a result, different sets of phosphorylation sites were modulated by PI3K inhibitors in cancer cells growing in tumors relative to when these cells were in culture. The p110δ PI3K-selective compound CAL-101 (Idelalisib) did not inhibit markers of PI3K activity in cancer or stromal cells; however, unexpectedly, it induced phosphorylation on SQ motifs in both subpopulations of tumor cells in vivo but not in vitro. Thus, the interaction between cancer cells and the stroma modulated the ability of PI3K inhibitors to induce the activation of apoptosis in solid tumors. Our study provides proof-of-principle of a proteomics workflow for measuring signaling specifically in cancer and stromal cells and for investigating how cancer biochemistry is modulated in vivo.
Solid tumors contain a heterogeneous population of cells. Transformed epithelial cells recruit different types of somatic cells to the tumor microenvironment where they influence varying aspects of cancer biology. The role of heterotypic communication between normal stromal cells and transformed cancer cells is well established (1, 2). Different somatic cell types, including fibroblasts, epithelial cells, and cells of the immune system—all of which are found in tumors—promote cancer cell development by means of gap-junction intercellular communication, direct cell-to-cell contacts, and by the release of growth factors, enzymes, and cytokines that act on neighboring malignant cells (3–6).
The tumor microenvironment determines the ability of cancer cells to survive in specific organs and their ability to proliferate and metastasize (7–9). Growth factors released from tumor-associated stromal cells also influence how cancer cells respond to drug administration (10). Therefore, the advancement of targeted cancer therapies requires an understanding of how the tumor microenvironment modulates the biochemistry of transformed cancer cells. In addition, targeting the tumor stroma is emerging as an intriguing concept for the development of anti-cancer therapies (11). It is therefore important to investigate specific effects of compounds in clinical development on stromal cells in addition to those exerted toward malignant cancer cells (12).
Here we investigated the effects that changes in growing conditions from a two-dimensional cell culture to an in vivo three-dimensional tumor environment had in modulating protein and phosphoprotein expression in human cancer cells. For this, we used mass spectrometry (MS) to specifically measure cancer and stromal proteomes and phosphoproteomes within mouse tumor xenografts.
We also investigated the effects that the pharmacological inhibitors of PI3K, namely GDC-0941 or CAL-101, would have on the phosphoproteomes of stromal cells relative to cancer cells in solid tumors. GDC-0941 is an inhibitor with specificity for class I PI3Ks, whereas CAL-101 specificity is restricted to the p110δ isoform of PI3K (13, 14), which in untransformed tissues is mainly found in leukocytes (15). The PI3K signaling pathway is often deregulated in different cancer types (16), including colorectal cancer (17), and both compounds used in this study are in different stages of clinical development (18–20). PI3K signaling has also been implicated in mediating the effects that the microenvironment has on cancer cells (21).
We found that in vivo growth conditions had profound effects on phosphoprotein expression, which was reflected on the phosphorylation sites modulated by PI3K inhibitors in vivo relative to in vitro and in their ability to induce apoptotic markers across these two cell culture conditions.
MATERIALS AND METHODS
Cell Culture
The colorectal cell-line DLD-1 was purchased from A.T.C.C. (supplied by LGC Standards, Teddington, U.K.) and cultured at 37 °C in a 5% CO2 incubator in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were treated with 1 μm GDC-0941, CAL-101, or vehicle for 2 h before lysing.
Mouse Xenografts
This study was carried out in accordance with the regulations of the Animals (Scientific Procedures) Act 1986. The protocol was approved by the local Ethical Review Committee and by the U.K. Home Office. DLD-1 cells (2 × 106) were injected subcutaneously in three different places into the flanks of 8-week old female Fox Chase SCID® Mice (Charles River Laboratories, Wilmington, MA). After 7 days postinjection, when mice with tumors greater than 75 mm, mice were divided into three groups and treated with GDC-0941 (100 mg/kg of body mass) in 0.5% methylcellulose and 0.2% polysorbate 80 (Tween 80) in de-ionized water (MCP buffer), CAL-101 (30 mg/Kg) in MCP buffer, or MCP buffer according to the same dose schedule. All treatments were intravenous. Mice were anesthetized with pentobarbital and killed after 2 h of treatment. Tumors were removed, weighed, and snap-frozen in liquid nitrogen until further analysis.
Sample Preparation for Proteomic and Phosphoprotoemic Analysis
Cells and tumors were lysed in a urea-based lysis buffer and proteins were digested using trypsin as reported previously (21, 22). Phosphopeptides were enriched from total peptides by TiO2 chromatography essentially as described previously (23) with the modifications described elsewhere (22).
Mass Spectrometry
Enriched phosphopeptides and peptides were analyzed by LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Hemel Hempstead, UK) coupled to EASY-nLC (Proxeon, ThermoScientific). Peptide separation was performed in a C18 Pepmap reverse phase column (75 μm I.D, 3 μm particle size; proxeon, Thermo-Fisher) using solution A (0.1% formic acid in liquid chromatography (LC)1-MS grade water) and solution B (0.1% formic acid in LC-MS ACN) as mobile phases. Gradient runs from 2–30% solution B in 100 min and from 30–60% in 10 mins followed by a final 10 min wash at 85% B.
Full MS scans were acquired in the Orbitrap mass analyzer over the range m/z 375–1500 with a mass resolution of 30,000. For unphosphorylated peptides, tandem MS (MS/MS) was acquired using top seven data-dependent acquisition using high energy collision dissociation (40%). For phosphopeptides, MS/MS was acquired using top 10 data dependent acquisition by collision induced dissociation (35%) and multistage activation. Gas phase fractionation method was applied to acquire MS/MS scans.
Peptide Identification by Database Search
MS/MS data were converted to mgf files using Mascot Distiller (version 2.2) and searched against the 2012_03 databases of UniProt-TrEMBL (104,945 and 60,427 entries for Homo sapiens and Mus musculus sequences, respectively) and UniProt SwissProt (20,249 Homo sapiens and 16,521 Mus musculus entries) and a decoy database using the Mascot search engine (version 2.2). The data was searched twice, restricting searches against human or mouse-specific sequences in each separate search. For phosphoproteomics, multistage activation data was searched with tolerance windows were 5 ppm and 600 mmu for parent and fragment ions, respectively. Tolerance windows for high energy collision dissociation data were 5 ppm and 50 mmu for parent and fragment ions, respectively. Allowed variable modifications were methionine oxidation, pyroglutamate at the N terminus and phosphorylation of serine, threonine, and tyrosine residues. Significance of peptide identification was assessed by comparing results returned by searches against random and forward databases. Fold discovery rates at several cut-off values of Mascot scores and mass tolerances were used to calculate an empirical value of probability of random identification. These probabilities of false discovery are listed for each phosphopeptide in the supplementary datasets. The probability of correct phosphorylation site assignment was determined by the Mascot delta score approach (24). Ambiguous phosphorylation site assignments (delta score < 15) are reported as gene name followed by start and end of the amino acid sequence.
Data Analysis and Bioinformatics
Relative quantification of peptides across experimental conditions was achieved by comparing peak heights of extracted ion chromatograms (automated by Pescal) as described previously (21, 25). The data were normalized to the sum of all intensities derived from a sample (columns). When comparing peptide or phosphopeptide signals across growth conditions, these were also divided by the average signals of such peptide across all the samples (rows). When comparing the effects of inhibitors on phosphorylation, phosphopeptide signals were divided by those of the untreated control samples. The p values of differences across conditions were obtained by means of a t test of log2 transformed fold changes and these were adjusted for multiple testing through the Benjamini-Hochberg procedure as described before (21).
To identify peptide sequences specific for human or mouse, peptides returned as positive identifications by Mascot were searched against the UniProt-TrEmbl database restricted to human or mouse sequences using a script written in Visual Basic to automate these searches. Failure to detect peptides from human or mouse searches in mouse or human databases, respectively, indicated that these peptides were specific for either species.
