Abstract
Antibodies are of importance for the field of proteomics, both as reagents for imaging cells, tissues, and organs and as capturing agents for affinity enrichment in mass-spectrometry-based techniques. It is important to gain basic insights regarding the binding sites (epitopes) of antibodies and potential cross-reactivity to nontarget proteins. Knowledge about an antibody's linear epitopes is also useful in, for instance, developing assays involving the capture of peptides obtained from trypsin cleavage of samples prior to mass spectrometry analysis. Here, we describe, for the first time, the design and use of peptide arrays covering all human proteins for the analysis of antibody specificity, based on parallel in situ photolithic synthesis of a total of 2.1 million overlapping peptides. This has allowed analysis of on- and off-target binding of both monoclonal and polyclonal antibodies, complemented with precise mapping of epitopes based on full amino acid substitution scans. The analysis suggests that linear epitopes are relatively short, confined to five to seven residues, resulting in apparent off-target binding to peptides corresponding to a large number of unrelated human proteins. However, subsequent analysis using recombinant proteins suggests that these linear epitopes have a strict conformational component, thus giving us new insights regarding how antibodies bind to their antigens.
Antibodies are used in proteomics both as imaging reagents for the analysis of tissue specificity (1) and subcellular localization (2) and as capturing agents for targeted proteomics (3), in particular for the enrichment of peptides for immunoaffinity methods such as Stable Isotope Standards and Capture by Anti-peptide Antibodies (4). In fact, the Human Proteome Project (5) has announced that one of the three pillars of the project will be antibody-based, with one of the aims being to generate antibodies to at least one representative protein from all protein-coding genes. Knowledge about the binding site (epitope) of an antibody toward a target protein is thus important for gaining basic insights into antibody specificity and sensitivity and facilitating the identification and design of antigens to be used for reagents in proteomics, as well as for the generation of therapeutic antibodies and vaccines (1, 6). With over 20 monoclonal-antibody-based drugs now on the market and over 100 in clinical trials, the field of antibody therapeutics has become a central component of the pharmaceutical industry (7). One of the key parameters for antibodies includes the nature of the binding recognition toward the target, involving either linear epitopes formed by consecutive amino acid residues or conformational epitopes consisting of amino acids brought together by the fold of the target protein (8).
A large number of methods have therefore been developed to determine the epitopes of antibodies, including mass spectrometry (9), solid phase libraries (10, 11), and different display systems (12–14) such as bacterial display (15) and phage display (16). The most common method for epitope mapping involves the use of soluble and immobilized (tethered) peptide libraries, often in an array format, exemplified by the “Geysen Pepscan” method (11) in which overlapping “tiled” peptides are synthesized and used for binding analysis. The tiled peptide approach can also be combined with alanine scans (17) in which alanine substitutions are introduced into the synthetic peptides and the direct contribution of each amino acid can be investigated. Maier et al. (18) described a high-throughput epitope-mapping screen of a recombinant peptide library consisting of a total of 2304 overlapping peptides of the vitamin D receptor, and recently Buus et al. (19) used in situ synthesis on microarrays to design and generate 70,000 peptides for epitope mapping of antibodies using a range of peptides with sizes from 4-mer to 20-mer.
So far it has not been possible to investigate on- and off-target binding in a proteome-wide manner, but the emergence of new methods for in situ synthesis of peptides on ultra-dense arrays has made this achievable. Here, we describe the design and use of peptide arrays generated with parallel in situ photolithic synthesis (20) of a total of 2.1 million overlapping peptides covering all human proteins with overlapping peptides. Miniaturization of the peptide arrays (21) has led to improved density of the synthesized peptides and consequently has improved the resolution and coverage of the epitope mapping. This has allowed us to study the specificity and cross-reactivity of both monoclonal and polyclonal antibodies across the whole “epitome” with the use of both proteome-wide arrays and focused-content peptide arrays covering selected antigen sequences to precisely map the contribution of each amino acid of the target protein for binding recognition of the corresponding antibodies. The results show the usefulness of proteome-wide epitope mapping, showing a path forward for high-throughput analysis of antibody interactions.
EXPERIMENTAL PROCEDURES
Array Design
Whole human proteome arrays were designed based on the human Consensus CDS (version 37.1) protein set provided by the National Center for Biotechnology Information (NCBI) (22). To essentially cover the proteome, 2.1 million 12-mer peptides overlapping by six amino acids were randomly distributed on the array. Focused-content arrays for more detailed epitope mapping and alanine scanning were designed with a mix of 12-mer and 15-mer peptides overlapping by 11 and 14 amino acids, respectively. Peptides of both lengths were designed to cover the entire sequences of the protein fragments used for immunization. Additional peptides corresponding to the 15-mer peptides, but with the middle amino acid substituted by an alanine, were also included in the design, and all peptides were randomly distributed.
Peptide Synthesis
Combinatorial synthesis of the peptide libraries was accomplished by means of light-directed array synthesis in a Roche-Nimblegen Maskless Array Synthesizer (20, 23–25) using an amino-functionalized 1-inch × 3-inch microscope slide as a substrate coupled with six-amino hexanoic acid as a spacer and amino acid derivatives carrying a photosensitive 2-(2-nitrophenyl)propyl-oxycarbonyl group at the α-amino function as in the study by Laursen and colleagues (26). Coupling of amino acids was done with pre-activation in 30 mm amino acid, 30 mm activator (HOBt/HBTU), and 60 mm ethyldiisopropylamine in N,N-dimethylformamide for 5 to 7 min before flushing of the substrate for 5 min. Washings were done with 1-methyl-2-pyrrolidinone, and site-specific cleavage of the 2-(2-nitrophenyl)propyl-oxycarbonyl group was accomplished by irradiation of an image created by a Digital Micromirror Device (Texas Instruments Inc., Dallas, TX; Super Extended Graphics Array Plus graphics format), projecting light with a 365-nm wavelength to a selection of ∼1.4 million features of (13 × 13) μm2 at a total dose of ∼10 J/cm2 in 1-methyl-2-pyrrolidinone. Final treatment of the slide with TFA/water/triisopropylsilane for 30 min cleaved off the side-chain protection of the amino acids.
Antibody Incubation and Scanning
De-protected slides were washed twice with TBSTT (20 mm Tris, 0.9% NaCl, pH 7.4, 0.1% Tween 20, 0.4% Triton X-100) in a LockMailer slide jar (Aldrich) by inverting the jar for 2 min. The slides were then washed twice in TBS (20 mm Tris, 0.9% NaCl, pH 7.4) for 2 min, rinsed quickly three times with de-ionized water, and dried. Mixer masks (Roche NimbleGen Inc., Madison, WI) were attached to the slides, and antibody samples diluted in binding buffer (10 mm Tris, 0.45% NaCl, pH 7.4, alkali soluble casein 0.5% (Novagen, EMD Chemicals, San Diego, CA)) were injected into the mixer compartments. The slides were incubated overnight in a NimbleGen Hybridization Station (Roche NimbleGen Inc.) according to the manufacturer's instructions. After the primary incubation, the slides were submerged in TBSTT and the mixers were removed. The slides were washed twice with TBSTT and twice with TBS as described above. Secondary DyLight649-conjugated anti-rabbit or Cy3-conjugated anti-mouse antibodies (Jackson ImmunoResearch, West Grove, PA) were diluted to 0.15 μg/ml in binding buffer in LockMailer jars, and the slides were incubated for 3 h on a shaking table. The slides were washed twice with TBSTT and twice with TBS as described above, quickly rinsed three times in de-ionized water, and dried. The slides were subsequently scanned at 2-μm resolution using a NimbleGen MS200 scanner (Roche NimbleGen Inc.).
