Abstract
The biological and clinical relevance of glycosylation is becoming increasingly recognized, leading to a growing interest in large-scale clinical and population-based studies. In the past few years, several methods for high-throughput analysis of glycans have been developed, but thorough validation and standardization of these methods is required before significant resources are invested in large-scale studies. In this study, we compared liquid chromatography, capillary gel electrophoresis, and two MS methods for quantitative profiling of N-glycosylation of IgG in the same data set of 1201 individuals. To evaluate the accuracy of the four methods we then performed analysis of association with genetic polymorphisms and age. Chromatographic methods with either fluorescent or MS-detection yielded slightly stronger associations than MS-only and multiplexed capillary gel electrophoresis, but at the expense of lower levels of throughput. Advantages and disadvantages of each method were identified, which should inform the selection of the most appropriate method in future studies.
Glycans are important structural and functional components of the majority of proteins, but because of their structural complexity and the absence of a direct genetic template our current understanding of the role of glycans in biological processes lags significantly behind the knowledge about proteins or DNA (1, 2). However, a recent comprehensive report endorsed by the US National Academies concluded that “glycans are directly involved in the pathophysiology of every major disease and that additional knowledge from glycoscience will be needed to realize the goals of personalized medicine” (3).
It is estimated that the glycome (defined as the complete set of all glycans) of a eukaryotic cell is composed of more than a million different glycosylated structures (1), which contain up to 10,000 structural glycan epitopes for interaction with antibodies, lectins, receptors, toxins, microbial adhesins, or enzymes (4). Our recent population-based studies indicated that the composition of the human plasma N-glycome varies significantly between individuals (5, 6). Because glycans have important structural and regulatory functions on numerous glycoproteins (7), the observed variability suggests that differences in glycosylation might contribute to a large part of the human phenotypic variability. Interestingly, when the N-glycome of isolated immunoglobulin G (IgG)1 was analyzed, it was found to be even more variable than the total plasma N-glycome (8), indicating that the combined analysis of all plasma glycans released from many different glycoproteins blurs signals of protein-specific regulation of glycosylation.
A number of studies have investigated the role of glycans in human disease, including autoimmune diseases and cancer (9, 10). However, most human glycan studies have been conducted with very small sample sizes. Given the complex causal pathways involved in pathophysiology of common complex disease, and thus the likely modest effect sizes associated with individual factors, the majority of these studies are very likely to be substantially underpowered. In the case of inflammatory bowel disease, only 20% of reported inflammatory bowel disease glycan associations were replicated in subsequent studies, suggesting that most are false positive findings and that there is publication bias favoring the publication of positive findings (11). This situation is similar to that which occurred in the field of genetic epidemiology in the past when many underpowered candidate gene studies were published and were later found to consist of mainly false positive findings (12, 13). It is essential, therefore, that robust and affordable methods for high-throughput analysis are developed so that adequately powered studies can be conducted and the publication of large numbers of small studies reporting false positive results (which could threaten the credibility of glycoscience) be avoided.
Rapid advances of technologies for high-throughput genome analysis in the past decade enabled large-scale genome-wide association studies (GWAS). GWAS has become a reliable tool for identification of associations between genetic polymorphisms and various human diseases and traits (14). Thousands of GWAS have been conducted in recent years, but these have not included the study of glycan traits until recently. The main reason was the absence of reliable tools for high-throughput quantitative analysis of glycans that could match the measurements of genomic, biochemical, and other traits in their cost, precision, and reproducibility. However, several promising high-throughput technologies for analysis of N-glycans were developed (8, 15–20) recently. Successful implementation of high-throughput analytical techniques for glycan analysis resulted in publication of four initial GWAS of the human glycome (21–24).
In this study, we compared ultra-performance liquid chromatography with fluorescence detection (UPLC-FLR), multiplex capillary gel electrophoresis with laser induced fluorescence detection (xCGE-LIF), matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), and liquid chromatography electrospray mass spectrometry (LC-ESI-MS) as tools for mid-to-high-throughput glycomics and glycoproteomics. We have analyzed IgG N-glycans by all four methods in 1201 individuals from European populations. The analysis of associations between glycans and ∼300,000 single-nucleotide genetic polymorphisms was performed and correlation between glycans and age was studied in all four data sets to identify the analytical method that shows the strongest potential to uncover biological mechanisms underlying protein glycosylation.
EXPERIMENTAL PROCEDURES
Study Participants
All research in this study involved adult human participants from the Croatian Adriatic islands of Vis and Korčula who were recruited within a larger genetic epidemiology program previously described (25). The study conforms to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the University of Split Medical School. All participants in this study have signed the appropriate informed consent. IgG was purified from the plasma of 1821 individuals using monolithic protein G 96-well plates at the Genos Glycoscience Laboratory in Zagreb. Aliquots of purified IgG were sent to the Leiden University Medical Center for MS analysis (MALDI-TOF-MS and LC-ESI-MS) of IgG glycopeptides and to the Max Planck Institute and glyXera in Magdeburg for xCGE-LIF analysis of IgG glycans. UPLC-FLR analysis was performed at Genos. Using all four methods, 1201 individuals were successfully analyzed.