For pathway analysis, we used an approach similar to the one reported elsewhere (26). Briefly, ontologies were obtained from SwissProt (release March 2012) and pathways from the NCI Pathway database (http://pid.nci.nih.gov/, accessed March 2012). The fold changes of peptides or phosphopeptides differentially regulated across conditions belonging to each of these pathways were then averaged using a script written in Visual Basic and significance assessed by means of a t test of log2 transformed data. Pathway analysis was also carried out using publically available tools (27).
Immunoblotting
Cells were homogenized in lysis buffer (1% Triton X-100, 50 mm Tris/HCl, pH 7.5, 150 mMNaCl, and 1 mMEDTA) supplemented with protease and phosphatase inhibitors. Proteins were separated by SDS/PAGE (8%, 10%, and 15% gels), transferred on to PVDF membranes and then probed with antibodies against p-Akt/PKB (S473), PKB/Akt, p42/44 MAP kinase, p-4E-BP1, p-S6 ribosomal (S235/236), S6 ribosomal, APC-2, CSK21, PDH, p-(S/T)-Q (ATR/ARM substrate), β-catenin, Akt1 (phospho S129), and vinculin.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (28) with the dataset identifier PXD000218 and DOI 10.6019/PXD000218. Results are also shown in supplementary Data Sets S1, S2, and S3 and representative XICs are shown in supplementary Data Sets S4 and S5.
RESULTS
Mouse tumor xenografts are often used as biological models in preclinical studies as this in vivo growth condition is thought have higher biological relevance than in vitro models in which cells are grown on plastic. However, the extent to which the biochemistry and signaling differ between cells growing in culture relative to the same cells growing in a tumor xenograft have not been investigated systematically. A problem is that antibodies often cross-react across species, thus complicating the verification of whether potential differences in protein and phosphorylation site abundance across cells grown in vitro relative to the same cells grown as a xenograft are because of true biological differences or because of cross-reactivity with mouse proteins originating from stromal cells. In contrast, the specificity of high resolution MS should, in principle, allow us to distinguish proteins originating from different species within the tumor xenograft, therefore allowing the assessment of biochemical differences between cells maintained in vivo relative to those grown under in vitro conditions.
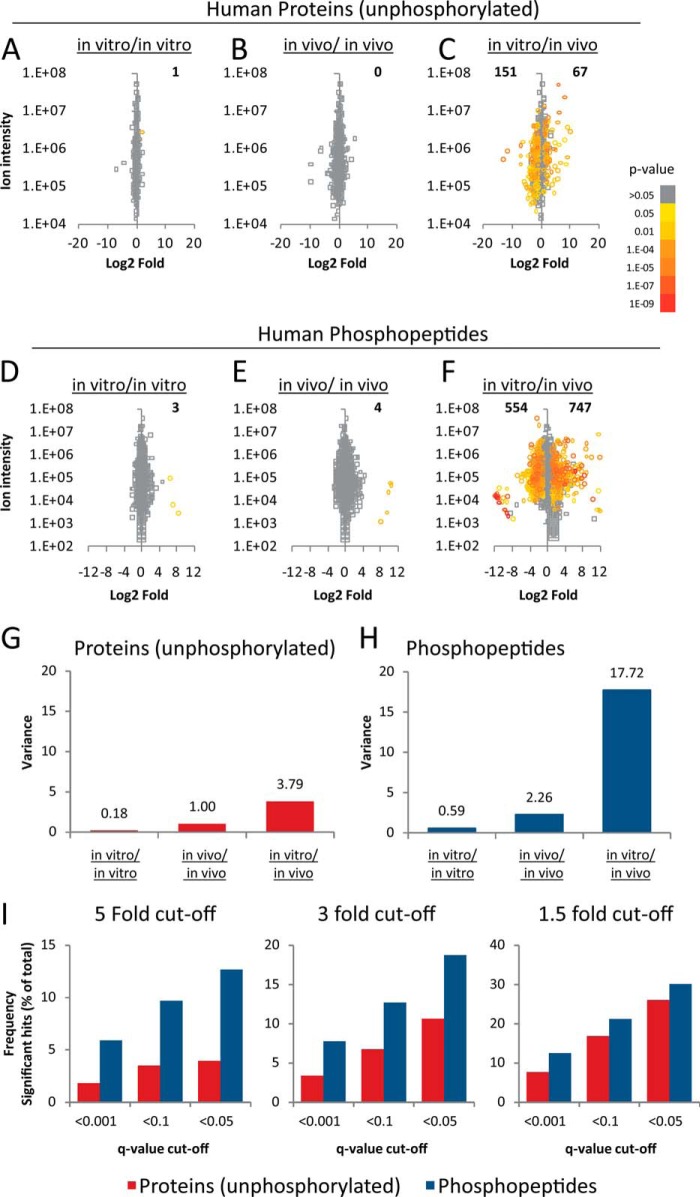
To investigate whether it was possible to measure signaling and biochemistry specific to human cells within tumor xenografts, we compared protein and phosphoprotein expression in the colorectal cancer cell-line DLD-1 grown under two different conditions (either in vivo or in vitro), with or without treatment with PI3K inhibitors (Fig. 1A). For this comparison, we used label-free MS techniques for accurate and precise quantification of proteins and phosphoproteins on a large scale (21, 22, 25, 29). These techniques are similar to those used in other laboratories (26, 30–33). In order to investigate biological variability within sample groups and to infer statistical significance of the data, six biological replicates were analyzed per condition. Several thousand proteins and phosphopeptides were identified (actual numbers being dependent on the statistical thresholds of identification). Comparing the outputs of peptides identifications based on searches against decoy protein databases showed that >90% peptides were identified with less than 1 in 100 probability of false discovery, all of them possessing a false discovery rate of < 5% (supplemental Fig. S1).
Fig. 1.
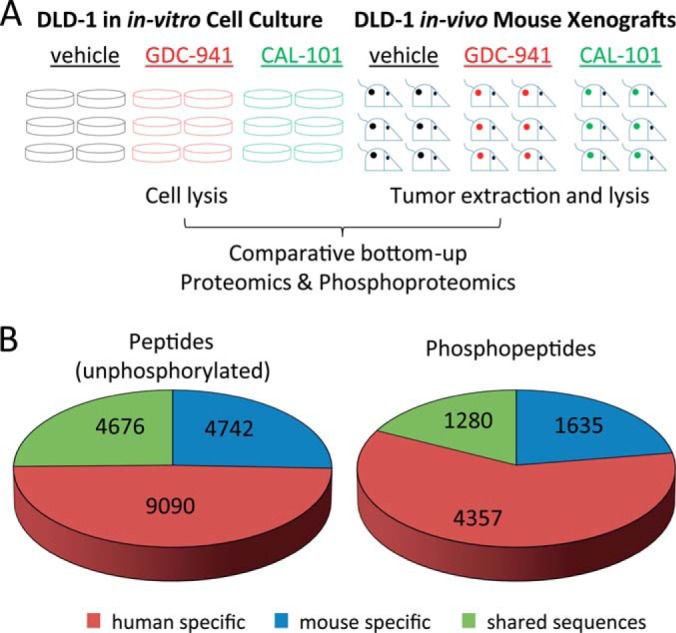
Strategy for the global analysis of proteomes and phosphoproteomes specific to cancer and stromal cells within tumors and overview of results. A, The DLD-1 cell-line was cultured in either in vitro cell culture conditions (on plastic) or in vivo mouse xenografts. Cells were treated by GDC-0941, CAL-101, or vehicle in six biological replicates and then processed for proteomic and phosphoproteomics analysis. B, Classification of peptides and phosphopeptides found in the study based on whether these contain species specific sequences.