Image Aligning and Data Filtering
The scan images were aligned and peptide feature mean fluorescence values were exported using the NimbleScan2 software (Roche NimbleGen Inc.). Before further analysis, confirmed false-positive signals caused by dirt on the arrays were removed.
Generation of Antibodies
Antigens were designed using the software PRESTIGE (27). Gene fragments were amplified from a pool of RNA isolated from human tissues, cloned into a vector, and expressed in Escherichia coli. To generate polyclonal antibodies, purified and validated recombinant protein fragments were used for immunization of New Zealand White rabbits, and the polyclonal rabbit sera were purified using their corresponding antigens as affinity ligands (28). The monoclonal antibody was generated as described elsewhere (29).
Antigen Array Analysis
The 274 protein fragments corresponding to the peptides bound by the anti-PODXL1 and anti-RBM3 antibodies were spotted on an epoxy-coated glass surface (CapitalBio, Bejing, China) using a non-contact printer (ArrayJet Marathon, Arrayjet Ltd., Roslin, UK). The microarray slides were incubated overnight at 37 °C and then blocked for 1 h in PBST (1× PBS, 0.1% Tween20) supplemented with 3% bovine serum albumin. The slides underwent two 5-min washes with PBST and one 5-min wash with 1× PBS before a final rinse with de-ionized water. The slides were dried and stored in the dark at 4 °C until use. The polyclonal PODXL antibody and the monoclonal antibody toward RBM3 were diluted 1:500 and 1:100 in PBST, respectively. The slides underwent two 5-min washes with PBST before incubation for 1 h with secondary antibodies (anti-rabbit-Alexa647 for the polyclonal antibody and anti-mouse-Alexa647 for the monoclonal antibody (Invitrogen)). After two 5-min washes with PBST, one 5-min wash with PBS, and a quick rinse in de-ionized water, the slides were dried before scanning with a G2565BA array scanner (Agilent Technologies, Santa Clara, CA). Image analysis and data extraction were performed using GenePix 5.1 software (Molecular Devices, Sunnyvale, CA).
Western Blot
Approximately 15 μg of total protein from cell lines lysates (RT-4, U251 MG, Caco-2, HEK293) or HEK293 overexpression lysates (PODXL LY401657, SCG2 LY418654, MKNK2 LY413712, RNF214 LY403969, GPR56 LY428998, PRY2 LY424151, CDK2AP2 LY417039, MS4A8B LY403116, STARD13 LY406044, all from Origene, Rockville, MD) were run on precast 4–20% CriterionTM TGXTM SDS-PAGE gradient gels (Bio-Rad Laboratories, Hercules, CA) under reducing conditions. Electroblotting of the separated proteins onto 0.2-μm PVDF membranes was performed using the Trans-Blot® TurboTM Transfer System (Bio-Rad Laboratories), according to the manufacturer's instructions. Membranes were blocked (5% dry milk, 0.5% Tween20, 1× TBS, 0.1 m Tris-HCl, 0.5 m NaCl, pH 7.5) for 1 h before being incubated with primary antibodies diluted 1:250 in blocking buffer for 1 h. The membranes were washed four times (5 min each time) in 1× TBS with 0.05% Tween20 before being incubated for 45 min with secondary HRP-conjugated swine anti-rabbit or goat anti-mouse antibody (DakoCytomation, Glostrup, Denmark) diluted 1:3000 and 1:6000 in blocking buffer, respectively. The membranes were again washed four times (5 min each time) before SuperSignal® West Dura Extended Duration Substrate (Pierce) was added and chemiluminescence detection was carried out using a Chemidoc charge-coupled device (CCD)-camera system (Bio-Rad Laboratories), according to the manufacturer's instructions.
RESULTS
Generation of Whole-proteome Peptide Microarrays Using Photolithography
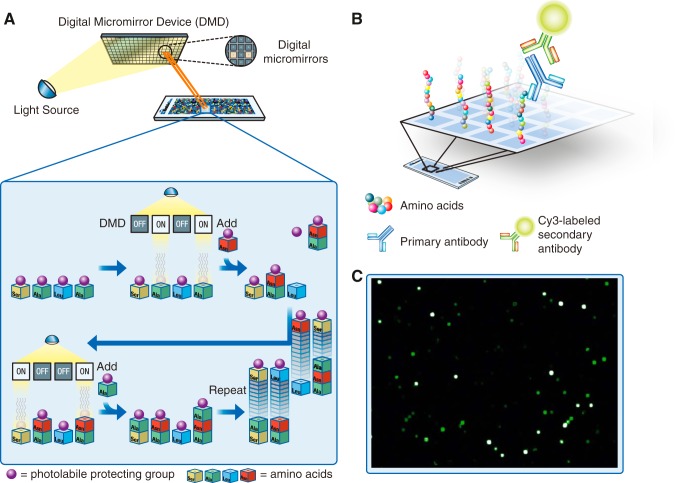
The principle of the synthesis of peptide arrays and their use for antibody binding analysis is outlined in Fig. 1. A UV-light source combined with digital micromirrors is used to selectively activate small squares of the array, and amino acids with photo-labile protective groups are then added to the whole array. The amino acids will only be incorporated into the previously activated peptides, and through repeated cycles of activation and coupling, all unique peptides on the array can be synthesized in parallel. Because of the digital nature of the synthesis technology, array peptide sequences can be readily changed, enabling the synthesis of both custom and fixed-content array designs. After synthesis the arrays are incubated with the antibodies of interest, and this primary binding is detected with a fluorophore-conjugated secondary antibody. The arrays are scanned with a high-resolution microarray slide scanner, and the fluorescence intensities of the peptide features are evaluated. To cover the entire human proteome, based on the consensus coding sequences (CCDS) definition of the human proteome provided by NCBI, arrays were designed with 2.1 million overlapping 12-mer peptides with a six-amino-acid lateral shift.
Fig. 1.
The principle of in situ peptide array synthesis and subsequent antibody binding analysis. A, digital micromirrors individually activate square features on the array by reflecting light on the photo-labile protecting groups of the previously incorporated amino acids. Repeated cycles of selective activation, addition of amino acids, and removal of excess amino acids enables parallel synthesis of peptides with unique sequences. B, schematic picture of incubation of the peptide array with the primary antibody and fluorophore-labeled secondary antibody. C, a scan image of a part of a planar ultra-dense peptide array in which the bright spots correspond to peptide features bound by antibodies.
Epitope Mapping of Antibodies Using Focused-content Peptide Microarrays
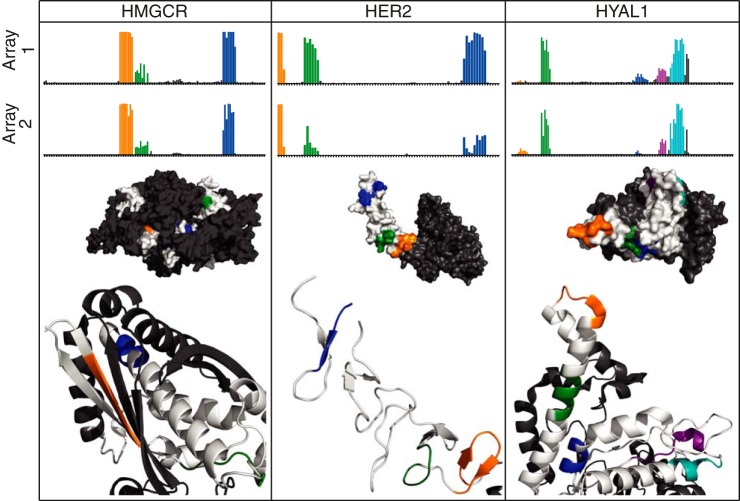
The study was initiated by the analysis of three polyclonal antibodies toward recombinant fragments of the human proteins HMGCR, HER2, and HYAL1. All targets had a previously determined three-dimensional structure. In this case, focused-content planar peptide arrays were designed with overlapping 12-mer peptides, covering the sequences of the protein fragments used for antibody generation, with a lateral shift of a single amino acid residue. The analysis was performed as technical replicates on two separate microarrays. The two technical replicates for the target HMGCR showed almost identical results with three distinct epitopes, two major epitopes (color-coded orange and blue in Fig. 2) and a minor epitope (green) close to the most N-terminal of the major epitopes. The three-dimensional structure of the native target showed that the three epitopes were parts of different structural elements, one α-helix, one β-pleated sheet, and one loop structure. The epitope mapping of the antibody toward the human epidermal growth factor receptor showed three distinct epitopes. The three-dimensional model of the native target showed that the epitopes were part of β-pleated sheets or loops. For the HYAL1 target, two major epitopes (green and cyan) were found using the microarray. The three-dimensional model of the native target showed that all five epitopes consisted of α-helical elements, although two of them also contained residues from an adjacent loop region.