Isolation of IgG
Immunoglobulin G was isolated from plasma by affinity chromatography using a 96-well protein G monolithic plate (BIA Separations, Ajdovščina, Slo). The protein G plate was first washed with 10 column volumes (CV) of ultrapure water and equilibrated with 10 CV of binding buffer (1× PBS, pH 7.4; Fisher Scientific, Pittsburgh, PA, USA). Plasma samples (50 μl) were diluted 10× with the binding buffer, applied to the plate and instantly washed five times with 5 CV of binding buffer to remove unbound proteins. IgGs were eluted from the protein G monoliths using 5 CV of 100 mm formic acid (FA; Fisher Scientific), pH 2.5, into a 96 deep well plate and immediately neutralized to pH 7.0 with 1 m ammonium bicarbonate (Fisher Scientific). After each sample application, the plate was regenerated with the following buffers: 10 CV of 10× PBS, followed by 10 CV of 0.1 m FA and afterward 10 CV of 1× PBS to re-equilibrate the monoliths. Each step of the isolation was done under vacuum (approx. 60 mmHg pressure reduction while applying the samples, 500 mmHg during elution and washing steps) using a manual set-up consisting of a multichannel pipette, a vacuum manifold (Beckman Coulter, La Brea, CA, USA), and a vacuum pump (Pall Life Sciences, Ann Arbor, MI, USA).
Hydrophilic Interaction Chromatography of IgG N-glycans-Sample Preparation and Analysis
Glycan Release and Labeling
Aliquots (1/5; 200 μl) of the protein G eluates were applied to a 96-well flat-bottomed microtiter plate, dried down in a vacuum concentrator and reduced by adding 2 μl of 5× sample buffer (125 μl of 0.5 m Tris (Sigma-Aldrich, St, Louis, MO, USA), pH 6.6, 200 μl of 10% SDS (Sigma-Aldrich), and 675 μl of water), 7 μl of water, and 1 μl of 0.5 m dithiothreitol (DTT; Sigma-Aldrich) and incubating at 65 °C for 15 min. Ultrapure water was used throughout. The samples were then alkylated by adding 1 μl of 100 mm iodoacetamide (IAA; Sigma-Aldrich) and incubated for 30 min in the dark at room temperature. Afterward, the samples were immobilized in a gel block by adding 22.5 μl of 30% (w/w) acrylamide/0.8% (w/v) bis-acrylamide stock solution (37.5:1, Protogel; Sigma-Aldrich), 11.25 μl of 1.5 m Tris, pH 8.8, 1 μl of 10% (w/v) SDS (Invitrogen, Carlsbad, CA, USA), 1 μl of 10% (w/v) ammonium peroxodisulphate (APS; Sigma-Aldrich), and 1 μl of N,N,N,N′-tetramethyl-ethylenediamine (TEMED; Invitrogen). The gel blocks were transferred to a Whatman protein precipitation plate and washed with 1 ml of acetonitrile with vortexing on a plate shaker for 10 min, followed by removal of the liquid on a vacuum manifold. The gel blocks were then washed twice with 1 ml of 20 mm sodium bicarbonate (NaHCO3; Sigma-Aldrich), pH 7.2, followed by 1 ml of acetonitrile (ACN; J.T.Baker, Phillipsburg, NJ, USA). N-glycans were released by adding 50 μl of 2.5 mU PNGase F (ProZyme, Leandro, CA, USA) in 20 mm NaHCO3, pH 7.2, to reswell the gel pieces. After 5 min another 50 μl of 20 mm NaHCO3 was added and the plates were subsequently sealed with adhesive film (USA Scientific, Ocala, FL, USA) and incubated overnight at 37 °C. The released N-glycans were collected into a 2-ml polypropylene 96-well plate (Waters, Milford, MA, USA) by washing the gel pieces with 3 × 200 μl of water, followed by 200 μl of ACN, 200 μl of water, and finally 200 μl of ACN. The released N-glycans were dried, redissolved in 20 μl of 1% FA, incubated at room temperature for 40 min, and dried again. N-glycans were labeled with 5 μl of 2-AB labeling solution (55 mg of anthranilamide, 66 mg of sodium cyanoborohydride, 330 μl of glacial acetic acid, and 770 μl of dimethyl sulfoxide (DMSO); all from Sigma-Aldrich), shaken for 5 min, incubated for 30 min at 65 °C, shaken again for 5 min, and further incubated for 90 min. Excess 2-AB was removed using solid-phase extraction with 1-cm square pieces of prewashed Whatman 3MM chromatography paper which was dried, folded into quarters and placed into a Whatman protein precipitation plate (prewashed with 200 μl of ACN followed by 200 μl of water). The 5 μl of 2-AB labeled IgG N-glycans were applied to the paper and left to dry and bind for 15 min. The excess 2-AB was washed off the paper by shaking with 1.6 ml of ACN for 15 min and then removing the ACN using a vacuum manifold; this step was repeated four times. The labeled N-glycans were eluted from the paper by shaking with 500 μl of water for 20 min and collected by vacuum into a 2-ml 96-well plate; this step was repeated two times. The eluted 2-AB IgG N-glycans were dried before resuspending in a known volume of water ready for analysis by UPLC-FLR.