Peptide sequences returned as positive identifications were blasted against the human and mouse Uniprot databases to identify peptides specific to human or mouse. Approximately 50% of sequences identified were human-specific (i.e. sequences that were not present in the mouse protein database), whereas ∼25% of peptides were mouse-specific (Fig. 1B). Because mouse and human sequences were derived from stromal and from malignant cells respectively, our data allowed specific investigation of protein and phosphoprotein expression in these two different cell populations within the tumor.
In Vivo Growth Conditions Extensively Modulate Phosphorylation Networks Despite Inducing Relatively Modest Changes in Protein Expression
We first compared the intensities of human-specific peptides across DLD-1 cells grown in vitro or in vivo. From a total of six replicate groups, we compared the average of intensities of three randomly chosen replicate control samples against the average of the other three. This analysis allowed the assessment of technical and biological variability within the data set because, in principle, differences in protein expression should be minimal within a given sample group. The results of these analyses are given in supplementary Datasets S1, S2, and S3 and summarized in Figs. 2A and 2B for the in vitro and in vivo data, respectively. Only one protein was significantly differentially expressed across >1500 protein data points at q < 0.05 (p value post Benjamini-Hochberg multiple testing correction), indicating minimal variability in protein expression within sample groups. We identified 761 proteins with at least three human-specific peptides and a comparison with protein abundances listed in the PaxDb (34) showed that these are among to the 25% most abundant proteins in humans. Of these, 151 proteins were found to show increased expression in cells grown in vivo whereas 67 proteins showed an increase in expression in vitro (Fig. 2C). Similarly, for the phosphoproteomics data, 3680 phosphopeptides with human-specific sequences could be quantified across the 36 experimental conditions, producing 132,480 quantitative data points. The intensities of just seven phosphopeptides were found to be significantly differentially expressed within the control sample groups from 7360 comparisons (Figs. 2D and 2E); i.e. less than 1 measurement in 1000 quantitative data points was significant within the control groups based on a q < 0.05, indicating minimal biological variability within sample groups. In contrast, using the same criterion, 554 and 747 phosphopeptides were increased or decreased, respectively, in cells grown in vivo relative to those grown in vitro (Fig. 2F).
Fig. 2.
In vivo growth conditions extensively modulated the proteomes and phosphoproteomes of cancer cells. A, Comparison of the proteomes of three randomly chosen biological replicates from control cells grown in vitro with the proteomes of other three replicates. B, Comparison of the proteomes of three randomly chosen biological replicates from control cells grown in vivo with the proteomes of other three replicates. C, Comparison of the proteomes of six biological replicates of cells grown in vivo with the proteomes of six biological replicates of cells grown in vitro. D, The phosphoproteomes of three randomly chosen biological replicates from control cells grown in vitro were compared with the phosphoproteomes of other three replicates. E, The phosphoproteomes of three randomly chosen biological replicates from cells grown in vivo were compared with the phosphoproteomes of other three replicates. F, The phosphoproteomes of six biological replicates of cells grown in vivo were compared with the phosphoproteomes of six biological replicates of cells grown in vitro. G, Variance of the protein log2 fold-changes shown in A, B, and C. H, Variance of the phosphoprotein log2 fold-changes shown in D, E, and F. I, Proportion of significant differences in the amounts of proteins and phosphorylation sites between cells grown in vivo or in vitro at different significance thresholds. Fold-difference significance thresholds are in log2 scale and include increased as well as decreased in one condition relative to the other. Number of significant hits was calculated as the percentage of significant changes at a given set significance thresholds relative to the total number of proteins or phosphopeptides identified and quantified.
Biological variability was greater in human cancer cells grown in tumors than in those grown on plastic, as demonstrated by the greater spread of data for the in vivo compared with the in vitro data (Figs. 2G and 2H). When comparing proteomes and phosphoproteomes between cells grown in vivo relative to those grown in vitro, we observed that the variances of fold-changes were 3.79 and 17.72 for the proteomics and phosphoproteomics data respectively (Figs. 2G and 2H). We consistently found a greater number of significant differences in phosphorylation sites than in proteins modulated by in vivo growth conditions at several significance thresholds. For example, Fig. 2I shows that 12.7% of the quantified phosphorylated peptides were modulated between growth conditions by at least fivefold (log2) and with q < 0.05, whereas, using the same significance threshold, only 3.9% of proteins were different between conditions. In other words, there was a fourfold difference in the number of phosphopeptides relative to proteins modulated by growing conditions. Thus, overall, our data indicate that in vivo growth conditions had a greater impact on the phosphoproteome than on the proteome of our cell-line model.
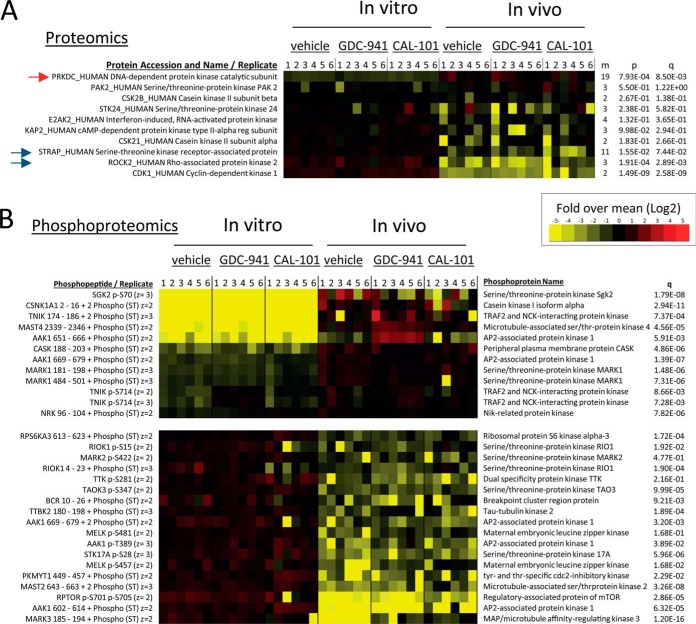
Differential Regulation of Kinase Expression and Phosphorylation in Cancer Cells Grown In Vivo or In Vitro
To investigate whether the large impact that in vivo growth conditions had on the phosphoproteome was because of differential expression of protein kinases across conditions, we mined our data to specifically assess the abundance and phosphorylation of this protein class. The proteomics data set contained seven protein kinases detected with at least three unphosphorylated peptides and ten of them with at least two (Fig. 3A). The expression of the catalytic subunit of DNA-dependent protein kinase was particularly high in cells grown in tumors (red arrow in Fig. 3A), whereas ROCK2 and CDK1 were decreased in abundance in this sample group (blue arrows in Fig. 3A). We also detected 78 phosphopeptide ions that specifically matched to human protein kinases. Fig. 3B summarizes the relative intensities of these phosphopeptides representing protein kinases found to be substantially different between cells grown in vivo and in vitro. Illustrative examples of phosphopeptides showing differences across conditions include those on SGK2 (at Ser70) and TNIK (at Ser714), which were increased in tumors, and on RPTOR (Raptor - a regulatory subunit of mTORC1) and RPS6KA3 (downstream of mTORC1), which were increased in cells grown in culture relative to tumors (Fig. 3B). These observations indicate that cells grown in tumors had markedly different patterns of phosphorylation on protein kinases (Fig. 3B) and on their substrates (Fig. 2) relative to the same cells grown in cell culture. Because, by definition, the abundance of phosphorylation sites are directly related to protein kinase activity, the data shown in Figs. 2 and 3 suggest that in vivo conditions extensively modulate kinase pathway activation in cancer cells.
Fig. 3.
In vivo growing conditions modulate protein kinase expression and phosphorylation. A, Protein kinases found by proteomics in cells grown in vivo and in vitro. m: number of peptides matched to the named protein; p: p value of differences between protein expression across cells grown in vivo or in vitro; q: adjusted p value using the Benjamini-Hochberg procedure. B, As in A, but the analysis was at the phosphopeptide level.