Fig. 2.
Epitope mapping of polyclonal antibodies toward three protein targets. Antibodies toward three targets (HMGCR, HER2, and HYAL1) were analyzed using planar peptide arrays (arrays 1 and 2) with synthetic 12-mer peptides covering the corresponding antigen sequences. Each bar on the x-axis corresponds to one of the overlapping peptides required to cover the antigen, and the height shows the relative antibody binding. Below, three-dimensional structures of the three protein targets with mapped epitopes on their molecular surface together with a zoomed view of the secondary structural features of the epitopes. The protein fragments used for antibody generation are shown in white, and the epitopes identified on the planar peptide arrays are shown in colors (yellow, green, blue, purple, and cyan) corresponding to the highlighted epitopes in the plots above.
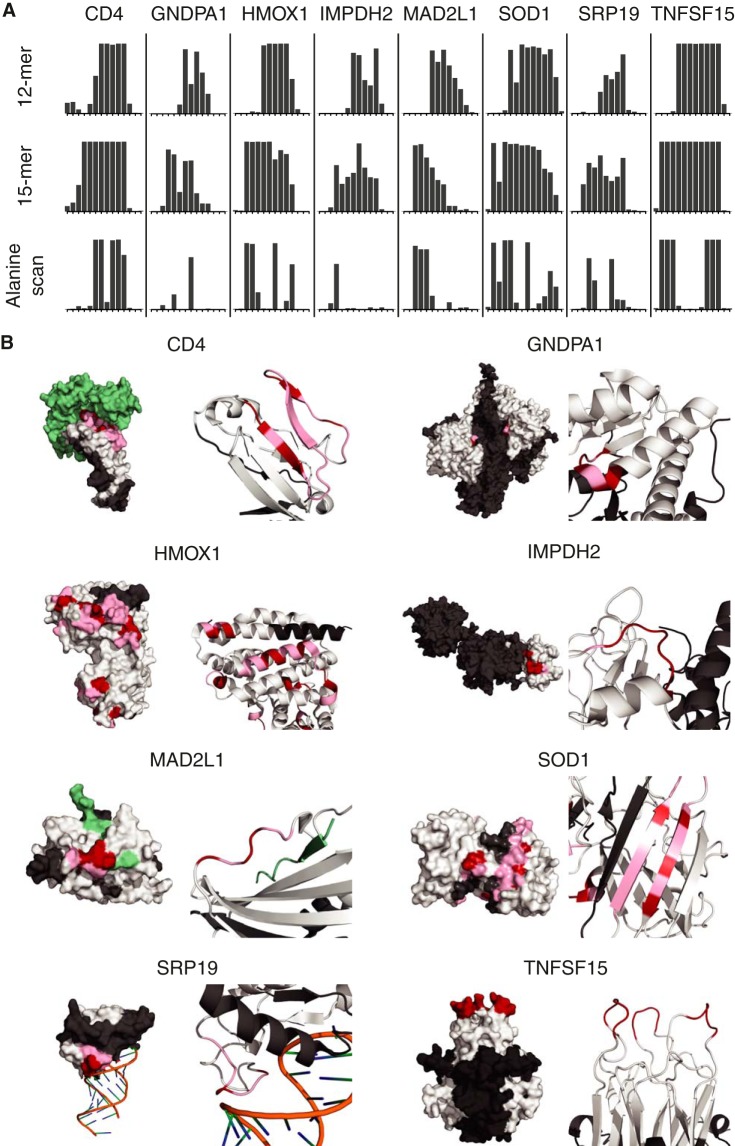
Comparative Epitope Mapping of Antibodies Using a Focused-content Microarray with 12- and 15-mer Peptides and Alanine Substitutions
Eight polyclonal antibodies toward additional human protein targets with known three-dimensional structures were studied using both 12-mer and 15-mer peptides to analyze the differences using different peptide lengths. In addition, an alanine substitution was introduced into the middle position of the 15-mer peptides to allow a precise contribution for binding of the middle residue in the sequence, similar to the alanine scan as described earlier (17). Planar peptide arrays covering the complete antigen sequence of the eight target sequences were designed and produced, and the results for one of the epitopes for each target are shown in Fig. 3A and supplemental Table S1. No differences between the 12-mer and the 15-mer peptide scans were observed for any of the eight targets, supporting similar results from Buus et al. (19) based on peptide scans using peptides of various lengths. By substituting the middle amino acid of each 15-mer with an alanine, we achieved even more detailed mapping. In the case of TNFSF15, the 15-mer mapping was interpreted as a minimal epitope five amino acids long, and the alanine substitutions not only confirmed the length, but also showed that all five amino acids were crucial for binding. For some epitopes (e.g. IMPDH2), almost all of the amino acids of the epitope are needed for binding, whereas for other epitopes (e.g. CD4) only a few of the amino acids are absolutely crucial.
Fig. 3.
Epitope mapping and alanine scanning of antibodies toward eight human protein targets. A, major epitope regions of antibodies epitope mapped on two planar arrays with overlapping peptides, 12 and 15 amino acids long, respectively, with a 1-amino-acid lateral shift covering the antigen sequences. Each bar on the x-axis corresponds to one of the overlapping peptides, and the height shows the relative antibody binding. Below, binding profiles of the antibodies to peptides corresponding to the 15-mer peptides, but with the middle amino acid substituted by an alanine (“alanine scan”). B, three-dimensional structures of the eight protein targets showing the molecular surface and the secondary structure of the epitope regions. The part of the protein used for antibody generation is shown in white, ligands are in green, the epitopes are highlighted in pink, and the residues essential for antibody binding, identified by alanine substitutions, are shown in red.
In Fig. 3B, the three-dimensional models of the native targets are shown with the consensus epitopes indicated, with the residues identified as most important by the alanine scans highlighted. For two of the targets (SOD1 and CD4), the epitopes were found in β-pleated sheets, whereas for two other targets (GNDPA1 and HMOX1), the epitopes consisted of α-helical elements. For the last four targets (IMPHD2, MAD2DL1, SRP19, and TNFSF15), the epitopes were confined to loop structures. Note that TNFSF15 consists of three identical subunits, and the epitope therefore occurs three times in the native protein. In all cases, the epitopes were situated on the surface of the native structure.