Hydrophilic Interaction Chromatography
2-AB labeled IgG N-glycans were separated by hydrophilic interaction chromatography on a Waters Acquity UPLC instrument consisting of a quaternary solvent manager, sample manager and a FLR fluorescence detector set with excitation and emission wavelengths of 330 and 420 nm, respectively. The instrument was under the control of Empower 2 software, build 2145 (Waters). Labeled N-glycans were separated on a Waters BEH Glycan chromatography column, 100 × 2.1 mm i.d., 1.7 μm BEH particles, with 100 mm ammonium formate, pH 4.4, as solvent A and ACN as solvent B. A linear gradient of 75%-62% ACN was used at flow rate of 0.4 ml/min in a 20 min analytical run. Samples were maintained at 5 °C prior to injection, and the separation temperature was 60 °C. The system was calibrated using an external standard of hydrolyzed and 2-AB labeled glucose oligomers from which the retention times for the individual glycans were converted to glucose units (GU). Data processing was performed using an automatic processing method with a traditional integration algorithm after which each chromatogram was manually corrected to maintain the same intervals of integration for all the samples. The chromatograms obtained were all separated in the same manner into 24 peaks and the amount of glycans in each peak was expressed as % of total integrated area.
Mass Spectrometry (MALDI-TOF-MS and nanoLC-ESI-MS) of IgG N-glycopeptides - Sample Preparation and Analysis
Trypsin Digestion and Reverse-phase Solid-phase Extraction (RP-SPE)
Aliquots (1/20; 50 μl) of the protein G eluates were applied to 96-well polypropylene V-bottom microtiter plates. TPCK trypsin (Sigma-Aldrich) was first dissolved in ice-cold 20 mm acetic acid (Merck, Darmstadt, Germany) to a final concentration of 0.4 μg/μl after which it was further diluted to 0.02 μg/μl with ice-cold ultrapure water. To each sample 20 μl of the diluted trypsin was added followed by overnight incubation at 37 °C.
For reverse-phase desalting and purification of glycopeptides, 5 mg of Chromabond C18ec beads (Marcherey-Nagel, Düren, Germany) were applied to each well of an OF1100 96-well polypropylene filter plate with a 10 μm polyethylene frit (Orochem Technologies Inc., Lombard, IL, USA). The RP stationary phase was activated with 3 × 200 μl 80% ACN containing 0.1% trifluoroacetic acid (TFA; Fluka, Steinheim, Germany) and conditioned with 3 × 200 μl 0.1% TFA. The IgG digests were diluted 10× in 0.1% TFA, loaded onto the C18 beads, and washed with 3 × 200 μl 0.1% TFA. The entire procedure was performed on a vacuum manifold (< 3 mmHg). IgG glycopeptides were eluted into a V-bottom microtiter plate by centrifugation at 500 rpm with 90 μl of 18% ACN containing 0.1% TFA. Eluates were dried by vacuum centrifugation, reconstituted in 20 μl MQ water and stored at −20 °C until analysis by MS.
MALDI-TOF-MS
Purified and desalted tryptic IgG glycopeptides (3 μl) were spotted onto MTP 384 polished steel target plates (Bruker Daltonics, Bremen, Germany) and allowed to dry at room temperature. Subsequently 1 μl of 5 mg/ml 4-chloro-α-cyanocinnamic acid (Cl-CCA; 95% purity; Bionet Research, Camelford, Cornwall, UK) in 50% ACN was applied on top of each sample and allowed to dry. Glycopeptides were analyzed on an UltrafleX II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) operated in the negative-ion reflectron mode, because negative-ion mode has been found well-suited for the analysis of IgG glycopeptides and specifically for sialylated glycopeptides (26), while reflectron mode greatly improves the resolution and sensitivity of the analysis. Ions between m/z 1000 and 3800 were recorded. To allow homogeneous spot sampling a random walk laser movement with 50 laser shots per raster spot was applied and each IgG glycopeptide sum mass spectrum was generated by accumulation of 2000 laser shots. Mass spectra were internally calibrated using a list of known glycopeptides. Data processing and evaluation were performed with FlexAnalysis Software (Bruker Daltonics) and Microsoft Excel, respectively. Structural assignment of the detected glycoforms was performed on the basis of literature knowledge of IgG N-glycosylation (27–32). The data were baseline subtracted and the intensities (peak heights) of a defined set of 27 glycopeptides (16 glycoforms for IgG1 and 11 for IgG2&3) were automatically defined for each spectrum as described before (33). See supplementary Table S1 for a complete list of the assigned peptides and corresponding MS signals.
In Caucasian populations, IgG2 and IgG3 have identical peptide moieties (E293EQFNSTFR301) of their tryptic Fc glycopeptides and were, therefore, not distinguished by the profiling method (34). Relative intensities of IgG Fc glycopeptides were obtained by integrating and summing four isotopic peaks followed by normalization to the total subclass specific glycopeptide intensities, as described previously (33).