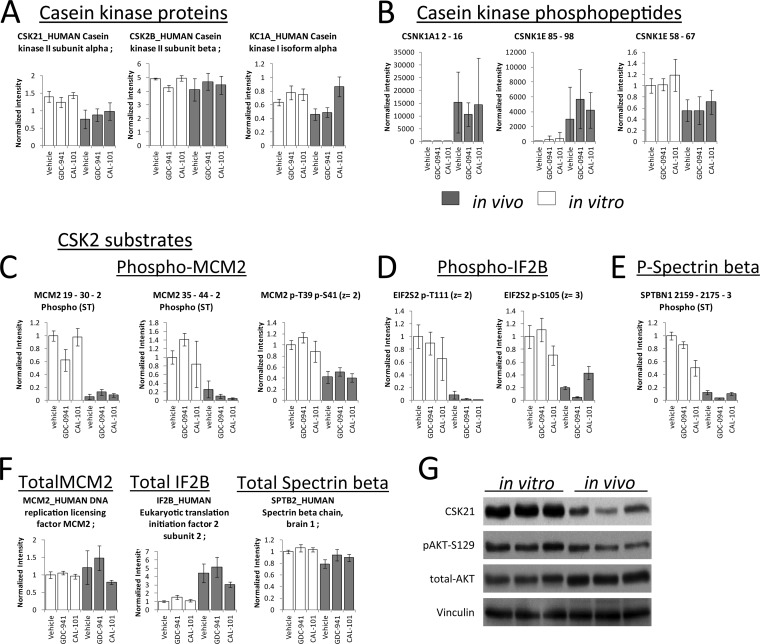
In Vivo Growth Conditions Regulate Casein Kinase Expression and Activity
We aimed to confirm some of the results regarding protein kinase expression and phosphorylation obtained by mass spectrometry. We observed a negative correlation between the signal intensities of un-phosphorylated peptides of different casein (CK) kinase isoforms with their corresponding phosphorylation sites (Fig. 4A and 4B). Total protein amounts of the CK2 alpha catalytic subunit were decreased in tumors by ∼twofold, whereas the amounts of the CK2 beta regulatory subunit were unaltered (Fig. 4A). This is an interesting observation, as it is known that CK2 activity is regulated by the ratio between the alpha and beta subunits (35–37) in addition to its absolute expression (38, 39). CK2 beta binds to CK2 alpha, resulting in the formation of an inactive polymeric complex; thus, an alteration in CK2 subunit ratios (Fig. 4A) suggested that this kinase was less active in tumors than in cell-lines. To investigate this possibility further, we examined the abundances of peptides containing phosphorylation sites reported to be direct substrates of CK2, representing MCM2, IF2B, and spectrin (40–42). We found that these were decreased in tumor cells (Fig. 4C, 4D, and 4E), indicating that CK2 activity was decreased in cells grown in vivo. The differences in phosphorylation on these sites were a consequence of kinase activity as opposed to the gene expression of MCM2, IF2B, or spectrin as similar MCM2 and spectrin protein amounts were observed across the conditions; IF2F even being increased in abundance in tumors (Fig. 4F).
Fig. 4.
Growing conditions modulated casein kinase expression, phosphorylation and activity. A, Fold-changes of casein kinase isoforms in cells grown in vitro or in vivo. B, Fold-changes of phosphorylation sites on casein kinase isoforms across cells grown under two different conditions. C, D, and E, Fold-changes of phosphorylation sites known to be substrates of CK2. F, Fold-changes in abundance of proteins known to be substrates of CK2. G, Western blot analysis of CK2 and its substrate Akt pSer129.
As a means to confirm the MS data, analysis of CK2 alpha by Western blot indicated that this protein was decreased in tumor cells relative to cell cultures (Fig. 4G). Similarly, there was a decrease on the WB signals of phospho Akt on Ser129 (Fig. 4G), a phosphorylation site that is often used as a marker of CK2 activity (43). Collectively, these data indicate that CK2 activities were decreased in cancer cells growing in vivo relative to those growing in vitro, thus validating the MS data and confirming that CK2 is one of the kinase pathways modulated by in vivo conditions.
Identification of Specific Pathways and Processes Differentially Regulated in Human Cancer Cells Grown in Different Conditions
To investigate biological processes that were differentially regulated in cells grown in vivo relative to in vitro, we utilized a computational approach for pathway analysis described previously (26). The results indicated that proteins and phosphoproteins belonging to many different pathways and ontologies were differentially expressed in cells grown in vitro or in vivo (supplemental Fig. S2). Illustrative examples include proteins in glycolysis, the TCA cycle and gluconeogenesis, which were increased in tumors, whereas translation factors were increased in cells grown in vitro (supplemental Fig. S2). Phosphorylation on these protein classes was also found to the greater in tumors (supplemental Fig. S2). There were often apparent differences between proteomics and phosphoproteomics pathway data in that proteins in the N-Cadherin, Wnt signaling, and FoxO family pathways were, on average, decreased in cells grown in tumors, whereas phosphopeptide signals associated with these pathways were generally increased. These differences are in line with the known mechanisms of Wnt/catenin signaling (discussed below).
Identification of Wnt/catenin Signaling as a Pathway Regulated by In Vivo Growth Conditions
Our results indicate that in vivo growth conditions decreased the activity of the Wnt/catenin pathway. On average, Wnt pathway proteins were decreased in cells grown in tumors, although the adenomatous polyposis coli (APC) protein 2, a member of this pathway, was increased in tumors ∼fivefold (supplemental Fig. S3). In contrast, different catenin isoforms were decreased in tumors between two- and sevenfold, whereas cadherin-1 was also decreased twofold (supplemental Fig. S3). These data were confirmed by Western blots for APC and β-catenin (supplemental Fig. S3), thus further validating the MS data and the results of the bioinformatic analysis. Several phosphopeptides within human catenin sequences were increased in cells grown in vivo relative to those grown in vitro (supplemental Fig. S3), although other catenin sites did not change or were even decreased (supplemental Fig. S3). Taken together, the data in supplemental Fig. S3 are in line with the known signaling mechanisms of the canonical Wnt/catenin pathway, in which an increase in APC expression results in enhanced phosphorylation of specific residues on catenin proteins, which are then targets for degradation (44). These data therefore indicate that in vivo growth conditions modulate Wnt/catenin pathway activity and serve to further validate the MS-based phosphoproteomic approach.