Proteome-wide Epitope Mapping of Three Polyclonal Antibodies
Ultra-dense peptide arrays with 2.1 million 12-mer peptides covering all human proteins were used for on- and off-target binding analysis of three polyclonal antibodies generated within the Human Protein Atlas project. In Fig. 4 the peptides with the highest binding intensities for each antibody are presented as amino acid sequences, and their binding intensity is relative to the peptide with the highest intensity on the corresponding array. For the antibody HPA003239 toward PCMT1 (Fig. 4A), peptides corresponding to three linear epitopes showed substantial binding, but high intensities were also seen for non-PCMT1 peptides. For the peptide showing the most binding, other peptides containing sequences similar to GAAAP (red) showed less binding intensity, but for the other two epitopes, GRLI (yellow) and VGSGS (purple), some off-target peptides actually had higher binding intensities. The proteome-wide epitope mapping of HPA005157 toward the protein TYK2 (Fig. 4B) revealed two dominating epitopes, VTGT (red) and APRF (yellow). Again, off-target peptides containing the epitope sequences were also bound by the antibody, and in the case of the APRF epitope, some even showed higher intensity than the corresponding TYK2 peptide. Interestingly, all but one bound nontarget peptide with the VTGT pattern had the epitope sequence C-terminally located on the peptide. A possible explanation is that the six-amino hexanoic acid used as a spacer between the C terminus and the microarray surface is very similar to the two consecutive glycines found after VTGT in the TYK2 sequence. Although most bound peptides showed sequence similarity to the TYK2 epitopes, two peptides with no apparent sequence similarity (black) also showed substantial binding. The antibody HPA020324 toward RRP1B showed very extensive binding to many nontarget peptides, with only a few key amino acids being similar to the RRP1B sequence (Fig. 4C). Many peptides share the PF-K pattern (red) in which many different combinations of amino acids are allowed in between. The second peptide (yellow) shared a TGPS-F pattern with the RRP1B sequence, but the RRP1B peptides ILVSPTGPSRVA and GPSRVAFDPEQ did not show any binding, indicating the importance of the phenylalanine and the initial threonine for proper binding. In the third epitope, TFGL were the most important amino acids, with serine and isoleucine being able to substitute threonine and leucine, respectively. The antibody's fourth epitope specificity (cyan) seemed to be very promiscuous in its binding, with only two key residues, leucine and lysine, with a three-amino-acid space in between being required for a strong peptide interaction. The combination of several epitopes defined by only two or three amino acids makes this antibody exhibit a lot of off-target peptide binding.
Fig. 4.
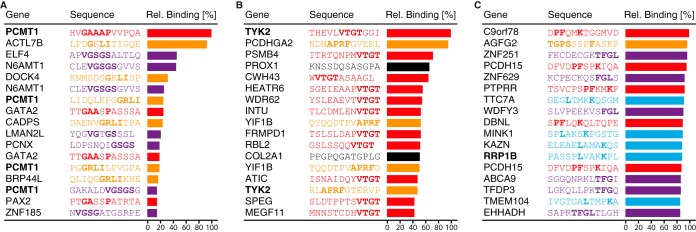
Proteome-wide off-target binding analysis of three polyclonal antibodies. Results from three polyclonal antibodies analyzed on ultra-dense peptide arrays with 12-mer peptides with a six-amino-acid lateral shift in total covering the entire human proteome. Gene origin, amino acid sequence, and relative binding intensity are shown for the peptides with the highest antibody binding for each array. A, peptides bound with high intensity by an anti-PCMT1 antibody revealed three epitopes, GAAAP (red), GRLI (yellow), and VGSGS (purple). B, an anti-TYK2 antibody shows binding to peptides containing the VTGT (red) or APRF (yellow) epitope sequences, but also to two peptides not sharing sequence similarity with the target protein (black). C, for an anti-RRP1B antibody, four epitope patterns were identified: PF-K (red), TGPS-F (yellow), TFGL (purple), and L-K (cyan). Many off-target peptides showed high antibody binding, and only one peptide originating form the RRP1B sequence was present among the top 17 peptides.
Proteome-wide Epitope Mapping of a PODXL Polyclonal Antibody
The extensive off-target binding displayed for the three antibodies prompted us to perform a more in-depth analysis of a polyclonal antibody in order to investigate on- and off-target binding using both peptides and protein fragments. We choose an antibody toward human PODXL, which is a sialomucin protein identified as an important component of glomerular podocytes in the human kidney. PODXL is a glycosylated membrane-bound protein with several isoforms with molecular weights predicted from the genome of 54 to 58 kDa. According to the literature (30), the major isoforms of this gene product exist as glycosylated products with higher molecular weights. We have shown that PODXL is differentially expressed in many human cancers (31–33), and a polyclonal antibody (HPA002110) has been generated by the Protein Atlas effort that functions well in immunohistochemistry (31). In order to investigate both on-target binding and off-target binding to other human proteins, the binding pattern of this antibody was analyzed using the proteome-wide peptide microarrays.
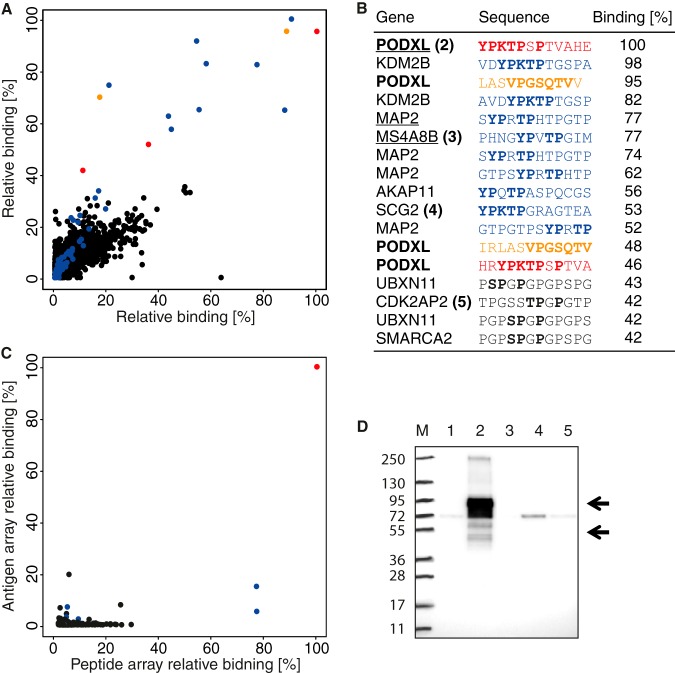
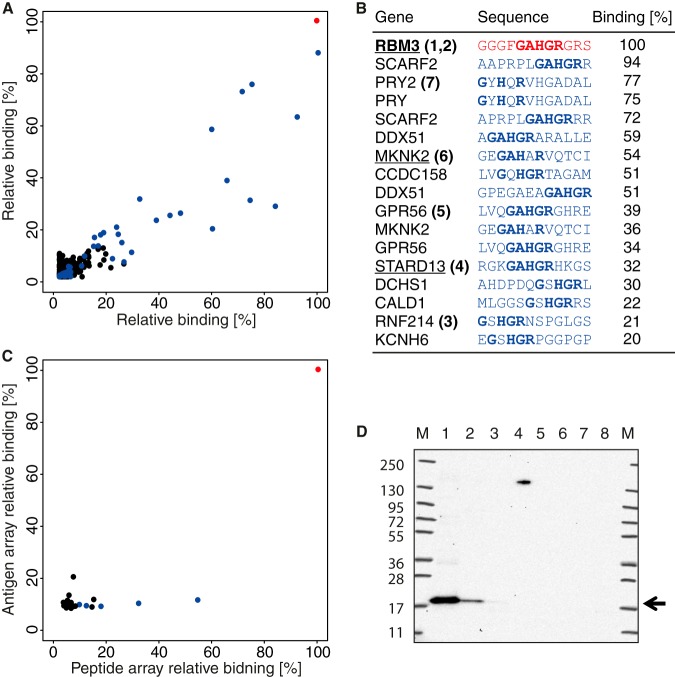
Epitope mapping was in this case performed using two separate technical replicates of the whole-proteome arrays, and the analysis of all 2.1 million peptides is shown in Fig. 5A, with the relative binding to each peptide in the two experiments plotted. Good reproducibility between the two whole-proteome arrays was obtained, and the results show that out of the 20 peptides with highest binding to the antibody, 4 were part of the target antigen sequence of PODXL, and 16 peptides were part of unrelated human proteins (Fig. 5B). The sequence of the PODXL peptides suggested that the antibody recognizes two distinct epitopes, indicated in orange and red, and that each epitope was present in two separate peptides on the whole-proteome array. Interestingly, all of the 16 off-target peptides shared residues present in one of the two PODXL epitopes (YPKTPSP), suggesting off-target binding to sequences similar to the target epitope. None of the highest cross-reactive peptides observed on the microarray seemed to involve the second VPGSQTV epitope.