Reverse Phase nano-LC-sheath-flow-ESI-MS
Purified and desalted tryptic IgG glycopeptides were also analyzed on an Ultimate 3000 HPLC system (Dionex Corporation, Sunnyvale, CA, USA), consisting of a degasser unit, binary loading pump, dual binary gradient pump, autosampler maintained at 5 °C and fitted with a 10 μl PEEK sample loop, and two column oven compartments set at 30 °C. To protect the trap and analytical column for particulates, samples were centrifuged at 4000 rpm for 5 min and passed through a 2 μm pore size stainless steel frit mounted between the autosampler transfer tubing and the trap column. Samples (250–5000 nl) were applied to a Dionex Acclaim PepMap100 C18 (5 mm × 300 μm i.d.) SPE trap column conditioned with 0.1% TFA (mobile phase A) for 1 min at 25 μl/min. After sample loading the trap column was switched in-line with the gradient and Ascentis Express C18 nano-LC column (50 mm × 75 μm i.d., 2.7 μm HALO fused core particles; Supelco, Bellefonte, USA) for 8 min while sample elution took place. This was followed by an off-line cleaning of the trap column with three full loop injections containing 5 μl 5% isopropanol (IPA) + 0.1% FA and 5 μl 50% IPA + 0.1% FA. On-column separation was achieved at 900 nl/min using the following gradient of mobile phase A and 95% ACN (Biosolve BV, Valkenswaard, the Netherlands; mobile phase B): 0 min 3% B, 2 min 5% B, 5 min 20% B, 6 min 30% B, 8 min 30% B, 9 min 0% B, and 14 min 0% B. The separation was coupled to a quadrupole-TOF-MS (micrOTOF-Q; Bruker Daltonics) equipped with a standard ESI source (Bruker Daltonics) and a sheath-flow ESI sprayer (capillary electrophoresis ESI-MS sprayer; Agilent Technologies, Santa Clara, USA). The column outlet tubing (20 μm i.d., 360 μm o.d.) was directly applied as sprayer needle. A 2 μl/min sheath-flow of 50% IPA, 20% propionic acid (PA) and 30% ultrapure water was applied by one of the binary gradient pumps to reduce the TFA gas phase ion pairing and assist with ESI spray formation. A nitrogen stream was applied as dry gas at 4 L/min with a nebulizer pressure of 0.4 bars to improve mobile phase evaporation. Glycan decay during ion transfer was reduced by applying 2 and 4 eV quadrupole ion energy and collision energy, respectively. Scan spectra were recorded from m/z 300 to 2000 with two averaged scans at a frequency of 1 Hz. Per sample the total analysis time was 16 min. The software used to operate the Ultimate 3000 HPLC system and the Bruker micrOTOF-Q were Chromeleon Client version 6.8 and micrOTOF control version 2.3, respectively.
Each LC-MS data set was calibrated internally using a list of known glycopeptides, exported to the open mzXML format by Bruker DataAnalysis 4.0 in batch mode (35) and aligned to a master data set of a typical sample (containing many of the (glyco)peptide species shared between multiple samples) using msalign2 (36) and a simple warping script in AWK (37). From each data set a list of 402 predefined features, defined as the peak maximum within mass window of + m/z 0.04 and a retention time window of +10 (38), were extracted using the in-house developed “Xtractor2D” software and merged to a complete data matrix as described previously (39). As input, Xtractor2D takes a data set in the mzXML format aligned to the master data set and a reference list with predefined features with m/z windows and retention times in seconds. The theoretical m/z values used to identify the glycopeptide features are calculated, and the retention times on the chromatographic time scale of the master data set are used for the alignment. Because of the use of TFA as ion pairing reagent, all glycopeptides belonging to the same IgG subclass have approximately the same retention time, regardless of the number of N-acetylneuraminic acid residues. The software and ancillary scripts are freely available at www.ms-utils.org/Xtractor2D. The complete sample-data matrix was finally evaluated using Microsoft Excel.
Structural assignment of the detected glycoforms was performed on the basis of literature knowledge of IgG N-glycosylation (27–32). Relative intensities of 20 IgG1, 20 IgG2/3, and 10 IgG4 glycopeptide species were obtained by integrating and summing the first three isotopic peaks of both doubly and triply charged glycopeptides species followed by background correction and normalization to the total IgG subclass specific glycopeptide intensities. The list of the assigned IgG1, IgG2 and 3, and IgG4 glycopeptides as well as the charge states corresponding m/z values is given in supplemental Table S1 as well as in (39). Nonfucosylated IgG4 species were not included in this list, because of spectral overlap with isomeric IgG1 species (listed in supplemental Table S1). These IgG4 species are not expected to influence the IgG1 glycopeptide abundance levels, because they elute after the IgG1 glycopeptides. There is also spectral overlap between several IgG2 and 3 and IgG4 glycopeptides, but because IgG4 elutes before IgG2 and 3 and is present at a much lower abundace, this is not expected to be a problem for the analysis of either of the glycopeptides.