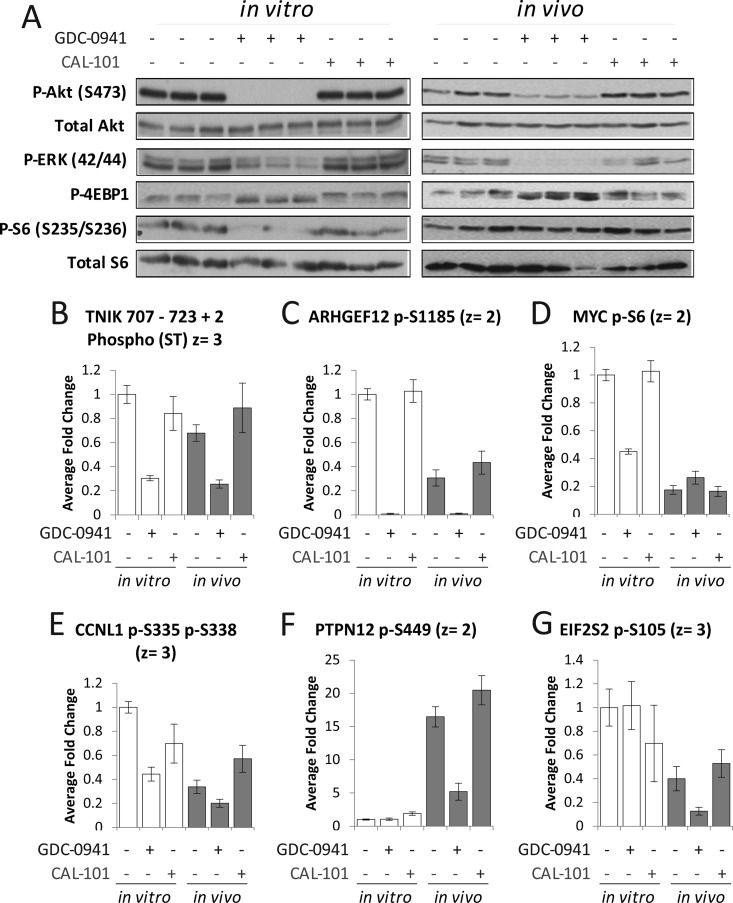
In Vivo Growth Conditions Affect the Responses of Cancer Cells to PI3K Inhibitors
Western-blot analyses indicated that the PI3K inhibitor GDC-0941 affected the phosphorylation of PKB/Akt and ERK in both cells grown in vitro and in vivo (Fig. 5A), although the extent of the inhibition was greater in vitro. These effects are likely to be driven by relative drug exposure. Nevertheless, Fig. 5A shows that GDC-0941 modulated PI3K/Akt signaling in both growing conditions to comparable extents. In contrast, CAL-101 did not have an effect on the phosphorylation of these pathway markers (Fig. 5A); this was expected as the expression of p110δ, the intended target of CAL-101, is restricted (in nontransformed cells) to leukocytes (15).
Fig. 5.
Growing conditions determine the phosphorylation sites that are modulated by PI3K inhibitors in cancer cells. A, Analysis of several markers of PI3K/Akt pathway activity as a function of treatment with GDC-0941 and CAL-101. B, C, Examples of sites modulated by GDC-0941 both in vivo and in vitro. D, E, F, G, Examples of sites specifically modulated in one growing condition but not the other. Values shown in bar charts are mean fold-change over control ± S.D. (n = 6).
MS-based phosphoproteomics data showed that the intensities of several phosphopeptide ions were significantly modulated by PI3K inhibitors (supplemental Fig. S4). The actual numbers of phosphopeptides affected by these treatments were dependent on the significance threshold applied; however, in general, GDC-0941 inhibited a greater number of phosphorylation sites than CAL-101 (supplemental Fig. S4). For example, at q < 0.01, GDC-0941 decreased the intensities of 73 phosphopeptide ions in vitro and 38 in vivo, whereas CAL-101 decreased the signal of 8 and 5, respectively (supplemental Fig. S4), thereby indicating that, in agreement with WB analyses (Fig. 5A), and as expected, GDC-0941 had a greater impact on the phosphoproteome than CAL-101.
Using supervised methods, we clustered phosphopeptides by their patterns of inhibition with the two PI3K compounds (supplemental Fig. S5). Several sites were specifically inhibited by GDC-0941 in cells grown either in vivo or in vitro, examples of which include a phosphopeptide derived from the protein kinase TNIK (Fig. 5B), previously implicated in Wnt signaling and in colon cancer growth (45), and on Ser1185 of the guanine exchange factor ARHGEF12 (Fig. 5C). In general, however, the effects of the inhibitors were substantially different between cells grown in tumors relative to those grown on plastic (supplemental Fig. S5). For example, phosphorylation sites derived from Myc (at Ser6) and Cyclin 1 (CCNL1 gene name), which were inhibited by GDC-0941 in vitro but not in vivo (Fig. 5D and 5E), and on the protein tyrosine phosphatase PTPN12 (at Ser449) and the transcription factor EIF2S2 (at Ser105), which were inhibited in vivo but not in vitro (Fig. 5F and 5G), despite the greater abundance of the latter site in cells grown in culture.
To investigate kinase groups differentially affected by PI3K inhibitors in human cancer cells grown in vitro or in vivo, we performed a phosphorylation motif analysis and kinase substrate enrichment analysis as described previously (46). Results of kinase substrate enrichment analysis showed that there were several kinase substrate groups that were preferentially inhibited in one growth condition relative to the other (supplemental Fig. S5), including those for CDK5 and MAP kinases, which were preferentially inhibited in vitro, whereas those for PKCD and RSK were preferentially inhibited in vivo (supplemental Fig. S6).
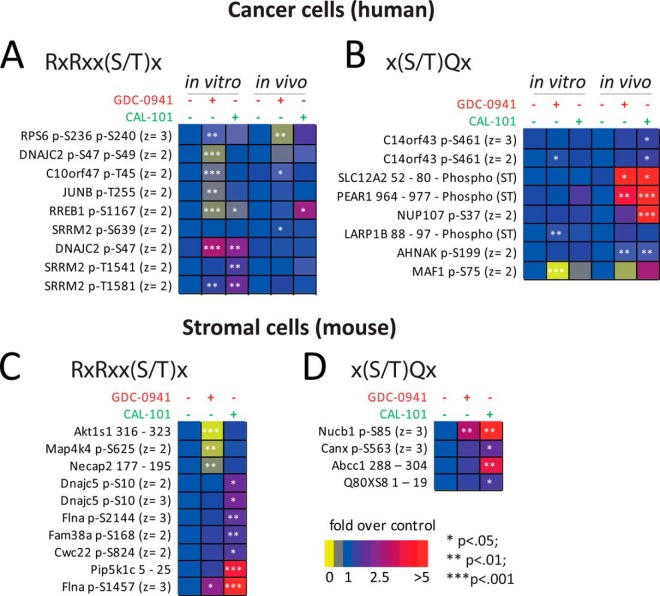
We also observed that phosphorylation sites representing the xRxRxxSx motif (where ‘x’ is any amino acid), which is the preferred recognition motif of PKB/Akt and related kinases (47), were generally inhibited by GDC-0941 in vivo and in vitro (Fig. 6A), whereas CAL-101 did not have an effect. In contrast, the effects of inhibitors were substantially different with regards to phosphorylation on motifs within xSQx sequences (Fig. 6B). Indeed, although the PI3K inhibitors did not have an effect on the phosphorylation of peptides with the xSQx motif in vitro, these phosphorylation sites were increased in vivo (Fig. 6B). The elevated expression of DNA-PK—which phosphorylates Ser/Thr in the context of SQ [ref (48)]—in cells grown in vivo relative to those in vitro (Fig. 3A, red arrow) may explain the dissimilar effects of PI3K inhibitors on the phosphorylation of this motif across cells grown in the two conditions. The increase in the phosphorylation of peptides containing the SQ motif as a result of PI3K inhibitor treatment is consistent with a previous study showing that inhibition of PI3K activates DNA-PK secondary to the induction of apoptosis caused by inhibiting this pathway (46).
Fig. 6.
PI3K inhibitors differentially modulated phosphorylation in cancer and tumor associated stromal cells. Fold-changes in the intensities of phosphopeptides containing human A, B, or mouse-specific sequences C, D, as a function of treatment with the PI3K inhibitors GDC-0941 and CAL-101. Phosphopeptides were classified based on their motif. Values shown represent mean fold-change over control (n = 6).