Fig. 5.
Proteome-wide off-target binding analysis of a polyclonal antibody toward PODXL. A, binding analysis using two identical ultra-dense peptide arrays with 12-mer peptides with a six-amino-acid lateral shift in total covering the entire human proteome. Peptides containing the YPKTPSPS and VPGSQTV epitopes are represented by red and orange dots, respectively, and peptides containing a pattern of the most important amino acids of the first epitope, YP-TP, are in blue. B, table of the 17 peptides with the highest mean relative binding on the whole-proteome peptide arrays. Amino acid patterns similar to the PODXL epitopes are shown in bold, and the digits after the gene names refer to lanes in the Western blot. Underlined genes have corresponding protein fragments present on the antigen array in C. C, comparison of antibody binding to 12-mer peptides (x-axis) and protein fragments containing the corresponding peptide sequences (y-axis) where binding is shown relative to the PODXL peptide and protein fragment showing the most binding (red). Peptides/antigens containing the YP-TP epitope pattern are shown in blue. D, off-target binding analysis of the polyclonal PODXL antibody using Western blot with a panel of HEK293 protein overexpression cell lysates corresponding to peptides bound on the whole-proteome array, marker (M), negative control (1), PODXL (2), MS4A8B (3), SCG2 (4), and CDK2AP2 (5).
To investigate the binding characteristics further, the Human Protein Atlas resource of more than 40,000 human recombinant cDNA clones (1) was explored for protein fragments (PrESTs) containing the off-target peptide sequences and shown to be binding to the antibody in the proteome-wide microarray. In all, 249 PrESTs were found in the collection. These cDNA clones were expressed recombinantly in E. coli, and the corresponding protein fragments were purified and spotted on a microarray for binding analysis. In Fig. 5C, the comparison between the relative binding of the peptide and the respective recombinant protein fragments is shown, and details of sequences and relative binding intensities are presented in supplemental Table S2. The results demonstrate low binding of the antibody to protein fragments other than its antigen, in contrast to the binding to corresponding peptides seen earlier. This lack of binding to the protein fragment was further supported by a Western blot analysis (Fig. 5D) using overexpression lysates of three of the protein targets (MS4A8B, SCG2, and CDK2AP2) containing the epitope sequence where the antibody bound the corresponding peptide on the microarray. Note that PODXL has been shown to have multiple isoforms and contains glycosylated residues, and therefore should give rise to several bands in the range from 54 to 95 kDa. Bands of expected sizes were detected in the cell lysate containing overexpressed PODXL, whereas for the other lysates only bands of sizes not corresponding to the recombinant off-target proteins were detected.
Proteome-wide Epitope Mapping of a Monoclonal Antibody toward Human RBM3
The proteome-wide analysis of binding was subsequently extended to a monoclonal antibody. RBM3 is a glycine-rich RNA- and DNA-binding protein and is one of the first proteins to be synthesized in a cold shock response (34). RBM3 is up-regulated in several different cancer forms, and the expression of RBM3 in the nucleus is a positive prognostic marker in, for example, breast cancer (35), ovarian cancer (36), malignant melanoma (37), and colorectal cancer (38). A monoclonal antibody (clone 6F11) was recently generated and shown to function well in Western blotting (38), and we therefore decided to analyze this monoclonal antibody for target and off-target binding using whole-proteome array analysis.
The epitope analysis of the monoclonal antibody was performed on two separate whole-proteome arrays, and the results of the technical replicates are summarized in Fig. 6A. The peptide with the highest relative binding was shown to correspond to a sequence of the target antigen, including a sequence GAHGR (Fig. 6B). Of the other 19 of the 20 highest binding peptides, none corresponded to the target protein. However, all 19 had parts of the GAHGR sequence included in their respective peptides, explaining the distinct cross-reactivity.
Fig. 6.
Proteome-wide off-target binding analysis of a monoclonal antibody toward RBM3. A, binding analysis using two identical high-density peptide arrays with 12-mer peptides with a six-amino-acid lateral shift in total covering the entire human proteome. The peptide containing the GFGAGHR epitope is show in red, and peptides containing a pattern of the most important amino acids of the epitope, G-H-R, are shown in blue. B, table of the 17 peptides with the highest mean relative binding on the whole-proteome peptide arrays. Amino acid patterns similar to the epitopes are shown in bold, and the digits after the gene names correspond to lanes in the Western blot. C, comparison of antibody binding to 12-mer peptides (x-axis) and protein fragments containing the corresponding peptide sequences (y-axis) where binding is shown relative to the RBM3 peptide and protein fragment showing the most binding (red). Peptides/antigens containing the G-H-R epitope pattern are shown in blue. D, off-target binding analysis of the monoclonal RBM3 antibody using Western blot with a panel of cell lines and HEK293 overexpression lysates of proteins corresponding to peptides bound on the whole-proteome array, marker (M), U-251 MG (1), R-T4 (2), RNF214 (3), STARD13 (4), GPR56 (5), MKNK2 (6), PRY2 (7), and negative control (8).
The Human Protein Atlas resource was again explored for protein fragments (PrESTs) containing the off-target peptide sequences, and a total of 25 PrESTs were found in the collection of encoded protein fragments encompassing the peptides found in the epitope mapping analysis. The protein fragments were expressed, purified, and spotted on a microarray for binding analysis. In Fig. 6C, a comparison between the relative binding to the peptide and the respective recombinant protein fragments is plotted with the relative binding intensities listed in supplemental Table S3. Similar to the polyclonal antibody described above, the results demonstrate low binding of the antibody to the protein fragment, in contrast to the binding to the peptide. This lack of binding to the protein fragment was supported by a Western blot analysis (Fig. 6D) using overexpression lysates of five of the protein targets (RNF214, STARD13, GPR56, MKNK2, and PRY2) known to contain the epitope and where the antibody binds to the peptide on the microarray. The Western blot revealed a band of the expected size (17 kDa) for two lysates from cell lines expressing RBM3 (U-251 MG and R-T4), but no bands of the expected size could be found for the other lysates, except for the lysate with overexpressed STARD13. A band of the expected size for STARD13 (125 kDa) was observed, suggesting that the GAHGR epitope of this protein is also recognized by the RBM3 monoclonal antibody.
Amino Acid Substitution Scans for Detailed Epitope Mapping
We decided to precisely map the residues involved in binding for the two antibodies analyzed above. For the polyclonal antibody toward PODXL, epitope-specific fractions of the polyclonal antibody were generated as described elsewhere (39). The two peptides corresponding to the main epitopes were synthesized and used as ligands in affinity chromatography, and epitope-specific antibodies were recovered from the polyclonal mix. These fractions were subsequently analyzed on a focused-content array containing 12-mer peptides corresponding to the previously identified epitopes and variants of these in which every amino acid was substituted for all of the other 19 amino acids. The results of the precise mappings of these two epitopes are shown in Fig. 7. The full amino acid substitution scan for the first PODXL epitope demonstrated binding to the sequence YPKTPSP, with the serine residue at position 6 contributing least to binding. Note that the histidine and arginine in positions 2 and 3 in THRYPKTPSPTV actually contributed negatively to the binding, as replacement of these amino acids with other amino acids in most cases yielded better binding. For the second epitope, the consensus sequence was found to be VPGSQTV, with the two flanking valine residues at positions 11 and 12 of the peptide LASVPGSQTVVV showing slightly negative contributions to binding. Thus, the precise mappings suggest that the numbers of interacting amino acid residues for the two epitopes of the PODXL polyclonal antibody are six and seven, respectively.