Multiplex Capillary Gel Electrophoresis with Laser-induced Fluorescence (xCGE-LIF) of IgG N-glycans - Sample Preparation and Analysis
Glycan Release and Labeling
Approximately 10 μg of the protein G monolithic plate IgG eluates were redissolved in 3 μl 1× PBS (Sigma-Aldrich) and dispensed in a 96-well microtiter plate (Greiner Bio-One, Solingen, Germany). IgG samples were denatured with the addition of 4 μl of 0.5% (w/v) SDS (AppliChem, Darmstadt, Germany) in 1× PBS and by incubation at 60 °C for 10 min. Subsequently, the remaining SDS was neutralized by adding 2 μl 4% (v/v) IGEPAL (Sigma-Aldrich) in 1× PBS. IgG N-glycans were released by adding 0.1 U PNGase F (BioReagent ≥ 95%, Sigma-Aldrich) in 1 μl 1× PBS. The 96-well microtiter plate was sealed with adhesive tape and the final sample volume of 10 μl was incubated for 3 h at 37 °C. After N-glycan release samples were dried in a vacuum centrifuge and stored until labeling at −80 °C.
Dried samples were redissolved by adding 2 μl of 1× PBS, 2 μl of 20 mm aminopyrene-1,3,6-trisulfonic acid (APTS; Darmstadt, Sigma-Aldrich) in 3.6 m citric acid monohydrate (CAaq; Merck-Millipore, Germany) and 2 μl of 0.2 m 2-picoline-borane (2-PB; Sigma-Aldrich) solution in DMSO (Sigma-Aldrich). Ultrapure water was used throughout. The 96-well microtiter plate was sealed using adhesive tape followed by shaking for 2 min at 900 rpm. Labeling was performed at 37 °C for 16 h. To stop the reaction, 100 μl 80% ACN (LC-MS Grade ≥ 99.5%, Sigma-Aldrich) was added and the plate was shaken for 2 min at 500 rpm. Post derivatization sample clean-up was performed by HILIC-SPE. To remove free APTS, reducing agent and other impurities, 200 μl of 100 mg/ml BioGel P10 (Bio-Rad, Munich, Germany) suspension in water/EtOH/ACN (70:20:10%, v/v) was applied to AcroPrep 96-well GHP Filter Plates (Pall Corporation, Dreieich, Germany). Solvent was removed by application of vacuum using a vacuum manifold (Merck-Millipore, Germany). All wells were prewashed with 5 × 200 μl water, followed by equilibration with 3 × 200 μl 80% ACN. The samples were applied to the wells of the GHP Filter Plate and shaken for 5 min at 500 rpm to enhance glycan binding. The plate was subsequently washed 5× with 200 μl 80% ACN containing 100 mm triethylamine (TEA; Sigma-Aldrich) adjusted to pH 8.5 with acetic acid (Sigma-Aldrich), followed by washing 3 × 200 μl 80% ACN. After addition of solvent, each washing step was followed by incubation for 2 min and removal of solvent by vacuum. For elution 1 × 100 μl (swelling of BioGel) and 2 × 200 μl of water were applied to each well followed by 5 min incubation at 500 rpm. The eluates were removed by vacuum and collected in a 96-well storage plate (Thermo Scientific, Germany). The combined eluates were either analyzed immediately by xCGE-LIF or stored at −20 °C until usage.
xCGE-LIF
For xCGE-LIF measurement, 1 μl of N-glycan eluate was mixed with 1 μl GeneScan 500 LIZ Size Standard (Invitrogen, Darmstadt, Germany; 1:50 dilution in Hi-Di Formamide) and 9 μl Hi-Di Formamide (Invitrogen). The mixture was transferred to a MicroAmp Optical 384-well Reaction Plate (Invitrogen), sealed with a 384-well plate septa (Invitrogen) and centrifuged at 1000 rpm for 1 min to avoid air bubbles at the bottom of the wells. The xCGE-LIF measurement was performed in a 3130xl Genetic Analyzer, equipped with a 50 cm 16-capillary array filled with POP-7 polymer (all from Invitrogen). After electrokinetic sample injection, samples were analyzed with a running voltage of 15 kV. Data were collected for 45 min. Raw data files were converted to .xml file format using DataFileConverter (Invitrogen) and subsequently analyzed using the MATLAB (The Mathworks, Inc., Natick, MA, USA) based glycan analysis tools glyXtool and glyXalign. GlyXtool was used for structural identification by patented migration time normalization to an internal standard and N-glycan database driven peak annotation (40). The data comparison was performed by glyXalign (41).
Genotype and Phenotype Quality Control
Individuals with a call rate less than 97% were removed, as well as SNPs with a call rate less than 98% (95% for CROATIA-Vis), minor allele frequency less than 0.02 or Hardy-Weinberg equilibrium p value less than 1 × 10−10. A total of 924 individuals from the CROATIA-Vis and 898 individuals from the CROATIA-Korčula cohort passed all genotype quality control thresholds.
IgG was purified from the plasma of 1821 individuals, out of which 1201 had their IgG glycans successfully measured by all four methods. Individuals who had not been successfully measured for all glycan traits using all four methods were removed in order to bias the comparison as little as possible. This left a total of 445 individuals from CROATIA-Vis and 655 individuals from CROATIA-Korčula for which genotype data was also available, providing a final meta-analysis sample size of 1100.