Effects of PI3K Inhibitors on the Phosphorylation of Proteins in Mouse Stromal Cells
As with the effects of PI3K inhibitors in human proteins (which are derived from cancer cells), these compounds also affected the phosphorylation of several peptides with mouse-specific sequences (derived from stromal cells, supplemental Fig. S7). Classification of these phosphopeptides based on motifs showed that GDC-0941 decreased the phosphorylation of some stromal peptides with the RxRxxSx motif, which is recognized by PKB/Akt downstream of PI3K (Fig. 6C). This included a site on PRAS40 (gene name Akt1S1) on T318, which is a protein constituent of the mTOR complex 1 and which is known to be downstream of PKB/Akt and PI3K (49). Not all phosphopeptides with this motif were inhibited by PI3K inhibitors in stromal cells and some of them even increased in phosphorylation as a result of treatment (Fig. 6C), which is consistent with the effect of the inhibitors in human (cancer) cells (Fig. 6A). This was particularly noticeable for CAL-101, which did not inhibit the phosphorylation on any of the peptides within the xRxRxxSx motif (Fig. 6C). Both GDC-0941 and CAL-101 increased the phosphorylation of sites in the context of the SQ motif in stromal cells (Fig. 6D) with the effects of CAL-101 being greater than those of GDC-0941.
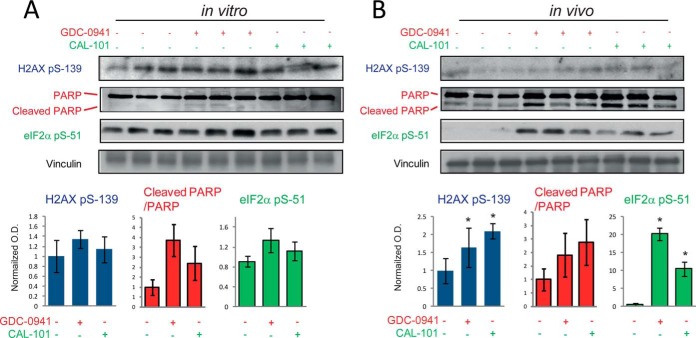
PI3K Inhibitors Modulate Markers of Apoptosis Differently In Vivo and In Vitro
The increase in the phosphorylation of sites in the context of the SQ motif may indicate that kinases with this substrate specificity are activated as a result of inhibitor treatment of tumors but not cells grown in culture. Kinases involved in DNA repair, such as DNA-PK and ATM, are known to phosphorylate Ser/Thr residues within this motif and are activated as a result of DNA cleavage that occurs during apoptosis (50). To investigate this possibility, we assessed H2AX phosphorylation on Ser139, a site within a SQ motif and a marker of DNA-PK activity and apoptosis (50), as a function of PI3K inhibitor treatment. Although pSer139 on H2AX was unchanged in cells grown in vitro (Fig. 7A), it increased ∼twofold as a function of GDC-0941 and CAL-101 treatment of tumors (Fig. 7B). Apoptosis, as assessed by PARP cleavage, increased when cells were treated with GDC-0941 but not with CAL-101 in vitro (Fig. 7A). However, signals of cleaved PARP increased as a result of treatment of tumors with both PI3K inhibitors (Fig. 7B). Similarly, the phosphorylation of eIF2α at Ser 51, which also occurs during the apoptotic process as a mechanism of inhibiting translation (51, 52), was increased 20-fold by GDC-0941 and 10-fold by CAL-101 in tumors, but not in cells grown in culture (green charts in right panels in Figs. 7A and 7B). Inhibition of translation is a key event during apoptosis (53). Thus, in agreement with the phosphoproteomics data showing an increase in the phosphorylation of SQ motifs as a function of inhibitor treatment in both cancer and stromal cells (Fig. 6B and 6D), the data in Fig. 7 indicate that PI3K inhibitors initiate the process of apoptosis in tumors to a greater extent than in cells grown in culture.
Fig. 7.
PI3K inhibitors induced apoptotic markers in xenografts to a greater extent than in cells grown in culture. Immunoblots against phospho-H2AX (Ser139) and cleaved PARP as a function of treatment with PI3K inhibitors in cells grown in culture A, or in tumor xenografts B, Bottom graphs show the normalized densitometry values of three biological replicates (values are mean ± S.E., n = 3). * p < 0.05 by the Wilcoxon test.
DISCUSSION
The role of heterotypic interactions and of individual components present in the tumor microenvironment in the development of cancer is well established (1, 3, 4, 6, 8, 54). However, how the combination of factors present in tumors in vivo modulate protein networks in cancer cells is less well understood, partly because of the difficulty in measuring cancer cell specific protein expression and signaling. Indeed, antibodies often cross-react with both human and mouse proteins, thus complicating their use in the investigation of cell-specific changes in protein expression that may occur when cells are grown in vivo instead of in vitro.
Our study explored the hypothesis that the specificity of MS would allow the investigation of protein expression and signaling events specifically occurring in cancer or stromal cells present in mouse tumor xenografts. Comparison of human and mouse genomes has revealed that 70.1% of amino acid sequences are conserved across these two organisms (55). Therefore, on average, a typical tryptic peptide of 12 residues in length would differ by 3 or 4 amino acids between human and mouse, thus making it possible to distinguish them by mass. We empirically determined that although about one quarter of tryptic peptides matched to sequences in both human and mouse databases, the other three quarters of peptides identified in the study were specific to either species (Fig. 1B). These data are in agreement with the observation that differences between the coding regions of human and mouse genomes are not uniformly distributed (55).
Using this approach we observed that growing cells in different environments had profound effects in protein and phosphoprotein expression (Fig. 2). Interestingly, we found that the in vivo growth conditions produced greater changes on the phosphoproteome than on the proteome of our cell model, in line with the modulation of kinase cascades being a key mechanism by which the tumor microenvironment modulates tumor biology. These observations also suggest that small changes in protein expression may result in large changes in protein kinase activities. The low correlation between enzyme amounts and their activity is well understood for metabolic pathways; theoretical and experimental evidence have shown that small changes in the expression of several enzymes in a given pathway can result in large changes in pathways fluxes (56). Similarly, kinase cascades amplify small signals into larger outputs that are detected as protein phosphorylation events (57, 58). In addition to their expression and extent of post-translational modification, the activity of protein kinases can be modulated by allosteric mechanisms regulated by protein-protein interactions. For example, the activity of CK2 is modulated by the ratio of catalytic (alpha) to regulatory (beta) subunits as the latter binds and inactivates the catalytic activity of CK2 alpha (36, 38). We found that this ratio of CK2 catalytic to regulatory subunit was modulated by in vivo growth conditions (Fig. 4). It is therefore conceivable that signals in the in vivo tumor environment could induce greater changes in protein phosphorylation (a measure of kinase pathway activity) than in protein expression, as observed in this study (Fig. 2). Our data suggesting that in vivo growing conditions cause cancer cells to rewire protein kinase networks extensively, despite inducing relatively modest changes in protein expression, may have implications for our understanding of how cancer cells respond to signaling inhibitors—this notion being discussed further below.
Among the many biological processes found to be differentially regulated between cells grown in cell culture or in tumors (shown in supplemental Fig. S3), we confirmed by WB that in vivo growth conditions regulated the Wnt/catenin pathway and casein kinase activity. Confirmation by WB was undertaken to give support to the relevance of MS data; however, it should be noted that antibodies used for this work cross-react with both human and mouse sequences, whereas MS allowed the distinction between human and mouse-specific sequences. Therefore, the overall magnitude in fold differences between conditions was expected to be different for the two analytical methods. The Wnt pathway is frequently deregulated in colorectal cancers, with APC being one of the most frequently mutated tumor suppressors in this cancer type (44). Because casein kinases have been implicated in the regulation of catenin expression and transcriptional activity (43, 59, 60), our observation that in vivo conditions decreased the expression of different catenin isoforms (Fig. 4) is consistent with the decrease in casein kinase activities found in cells grown in vivo (Fig. 3). Our results are also consistent with recent reports showing that Wnt signaling is modulated by the tumor microenvironment (61). Pathway and ontology analyses (supplemental Fig. S2) also suggest that in vivo conditions modulate many other pathways and biological functions, including an overall reduction of apoptosis and translation in xenografts relative to cells growing on plastic.