Fig. 7.
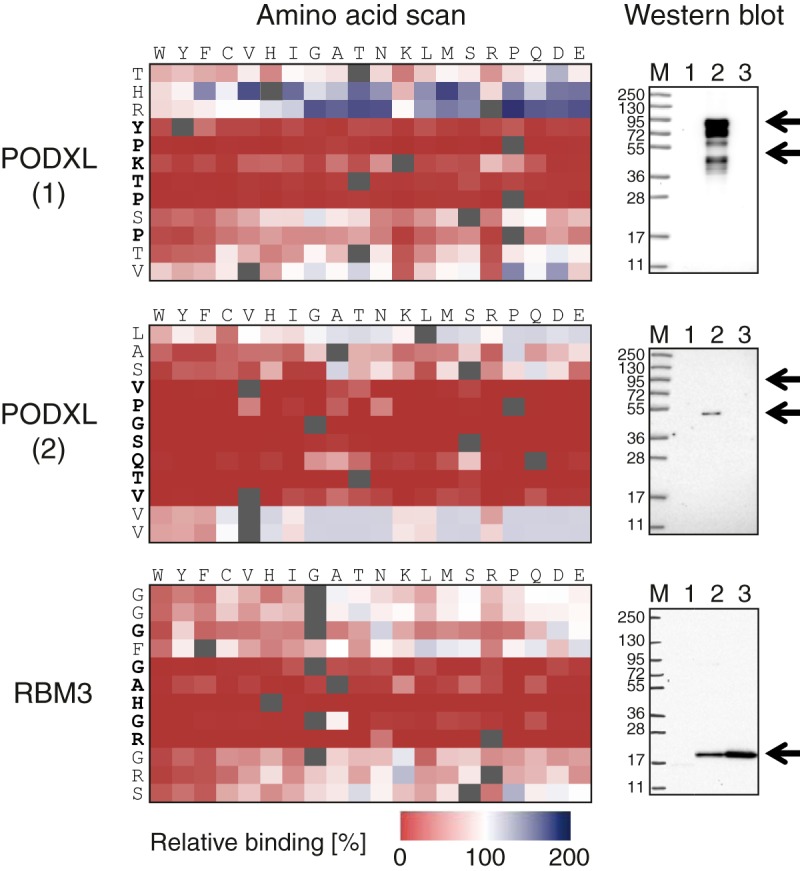
Full amino acid substitution scan and Western blot analysis of antibodies toward the human proteins PODXL and RBM3. Amino acid scans of two epitopes for the polyclonal PODXL antibody and a single epitope for the monoclonal RBM3 antibody, where each tile of the matrices corresponds to the substitution of one amino acid of the 12-mer peptides (rows) to one of the other 19 amino acids (columns). The change in antibody binding of each amino acid substitution relative to the binding of the original peptide is indicated by the color of the tiles, with gradients from white to red and white to green showing decreasing and increasing binding ability, respectively. Western blots using two epitope-specific antibody fractions isolated from the polyclonal PODXL antibody. Lane M: marker; Lane 1: HEK293; Lane 2: PODXL overexpression lysate; Lane 3: Caco-2. Western blot using the monoclonal antibody toward RBM3. Lane M: marker; Lane 1: HEK293; Lane 2: RT4; Lane 3: U251.
A Western blot analysis of the two epitope-specific antibodies (Fig. 7) showed a single band with a molecular weight of about 55 kDa for the antibodies toward the VPGSQTV epitope, whereas several bands of higher molecular weight were shown for the antibodies toward the other epitope. The latter is expected, as it has been suggested that PODXL has several glycosylated isoforms. The results therefore suggest that one of the epitopes (VPGSQTV) is only exposed on the non-glycosylated isoform of PODXL (lower arrow in Western blot), whereas the other epitope (YPKTPSP) is accessible also by the glycosylated isoforms of PODXL. This demonstrates the usefulness of epitope mapping coupled with functional analysis to explore the functionality of various antibodies binding to different regions of the target protein.
A similar detailed mapping was performed for the monoclonal antibody toward RBM3. The result seen in Fig. 7 shows that the sequence GAHGR contained the most important residues for binding of this monoclonal antibody, supporting the data from the whole-proteome analysis, which showed that all cross-reacting peptides from unrelated proteins contained parts of this sequence.
DISCUSSION
In this paper, we show the benefits of accessing the complete human “epitome” through high-density microarrays with in situ synthesized peptides. Target and off-target binding can be studied on a proteome-wide scale to investigate the cross-reactivity of both polyclonal and monoclonal antibodies. The fact that several millions of peptides can be synthesized in parallel makes it possible to cover all human protein-coding regions with overlapping peptides on a single microarray. We have used peptide arrays based on 2.1 million 12-mer peptides, each overlapping with six amino acids, to cover the human proteome, but it is not unlikely that higher density slides could be designed, allowing a shorter window between the overlapping peptides and giving an even more detailed binding analysis.
In this study we have also designed focused-content microarrays with peptides containing lateral shifts of only one amino acid, giving us a higher mapping resolution for the target antigen. The results are easily interpreted thanks to the single-residue lateral shift where the overlap of the bound peptides gives the minimal epitope required for binding. In addition, other focused-content arrays were designed with amino acid substitutions across all residues involved in binding, allowing the measurement of the contribution of the individual amino acids to antibody binding.
The polyclonal antibodies analyzed in this study were generated in a standardized manner within the framework of the Human Protein Atlas program (1), involving immunization of animals with recombinant proteins selected for their low sequence identity to other human proteins. The results for all the analyzed antibodies, covering a total of 13 human protein targets, support earlier suggestions (15) that the polyclonal antibodies are more “oligoclonal” than polyclonal. Thus, a large portion of the antigen is “epitope silent,” and the B-cell immunodominant regions consist of few epitopes. This supports earlier results from suspension bead arrays and bacterial surface displays (15, 40) suggesting that an immunization scheme based on recombinant protein fragments generates only two to three distinct linear epitopes per 100 amino acids in average.
The epitopes found here were all relatively short, confined to five to seven residues, supporting the findings of Sivalingam and Shepherd (8), who hypothesized, based on extensive literature studies, that antibodies toward B-cell epitopes need to detect only a single patch of key binding residues. The limited number of binding residues on the average epitope explains the extensive number of cross-reactive peptides across the proteome with the off-target peptides comprising subelements of the cognate epitope sequence of the target antigen. However, the subsequent analysis using recombinant protein fragments or full-length protein lysates demonstrated that the cross-reactive epitopes were not recognized when displayed in the context of a recombinant protein. Thus, the binding specificities for these linear epitopes depend on both the specific amino acid residues and their display in three-dimensional space. A likely explanation for this is that the peptides displayed on a microarray can adapt to almost any conformation (“induced fit”) upon binding to the antibody. This allows the off-target peptides with sequences similar to the epitope to interact with the antibody, even though the same peptide when presented as part of a protein fragment or full-length protein is not displayed in the correct conformation for binding.