Genome Wide Association Analysis
Each trait was adjusted for sex, age, and the first three principal components obtained from the population-specific identity-by-state (IBS) derived distances matrix. The residuals were transformed to ensure their normal distribution using quantile normalization. The “mmscore” function of GenABEL-package (42) (component of the GenABEL suite, http://www.genabel.org) was used for the association test under an additive model. This score test for family based association takes into account relationship structure and allowed unbiased estimations of SNP allelic effect when relatedness is present between examinees. The relationship matrix used in this analysis was generated by the “IBS” function of GenABEL (using weight = “freq” option), which uses genomic data to estimate the realized pair-wise kinship coefficient. All lambda values for the population-specific analyses were below 1.05 showing that this method efficiently accounts for family structure. Meta-analysis was performed using the inverse variance method implemented with the MetABEL package for R (42). The threshold for a SNP reaching genome wide significance was set at p < 5 × 10−8.
Correlations with Age
All glycan traits from the minimal data set were adjusted for sex and relatedness using the “polygenic” function of the GenABEL package for R (42). The resulting pgresiduals, that is, corrected glycan traits were used to calculate Spearman's rank correlation coefficients with age using the “cor.test” function implemented in stats package for R (43). Correlation coefficients were computed using the same 1100 individuals used for GWAS as the genetic data was required to account for relatedness within the population. To account for multiple testing, the significance level was Bonferroni adjusted (94 tests) and set at p ≤ 5.3 × 10−4.
Correlations with Other Methods
All glycan traits from the minimal data set were adjusted for sex, age, and relatedness using the “polygenic” function of the GenABEL package for R (42). The resulting pgresiduals, that is, corrected glycan traits were used to calculate Pearson's product-moment correlation coefficients and corresponding p values using the “cor.test” function in the stats package for R (43). Correlation coefficients were computed using the same 1100 individuals used for GWAS as the genetic data was required to account for relatedness within the population. The correlations were then compared for all the glycan traits from the minimal data set measured by the four different methods.
RESULTS
IgG N-glycosylation profiling was performed for 1201 individuals using four different analytical approaches: UPLC-FLR, xCGE-LIF, MALDI-TOF-MS, and LC-ESI-MS. An important difference between UPLC-FLR and xCGE-LIF, on one side, and MS-based methods, on the other side, is that UPLC-FLR and xCGE-LIF analyze IgG glycosylation at the level of released glycans (and therefore include glycans on both Fab and Fc parts of IgG), whereas MS-based methods included in this study analyze glycopeptides. Although in-depth analysis of released glycans may provide a detailed picture of the glycan structure, no information on the original glycan attachment site is provided. Such site-specific information can be obtained by the direct analysis of glycopeptides. Because different IgG subclasses have different amino acid sequences around the glycosylation site, by analyzing glycans at the glycopeptide level MS-based methods measure subclass-specific Fc glycosylation. However, unlike the used MS-based methods, UPLC-FLR and xCGE-LIF provide branch-specific information, that is, separation between the 3-arm and 6-arm isomers of glycan species (e.g. FA2[3]G1 and FA2[6]G1) because of a slightly higher retention of the 3-arm isomer. Another important difference between the used methods is the way they generate quantitative information. UPLC-FLR and xCGE-LIF have the advantage that only the fluorescent dye, attached to the reducing end of a glycan, is being detected. Because the structural diversity in glycans is confined to their nonreducing ends, it is safe to assume that each glycan structure will fluoresce with the same quantum yield. With the MS-based methods this is more complex, because the specific response factor of each glycopeptide is affected by both its own structure and by co-eluting peptides (44), thus the relative intensities of different glycans/glycopeptides cannot be directly compared.
Representative analyses of IgG glycosylation using UPLC-FLR, xCGE-LIF, MALDI-TOF-MS, and LC-ESI-MS are shown in Fig. 1. Details of the analytical procedures are presented in the Experimental Procedures section. In addition to the directly measured glycan structures, a number of derived traits that represent common biologically meaningful features (e.g. galactosylation, fucosylation, etc.) shared among several measured glycans were calculated as described previously (8, 33). A full list of traits and a description of how they were calculated is available in supplemental Table S1. Descriptive statistics of IgG glycosylation measured by four methods is provided in supplemental Table S2.
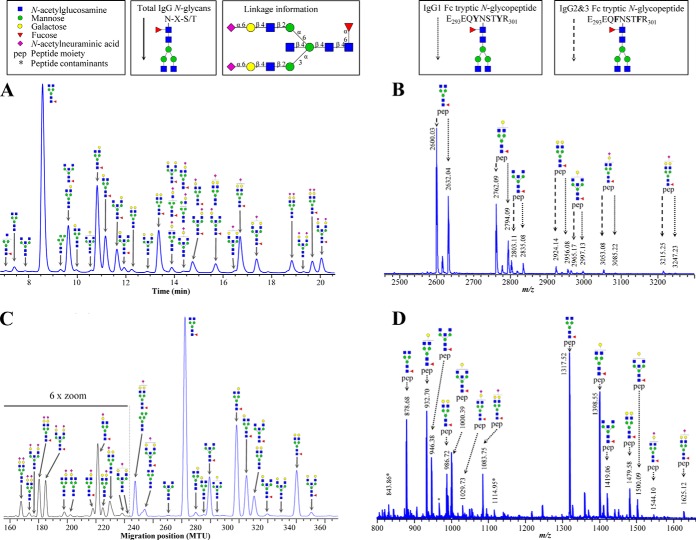
Fig. 1.