Our approach also made possible the assessment of how signaling inhibitors can modulate signaling specifically in cancer and stromal cells. We found that in vivo growth conditions rewired the basal kinase network extensively (Figs. 2, 3, and 4), an observation that was consistent with the differential impact on pathways in response to compound treatment across the two different growth conditions (Figs. 5, 6, and 7). Indeed, PI3K inhibitors reduced the intensities of different sets of phosphorylation sites in cells grown in vivo relative to those grown in vitro (Figs. 5, 6, and supplemental Fig. S5), thus indicating that growing conditions influence how signaling inhibitors modulate tumor biology.
GDC-0941 inhibits all class I PI3K isoforms, whereas CAL-101 is selective for p110δ; therefore, it was expected that the former inhibitor would modulate a greater number of sites than the latter in both cancer and stromal cells. Because, in untransformed cells, the expression of the p110δ isoform of PI3K is restricted to leukocytes, CAL-101 is in clinical trials to treat several types of hematological malignancies (62). Consistent with the expression pattern of p110δ, CAL-101 did not inhibit markers of PI3K/Akt pathway activation in either cancer or tumor-associated stromal cells, as assessed by both MS and WB methods (Figs. 5 and 6), yet this compound elicited an unexpected increase in the phosphorylation of sites within the SQ motif in both cancer cells (Fig. 6B) and tumor-associated stroma (Fig. 6D)—phosphorylation on SQ motifs being associated with the induction of apoptosis and DNA damage (50, 63). Induction of apoptosis in our model was confirmed by measuring several markers of apoptosis (Figs. 7), which were found to be increased when tumors (but not cells grown in culture) were treated with CAL-101 and GDC-0941. These data indicate that CAL-101 can modulate the biochemistry of both cancer and tumor-associated stromal cells in vivo to a greater extent than in vitro and suggest that this compound may have activity in a broader range of malignancies than previously appreciated. The mechanism of action of CAL-101 in these cell types seems to be independent of PI3K inhibition, and instead dependent on the growth of cancer cells in vivo.
Supplementary Material
Acknowledgments
We thank Mr. Wayne Pearce for assistance with mouse experiments and Dr. Hagen Kulbe and Dr. Khaled Ali for discussions and advice.
Footnotes
Author contributions: P.R.C. designed research; V.R., I.V., E.W., N.T., and P.R.C. performed research; N.T. contributed new reagents or analytic tools; P.R.C. analyzed data; P.R.C. wrote the paper.
Author contributions: V.R. performed wet-lab experiments, analyzed the data and edited the paper; I.V. performed mass spectrometry experiments, analyzed the data and edited the paper; E.W. performed experiments and edited the paper; N.T. contributed reagents and edited the paper; P.R.C. conceived study, performed bioinformatics experiments, analyzed and interpreted data, prepared figures, and wrote the paper.
* This work was supported by Bart's and The London Charity (Grant 297/997) and by Activiomics Ltd.
 This article contains supplemental Figs. S1 to S7 and Data Sets S1 to S5.
This article contains supplemental Figs. S1 to S7 and Data Sets S1 to S5.
Disclosures: I.V. and N.T. were employees of Activiomics Ltd while this work was undertaken and are now employees of Retroscreen Virology. P.R.C. consults for Retroscreen Virology.
1 The abbreviations used are:
- LC
- liquid chromatography
- MS-MS
- tandem MS.
REFERENCES
- 1. Elenbaas B., Weinberg R. A. (2001) Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp. Cell Res. 264, 169–184 [DOI] [PubMed] [Google Scholar]
- 2. Fang H., Declerck Y. A. (2013) Targeting the tumor microenvironment: from understanding pathways to effective clinical trials. Cancer Res. 73, 4965–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krutovskikh V. A., Piccoli C., Yamasaki H. (2002) Gap junction intercellular communication propagates cell death in cancerous cells. Oncogene 21, 1989–1999 [DOI] [PubMed] [Google Scholar]
- 4. Buess M., Nuyten D. S., Hastie T., Nielsen T., Pesich R., Brown P. O. (2007) Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer. Genome Biol. 8, R191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 6. Liotta L. A., Kohn E. C. (2001) The microenvironment of the tumour-host interface. Nature 411, 375–379 [DOI] [PubMed] [Google Scholar]
- 7. Fidler I. J., Poste G. (2008) The “seed and soil” hypothesis revisited. Lancet Oncol. 9, 808. [DOI] [PubMed] [Google Scholar]
- 8. Lowery F. J., Yu D. (2012) Growth factor signaling in metastasis: current understanding and future opportunities. Cancer Metast. Rev. 31, 479–491 [DOI] [PubMed] [Google Scholar]
- 9. Park E. S., Kim S. J., Kim S. W., Yoon S. L., Leem S. H., Kim S. B., Kim S. M., Park Y. Y., Cheong J. H., Woo H. G., Mills G. B., Fidler I. J., Lee J. S. (2011) Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc. Natl. Acad. Sci. U.S.A. 108, 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z. R., Du J., Davis A., Mongare M. M., Gould J., Frederick D. T., Cooper Z. A., Chapman P. B., Solit D. B., Ribas A., Lo R. S., Flaherty K. T., Ogino S., Wargo J. A., Golub T. R. (2012) Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofmeister V., Schrama D., Becker J. C. (2008) Anti-cancer therapies targeting the tumor stroma. Cancer Immunol. Immun. 57, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chin Y. R., Toker A. (2009) Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell. Signal. 21, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Folkes A. J., Ahmadi K., Alderton W. K., Alix S., Baker S. J., Box G., Chuckowree I. S., Clarke P. A., Depledge P., Eccles S. A., Friedman L. S., Hayes A., Hancox T. C., Kugendradas A., Lensun L., Moore P., Olivero A. G., Pang J., Patel S., Pergl-Wilson G. H., Raynaud F. I., Robson A., Saghir N., Salphati L., Sohal S., Ultsch M. H., Valenti M., Wallweber H. J., Wan N. C., Wiesmann C., Workman P., Zhyvoloup A., Zvelebil M. J., Shuttleworth S. J. (2008) The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J. Med. Chem. 51, 5522–5532 [DOI] [PubMed] [Google Scholar]
- 14. Herman S. E., Gordon A. L., Wagner A. J., Heerema N. A., Zhao W., Flynn J. M., Jones J., Andritsos L., Puri K. D., Lannutti B. J., Giese N. A., Zhang X., Wei L., Byrd J. C., Johnson A. J. (2010) Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood 116, 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., Zvelebil M. J., Higashi K., Volinia S., Downward J., Waterfield M. D. (1997) P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 94, 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong K. K., Engelman J. A., Cantley L. C. (2010) Targeting the PI3K signaling pathway in cancer. Curr. Opin. Gen. Dev. 20, 87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Velculescu V. E. (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 18. Robak P., Robak T. (2012) A targeted therapy for protein and lipid kinases in chronic lymphocytic leukemia. Curr. Med. Chem. 19, 5294–5318 [DOI] [PubMed] [Google Scholar]
- 19. Hoeflich K. P., Merchant M., Orr C., Chan J., Den Otter D., Berry L., Kasman I., Koeppen H., Rice K., Yang N. Y., Engst S., Johnston S., Friedman L. S., Belvin M. (2012) Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 72, 210–219 [DOI] [PubMed] [Google Scholar]
- 20. Lannutti B. J., Meadows S. A., Herman S. E., Kashishian A., Steiner B., Johnson A. J., Byrd J. C., Tyner J. W., Loriaux M. M., Deininger M., Druker B. J., Puri K. D., Ulrich R. G., Giese N. A. (2011) CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117, 591–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alcolea M. P., Casado P., Rodriguez-Prados J. C., Vanhaesebroeck B., Cutillas P. R. (2012) Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol. Cell. Proteomics: MCP 11, 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montoya A., Beltran L., Casado P., Rodriguez-Prados J. C., Cutillas P. R. (2011) Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods 54, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 24. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics 10, M110 003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cutillas P. R., Vanhaesebroeck B. (2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell. Proteomics 6, 1560–1573 [DOI] [PubMed] [Google Scholar]