These results supports the hypothesis presented by Lerner et al. (41) that synthesized peptides exist only a fraction of their time in their native conformation, limiting the likelihood of eliciting successful antibody reactions with the native protein when performing immunizations with peptides. This has implications for the generation of antibodies when the generated antibody not only needs to recognize a peptide from trypsin cleavage, but also needs to recognize the native protein in plasma, cells, or tissues. If one uses synthetic peptides as antigens, it is not unlikely that the generated antibodies might be functional only in applications with denatured proteins. Therefore, if the aim is not only to recognize trypsin-cleaved peptides, it might be desirable to design the synthetic peptides to an intrinsically unstructured part of the target protein (42). In this respect, it is interesting to note that antibodies toward synthetic peptides often are generated toward N- or C-terminal regions of the target protein with a relatively high likelihood of being nonstructured (43).
For immuno-proteomics, the results presented here also have implications for the use of antibodies for the capture of peptides. The fact that immunization using a recombinant protein fragment (PrEST) yielded several distinct linear epitopes involving relatively few amino acids suggests that the antibodies generated in this manner, primarily to be used for bioimaging of the target protein, could be promising reagents for affinity capture of peptides generated by trypsin digestion.
An interesting application for the whole-proteome peptide arrays would be the detection of autoantibodies in serum or plasma samples from patients suffering from autoimmune diseases, as a way of identifying new possible biomarkers for diagnostics. An important lesson from the work presented here is that peptides identified using the proteome-wide approach should subsequently be validated by binding analysis toward the corresponding recombinant proteins. The existence of large repositories of human genes and gene fragments in expression vectors (44, 45) will facilitate such validation schemes, and a convenient path for systematic autoimmune studies can thus be envisioned based on proteome-wide peptide arrays for screening followed by recombinant protein arrays for validation.
In summary, we have described a new method for epitope mapping and cross-reactivity analysis of antibodies using proteome-wide peptide arrays. The analysis using both monoclonal and polyclonal antibodies revealed that in addition to the expected distinct binding to sequences corresponding to the cognate target, significant signals were frequently observed to peptides comprising sub-elements of the cognate epitope sequence. However, subsequent analysis using recombinant proteins suggested that these linear epitopes have a strict conformational component, giving us new insights regarding the nature of antibody–antigen binding.
Supplementary Material
Acknowledgments
We acknowledge the entire staff of the Human Protein Atlas program and the Science for Life Laboratory for valuable contributions.
Footnotes
Author contributions: B.F., B.B., K.S., J.B., T.J.A., T.A.R., P.N., J.R., and M.U. designed research; B.F., B.B., F.J., P.N., and J.R. performed research; K.S., J.B., T.J.A., and T.A.R. contributed new reagents or analytic tools; B.F., B.B., F.J., P.N., E.P.H., J.R., and M.U. analyzed data; B.F., B.B., K.S., J.B., P.N., E.P.H., J.R., and M.U. wrote the paper.
* Funding was provided by the Knut and Alice Wallenberg Foundation, the ProNova center through a grant from VINNOVA, and PepChipOmics, a 7th Framework grant by the European Directorate.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PODXL
- podocalyxin-like protein
- RBM3
- RNA-binding motif protein 3
- PrEST
- protein epitope signature tag
- HEK293
- human embryonic kidney cell line.
REFERENCES
- 1. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Bjorling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 2. Stadler C., Rexhepaj E., Singan V. R., Murphy R. F., Pepperkok R., Uhlen M., Simpson J. C., Lundberg E. (2013) Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods 10, 315–323 [DOI] [PubMed] [Google Scholar]
- 3. Gillette M. A., Carr S. A. (2013) Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 10, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 5. Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C., Corthals G. L., Costello C. E., Deutsch E. W., Domon B., Hancock W., He F., Hochstrasser D., Marko-Varga G., Salekdeh G. H., Sechi S., Snyder M., Srivastava S., Uhlen M., Hu C. H., Yamamoto T., Paik Y. K., Omenn G. S. (2011) The human proteome project: current state and future direction. Mol. Cell. Proteomics 10, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Irving M. B., Pan O., Scott J. K. (2001) Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr. Opin. Chem. Biol. 5, 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sivalingam G. N., Shepherd A. J. (2012) An analysis of B-cell epitope discontinuity. Mol. Immunol. 51, 304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baerga-Ortiz A., Hughes C. A., Mandell J. G., Komives E. A. (2002) Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 11, 1300–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232 [Google Scholar]
- 11. Geysen H. M., Meloen R. H., Barteling S. J. (1984) Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. U.S.A. 81, 3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christmann A., Wentzel A., Meyer C., Meyers G., Kolmar H. (2001) Epitope mapping and affinity purification of monospecific antibodies by Escherichia coli cell surface display of gene-derived random peptide libraries. J. Immunol. Methods 257, 163–173 [DOI] [PubMed] [Google Scholar]
- 13. Cochran J. R., Kim Y. S., Olsen M. J., Bhandari R., Wittrup K. D. (2004) Domain-level antibody epitope mapping through yeast surface display of epidermal growth factor receptor fragments. J. Immunol. Methods 287, 147–158 [DOI] [PubMed] [Google Scholar]
- 14. Petersen G., Song D., Hügle-Dörr B., Oldenburg I., Bautz E. K. F. (1995) Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol. Gen. Genet. 249, 425–431 [DOI] [PubMed] [Google Scholar]
- 15. Rockberg J., Lofblom J., Hjelm B., Uhlen M., Stahl S. (2008) Epitope mapping of antibodies using bacterial surface display. Nat. Methods 5, 1039–1045 [DOI] [PubMed] [Google Scholar]
- 16. Larman H. B., Zhao Z., Laserson U., Li M. Z., Ciccia A., Gakidis M. A., Church G. M., Kesari S., Leproust E. M., Solimini N. L., Elledge S. J. (2011) Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 29, 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham B. C., Wells J. A. (1989) High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 18. Maier R. H., Maier C. J., Rid R., Hintner H., Bauer J. W., Onder K. (2010) Epitope mapping of antibodies using a cell array-based polypeptide library. J. Biomol. Screen. 15, 418–426 [DOI] [PubMed] [Google Scholar]
- 19. Buus S., Rockberg J., Forsström B., Nilsson P., Uhlen M., Schafer-Nielsen C. (2012) High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol. Cell. Proteomics 11, 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. (1991) Light-directed, spatially addressable parallel chemical synthesis. Science 251, 767–773 [DOI] [PubMed] [Google Scholar]
- 21. Pellois J. P., Zhou X., Srivannavit O., Zhou T., Gulari E., Gao X. (2002) Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 20, 922–926 [DOI] [PubMed] [Google Scholar]