Representative data from IgG glycosylation analysis of the same individual by (A) UPLC-FLR (continuous lines - total IgG N-glycans), (B) MALDI-TOF-MS (dotted lines - tryptic IgG1 Fc N-glycopeptide [M-H]− ions, striated lines - IgG2&3 Fc N-glycopeptide [M-H]− ions), (C) xCGE-LIF (continuous lines - total IgG N-glycans), and (D) LC-ESI-MS (dotted lines - tryptic IgG1 Fc N-glycopeptide [M+2H]2+ and [M+3H]3+ ions). Structural schemes are given in terms of pep (peptide moiety), blue square (N-acetylglucosamine), red triangle (fucose), green circle (mannose), yellow circle (galactose), and purple diamond (N-acetylneuraminic acid). Linkage information is given to indicate separation of linkage isomers by UPLC-FLR and xCGE-LIF. Glycan structures are assigned to most of the signals. The complete list of the assigned IgG N-glycans (UPLC-FLR and xCGE-LIF) and IgG N-glycopeptides (MALDI-TOF-MS and LC-ESI-MS) with the charge states corresponding m/z values is given in supplementary Table S1.
Because of the different level at which glycosylation was analyzed (released glycans versus glycopeptides), the information provided by the four used methods is similar, but not identical. To enable meta-analysis of data measured by different methods, we defined a shared set of glycan features common to all four methods (Table I).
Table I. Minimal shared dataset and median values for these IgG glycan structures and traits measured by four different methods.
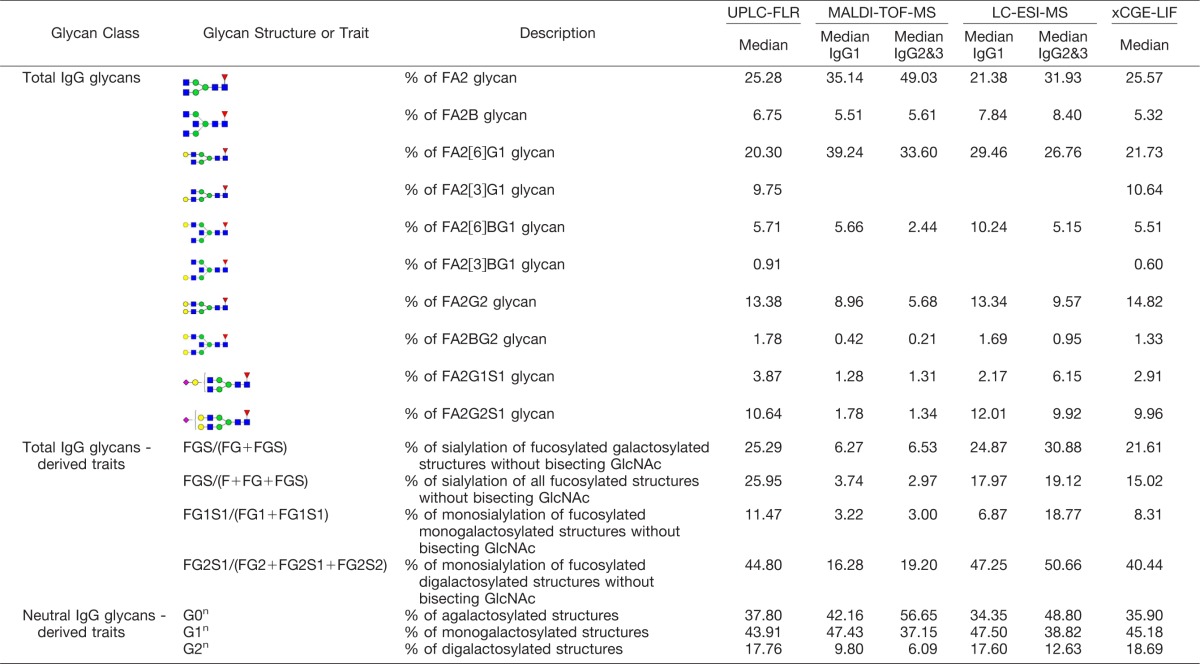
The glycome composition determined by MALDI-TOF-MS deviated pronouncedly from the results of other three methods, which produced more similar results. However, even methods based on fluorescent dye quantification (UPLC-FLR and xCGE-LIF) gave slightly different values for some glycan traits describing sialylation, for example FGS/(F+FG+FGS) and FG1S1/(FG1+FG1S1) (Table I). This indicates that in addition to different response factors in MS-based methods (which distort quantification), sample preparation and clean-up procedures (which can lead to selective loss or enrichment of some glycans) can also significantly distort final results.