- 26. Wisniewski J. R., Ostasiewicz P., Dus K., Zielinska D. F., Gnad F., Mann M. (2012) Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Sys. Biol. 8, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 28. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casado P., Cutillas P. R. (2011) A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments. Mol. Cell. Proteomics 10, M110 003079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stulemeijer I. J., Joosten M. H., Jensen O. N. (2009) Quantitative phosphoproteomics of tomato mounting a hypersensitive response reveals a swift suppression of photosynthetic activity and a differential role for hsp90 isoforms. J. Proteome Res. 8, 1168–1182 [DOI] [PubMed] [Google Scholar]
- 32. Schilling B., Rardin M. J., MacLean B. X., Zawadzka A. M., Frewen B. E., Cusack M. P., Sorensen D. J., Bereman M. S., Jing E., Wu C. C., Verdin E., Kahn C. R., Maccoss M. J., Gibson B. W. (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics 11, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rardin M. J., Newman J. C., Held J. M., Cusack M. P., Sorensen D. J., Li B., Schilling B., Mooney S. D., Kahn C. R., Verdin E., Gibson B. W. (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 110, 6601–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S. P., Hengartner M. O., von Mering C. (2012) PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell Proteomics 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lolli G., Pinna L. A., Battistutta R. (2012) Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization. ACS Chem. Biol. 7, 1158–1163 [DOI] [PubMed] [Google Scholar]
- 36. Deshiere A., Duchemin-Pelletier E., Spreux E., Ciais D., Combes F., Vandenbrouck Y., Coute Y., Mikaelian I., Giusiano S., Charpin C., Cochet C., Filhol O. (2013) Unbalanced expression of CK2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by Snail1 induction. Oncogene 32, 1373–1383 [DOI] [PubMed] [Google Scholar]
- 37. Korn I., Jacob G., Allende C. C., Allende J. E. (2001) The activity of CK2 in the extracts of COS-7 cells transfected with wild type and mutant subunits of protein kinase CK2. Mol. Cell. Biochem. 227, 37–44 [PubMed] [Google Scholar]
- 38. Battistutta R., Lolli G. (2011) Structural and functional determinants of protein kinase CK2alpha: facts and open questions. Mol. Cell. Biochem. 356, 67–73 [DOI] [PubMed] [Google Scholar]
- 39. Niefind K., Issinger O. G. (2010) Conformational plasticity of the catalytic subunit of protein kinase CK2 and its consequences for regulation and drug design. Biochim. Biophys. Acta 1804, 484–492 [DOI] [PubMed] [Google Scholar]
- 40. Llorens F., Duarri A., Sarro E., Roher N., Plana M., Itarte E. (2006) The N-terminal domain of the human eIF2beta subunit and the CK2 phosphorylation sites are required for its function. Biochem. J. 394, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281, 10281–10290 [DOI] [PubMed] [Google Scholar]
- 42. Bignone P. A., King M. D., Pinder J. C., Baines A. J. (2007) Phosphorylation of a threonine unique to the short C-terminal isoform of betaII-spectrin links regulation of alpha-beta spectrin interaction to neuritogenesis. J. Biol. Chem. 282, 888–896 [DOI] [PubMed] [Google Scholar]
- 43. Ponce D. P., Maturana J. L., Cabello P., Yefi R., Niechi I., Silva E., Armisen R., Galindo M., Antonelli M., Tapia J. C. (2011) Phosphorylation of AKT/PKB by CK2 is necessary for the AKT-dependent up-regulation of beta-catenin transcriptional activity. J. Cell. Physiol. 226, 1953–1959 [DOI] [PubMed] [Google Scholar]
- 44. Clevers H. (2006) Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 45. Shitashige M., Satow R., Jigami T., Aoki K., Honda K., Shibata T., Ono M., Hirohashi S., Yamada T. (2010) Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res. 70, 5024–5033 [DOI] [PubMed] [Google Scholar]
- 46. Casado P., Rodriguez-Prados J. C., Cosulich S. C., Guichard S., Vanhaesebroeck B., Joel S., Cutillas P. R. (2013) Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Sig. 6, rs6. [DOI] [PubMed] [Google Scholar]
- 47. Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. (1996) Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS letters 399, 333–338 [DOI] [PubMed] [Google Scholar]
- 48. Watanabe F., Teraoka H., Iijima S., Mimori T., Tsukada K. (1994) Molecular properties, substrate specificity and regulation of DNA-dependent protein kinase from Raji Burkitt's lymphoma cells. Biochim. Biophys. Acta 1223, 255–260 [DOI] [PubMed] [Google Scholar]
- 49. Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 50. Mukherjee B., Kessinger C., Kobayashi J., Chen B. P., Chen D. J., Chatterjee A., Burma S. (2006) DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair 5, 575–590 [DOI] [PubMed] [Google Scholar]
- 51. Saelens X., Kalai M., Vandenabeele P. (2001) Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J. Biol. Chem. 276, 41620–41628 [DOI] [PubMed] [Google Scholar]
- 52. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 53. Wiita A. P., Ziv E., Wiita P. J., Urisman A., Julien O., Burlingame A. L., Weissman J. S., Wells J. A. (2013) Global cellular response to chemotherapy-induced apoptosis. eLife 2, e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McAllister S. S., Weinberg R. A. (2010) Tumor-host interactions: a far-reaching relationship. J. Clin. Oncol. 28, 4022–4028 [DOI] [PubMed] [Google Scholar]
- 55. Emes R. D., Goodstadt L., Winter E. E., Ponting C. P. (2003) Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 12, 701–709 [DOI] [PubMed] [Google Scholar]
- 56. Cascante M., Boros L. G., Comin-Anduix B., de Atauri P., Centelles J. J., Lee P. W. (2002) Metabolic control analysis in drug discovery and disease. Nat. Biotechnol. 20, 243–249 [DOI] [PubMed] [Google Scholar]
- 57. Cobb M. H. (1999) MAP kinase pathways. Prog. Biophys. Mol. Bio. 71, 479–500 [DOI] [PubMed] [Google Scholar]
- 58. Ferrell J. E., Jr. (1996) Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 21, 460–466 [DOI] [PubMed] [Google Scholar]
- 59. Casagolda D., Del Valle-Perez B., Valls G., Lugilde E., Vinyoles M., Casado-Vela J., Solanas G., Batlle E., Reynolds A. B., Casal J. I., de Herreros A. G., Dunach M. (2010) A p120-catenin-CK1epsilon complex regulates Wnt signaling. J. Cell Sci. 123, 2621–2631 [DOI] [PubMed] [Google Scholar]
- 60. Tapia J. C., Torres V. A., Rodriguez D. A., Leyton L., Quest A. F. (2006) Casein kinase 2 (CK2) increases survivin expression via enhanced beta-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc. Natl. Acad. Sci. U.S.A. 103, 15079–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vermeulen L., De Sousa E. M. F., van der Heijden M., Cameron K., de Jong J. H., Borovski T., Tuynman J. B., Todaro M., Merz C., Rodermond H., Sprick M. R., Kemper K., Richel D. J., Stassi G., Medema J. P. (2010) Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12, 468–476 [DOI] [PubMed] [Google Scholar]
- 62. Fruman D. A., Rommel C. (2011) PI3Kdelta inhibitors in cancer: rationale and serendipity merge in the clinic. Cancer Disc. 1, 562–572 [DOI] [PubMed] [Google Scholar]
- 63. Chen B. P., Chan D. W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D. J. (2005) Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 280, 14709–14715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (28) with the dataset identifier PXD000218 and DOI 10.6019/PXD000218. Results are also shown in supplementary Data Sets S1, S2, and S3 and representative XICs are shown in supplementary Data Sets S4 and S5.