- 22. Pruitt K. D., Harrow J., Harte R. A., Wallin C., Diekhans M., Maglott D. R., Searle S., Farrell C. M., Loveland J. E., Ruef B. J., Hart E., Suner M.-M., Landrum M. J., Aken B., Ayling S., Baertsch R., Fernandez-Banet J., Cherry J. L., Curwen V., DiCuccio M., Kellis M., Lee J., Lin M. F., Schuster M., Shkeda A., Amid C., Brown G., Dukhanina O., Frankish A., Hart J., Maidak B. L., Mudge J., Murphy M. R., Murphy T., Rajan J., Rajput B., Riddick L. D., Snow C., Steward C., Webb D., Weber J. A., Wilming L., Wu W., Birney E., Haussler D., Hubbard T., Ostell J., Durbin R., Lipman D. (2009) The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19, 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh-Gasson S., Green R. D., Yue Y., Nelson C., Blattner F., Sussman M. R., Cerrina F. (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17, 974–978 [DOI] [PubMed] [Google Scholar]
- 24. Northen T. R., Brune D. C., Woodbury N. W. (2006) Synthesis and characterization of peptide grafted porous polymer microstructures. Biomacromolecules 7, 750–754 [DOI] [PubMed] [Google Scholar]
- 25. Shin D. S., Lee K. N., Yoo B. W., Kim J., Kim M., Kim Y. K., Lee Y. S. (2010) Automated maskless photolithography system for peptide microarray synthesis on a chip. J. Comb. Chem. 12, 463–471 [DOI] [PubMed] [Google Scholar]
- 26. Bhushan K. R., DeLisi C., Laursen R. A. (2003) Synthesis of photolabile 2-(2-nitrophenyl)propyloxycarbonyl protected amino acids. Tetrahedron Lett. 44, 8585–8588 [Google Scholar]
- 27. Berglund L., Bjorling E., Jonasson K., Rockberg J., Fagerberg L., Al-Khalili Szigyarto C., Sivertsson A., Uhlen M. (2008) A whole-genome bioinformatics approach to selection of antigens for systematic antibody generation. Proteomics 8, 2832–2839 [DOI] [PubMed] [Google Scholar]
- 28. Agaton C., Falk R., Hoiden Guthenberg I., Gostring L., Uhlen M., Hober S. (2004) Selective enrichment of monospecific polyclonal antibodies for antibody-based proteomics efforts. J. Chromatogr. A 1043, 33–40 [DOI] [PubMed] [Google Scholar]
- 29. Ehlen A., Brennan D. J., Nodin B., O'Connor D. P., Eberhard J., Alvarado-Kristensson M., Jeffrey I. B., Manjer J., Brandstedt J., Uhlen M., Ponten F., Jirstrom K. (2010) Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J. Transl. Med. 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriguez R. B., Butta N., Larrucea S., Alonso S., Parrilla R. (2006) Production and characterization of murine monoclonal antibodies against human podocalyxin. Tissue Antigens 68, 407–417 [DOI] [PubMed] [Google Scholar]
- 31. Larsson A., Johansson M. E., Wangefjord S., Gaber A., Nodin B., Kucharzewska P., Welinder C., Belting M., Eberhard J., Johnsson A., Uhlen M., Jirstrom K. (2011) Overexpression of podocalyxin-like protein is an independent factor of poor prognosis in colorectal cancer. Br. J. Cancer 105, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kershaw D. B., Thomas P. E., Wharram B. L., Goyal M., Wiggins J. E., Whiteside C. I., Wiggins R. C. (1995) Molecular cloning, expression, and characterization of podocalyxin-like protein 1 from rabbit as a transmembrane protein of glomerular podocytes and vascular endothelium. J. Biol. Chem. 270, 29439–29446 [DOI] [PubMed] [Google Scholar]
- 33. Casey G., Neville P. J., Liu X., Plummer S. J., Cicek M. S., Krumroy L. M., Curran A. P., McGreevy M. R., Catalona W. J., Klein E. A., Witte J. S. (2006) Podocalyxin variants and risk of prostate cancer and tumor aggressiveness. Hum. Mol. Genet. 15, 735–741 [DOI] [PubMed] [Google Scholar]
- 34. Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J. H., Itoh K., Matsuda T., Fujita J. (1997) Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 236, 804–807 [DOI] [PubMed] [Google Scholar]
- 35. Jogi A., Brennan D. J., Ryden L., Magnusson K., Ferno M., Stal O., Borgquist S., Uhlen M., Landberg G., Pahlman S., Ponten F., Jirstrom K. (2009) Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod. Pathol. 22, 1564–1574 [DOI] [PubMed] [Google Scholar]
- 36. Ehlen O., Nodin B., Rexhepaj E., Brandstedt J., Uhlen M., Alvarado-Kristensson M., Ponten F., Brennan D. J., Jirstrom K. (2011) RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl. Oncol. 4, 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jonsson L., Bergman J., Nodin B., Manjer J., Ponten F., Uhlen M., Jirstrom K. (2011) Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: an analysis of 215 cases from the Malmo Diet and Cancer Study. J. Transl. Med. 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hjelm B., Brennan D. J., Zendehrokh N., Eberhard J., Nodin B., Gaber A., Pontén F., Johannesson H., Smaragdi K., Frantz C., Hober S., Johnson L. B., Påhlman S., Jirström K., Uhlen M. (2011) High nuclear RBM3 expression is associated with an improved prognosis in colorectal cancer. Proteomics Clin. Appl. 5, 624–635 [DOI] [PubMed] [Google Scholar]
- 39. Hjelm B., Forsström B., Igel U., Johannesson H., Stadler C., Lundberg E., Ponten F., Sjöberg A., Rockberg J., Schwenk J. M., Nilsson P., Johansson C., Uhlén M. (2011) Generation of monospecific antibodies based on affinity capture of polyclonal antibodies. Protein Sci. 20, 1824–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hjelm B., Fernandez C. D., Lofblom J., Stahl S., Johannesson H., Rockberg J., Uhlen M. (2010) Exploring epitopes of antibodies toward the human tryptophanyl-tRNA synthetase. Nat. Biotechnol. 27, 129–137 [DOI] [PubMed] [Google Scholar]
- 41. Lerner R. A., Green N., Alexander H., Liu F. T., Sutcliffe J. G., Shinnick T. M. (1981) Chemically synthesized peptides predicted from the nucleotide sequence of the hepatitis B virus genome elicit antibodies reactive with the native envelope protein of Dane particles. Proc. Natl. Acad. Sci. U.S.A. 78, 3403–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 43. Trier N. H., Hansen P. R., Houen G. (2012) Production and characterization of peptide antibodies. Methods 56, 136–144 [DOI] [PubMed] [Google Scholar]
- 44. Team M. G. C. P., Temple G., Gerhard D. S., Rasooly R., Feingold E. A., Good P. J., Robinson C., Mandich A., Derge J. G., Lewis J., Shoaf D., Collins F. S., Jang W., Wagner L., Shenmen C. M., Misquitta L., Schaefer C. F., Buetow K. H., Bonner T. I., Yankie L., Ward M., Phan L., Astashyn A., Brown G., Farrell C., Hart J., Landrum M., Maidak B. L., Murphy M., Murphy T., Rajput B., Riddick L., Webb D., Weber J., Wu W., Pruitt K. D., Maglott D., Siepel A., Brejova B., Diekhans M., Harte R., Baertsch R., Kent J., Haussler D., Brent M., Langton L., Comstock C. L., Stevens M., Wei C., van Baren M. J., Salehi-Ashtiani K., Murray R. R., Ghamsari L., Mello E., Lin C., Pennacchio C., Schreiber K., Shapiro N., Marsh A., Pardes E., Moore T., Lebeau A., Muratet M., Simmons B., Kloske D., Sieja S., Hudson J., Sethupathy P., Brownstein M., Bhat N., Lazar J., Jacob H., Gruber C. E., Smith M. R., McPherson J., Garcia A. M., Gunaratne P. H., Wu J., Muzny D., Gibbs R. A., Young A. C., Bouffard G. G., Blakesley R. W., Mullikin J., Green E. D., Dickson M. C., Rodriguez A. C., Grimwood J., Schmutz J., Myers R. M., Hirst M., Zeng T., Tse K., Moksa M., Deng M., Ma K., Mah D., Pang J., Taylor G., Chuah E., Deng A., Fichter K., Go A., Lee S., Wang J., Griffith M., Morin R., Moore R. A., Mayo M., Munro S., Wagner S., Jones S. J., Holt R. A., Marra M. A., Lu S., Yang S., Hartigan J., Graf M., Wagner R., Letovksy S., Pulido J. C., Robison K., Esposito D., Hartley J., Wall V. E., Hopkins R. F., Ohara O., Wiemann S. (2009) The completion of the Mammalian Gene Collection (MGC). Genome Res. 19, 2324–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.