At the moment there is no “gold standard” method to analyze protein glycosylation with absolute precision, thus it is not possible to decide which of the methods we used most accurately reflects the real biological situation. Aiming to evaluate the precision of the four methods, we analyzed associations with individual genetic polymorphisms and correlations with age under the assumption that the most precise method will show the strongest associations with the biology underlying IgG glycosylation. Because glycome composition was shown to be under strong genetic influence (5, 8), we believe that a genome wide association approach is a good tool to comparatively assess the power of detecting associations between genetic polymorphisms and IgG N-glycans measured by each of the four methods. In order to have an unbiased approach GWAS was performed on the minimal shared data set using only data from individuals whose glycosylation traits were successfully measured by all four methods (n = 1201 glycomes, 1100 of them with complete genetic data). Genome wide significant association with SNPs in two genomic loci were obtained using all four methods. Six glycan traits showed significant genome wide association in at least one of the data sets generated by the various analytical methods; LC-ESI-MS analysis uncovered all six of these glycan traits, UPLC-FLR and xCGE-LIF determined five, and four of the traits were found with MALDI-TOF-MS. Glycan structures measured by MALDI-TOF-MS seemed to fare the worst in the GWAS comparison which also corresponded with lower correlation coefficients between MALDI-TOF-MS and other used methods for the glycan traits from the minimal data set (supplemental Table S4). All the observed associations replicated those from a recently published IgG glycome GWA study (24). However, because of the lower sample size in this study, not all associations from the previous paper could be replicated. SNPs with the most significant p values at each of the loci are listed in Table II. The full list of all associations with all glycans measured by all the methods is available in supplemental Table S3.
Table II. p values for significant associations between genetic polymorphisms and glycan structures or traits obtained by different methods. Bold text indicates that the p value reaches genome wide significance (p < 5 × 10−8).
| Glycan Structure Or Trait | Genes in Associated Region | SNP | Association p-Value |
|||||
|---|---|---|---|---|---|---|---|---|
| UPLC-FLR | MALDI-TOF-MS (IgG1) | MALDI-TOF-MS (IgG2&3) | LC-ESI-MS (IgG1) | LC-ESI-MS (IgG2&3) | xCGE-LIF | |||
| FA2BG1a | SMARCB1; DERL3 | rs9620326 | 1.47E-10 | 1.15E-07 | 1.70E-06 | 1.63E-08 | 4.11E-10 | 1.11E-10 |
| 1.54E-04 | 7.46E-06 | |||||||
| FA2G1S1 | ST6GAL1 | rs6764279 | 2.80E-22 | 0.2556 | 4.36E-10 | 1.13E-28 | 1.15E-27 | 1.60E-18 |
| FGS/(FG+FGS) | ST6GAL1 | rs6764279 | 1.14E-20 | 0.0154 | 1.86E-12 | 4.87E-12 | 1.64E-25 | 4.83E-18 |
| FGS/(F+FG+FGS) | ST6GAL1 | rs6764279 | 3.25E-04 | 0.1008 | 1.97E-04 | 1.21E-05 | 1.44E-09 | 3.82E-07 |
| FG1S1/(FG1+FG1S1) | ST6GAL1 | rs6764279 | 1.50E-22 | 0.3941 | 9.60E-21 | 2.51E-33 | 1.31E-40 | 5.61E-22 |
| FG2S1/(FG2+FG2S1+FG2S2) | ST6GAL1 | rs6764279 | 1.54E-36 | 1.26E-11 | 3.49E-23 | 4.67E-26 | 1.37E-32 | 1.71E-37 |
a This glycan structure is measured as two isomers with UPLC-FLR and xCGE-LIF (FA2[6]BG1, with galactose on the 6-arm and FA2[3]BG1, with galactose on the 3-arm), but as only one mass in the MS methods.
Glycosylation of IgG strongly correlates with age (8), and thus the strength of correlation of IgG glycans with age could also be used to compare the precision of different analytical methods. The results presented in Table III show that for the majority of glycans in the minimal shared data set all four methods show comparable strengths of correlation, with UPLC-FLR showing somewhat stronger correlation coefficients and lower p values. Table III presents only results from CROATIA-Vis, however, these replicated in CROATIA-Korcula and full results are present in supplemental Table S5.
Table III. Correlation of age and glycan structures or traits measured by different methods in Vis cohort. Presented p values are corrected for multiple testing using Bonferroni correction. Significance level is set at p ≤ 5.3 × 10−4 (94 tests).
| Glycan Class | Glycan Structure or Trait | UPLC-FLR (Total IgG) |
MALDI-TOF-MS |
LC-ESI-MS |
xCGE-LIF (Total IgG) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgG1 |
IgG2&3 |
IgG1 |
IgG2&3 |
||||||||||
| R | p | R | p | R | p | R | p | R | p | R | p | ||
| Total IgG glycans | FA2 | 0.598 | 1.28E-46 | 0.560 | 1.95E-39 | 0.626 | 2.01E-52 | 0.554 | 1.74E-38 | 0.621 | 2.57E-51 | 0.575 | 3.25E-42 |
| FA2B | 0.562 | 6.91E-40 | 0.472 | 2.70E-26 | 0.451 | 1.26E-23 | 0.557 | 5.65E-39 | 0.536 | 1.49E-35 | 0.555 | 1.15E-38 | |
| FA2G1a | −0.347 | 4.57E-13 | −0.499 | 5.44E-30 | −0.585 | 4.47E-44 | −0.117 | 1.00E+00 | −0.405 | 1.56E-18 | −0.255 | 1.83E-06 | |
| −0.107 | 1.00E+00 | −0.035 | 1.00E+00 | ||||||||||
| FA2BG1a | 0.026 | 1.00E+00 | −0.085 | 1.00E+00 | −0.396 | 1.49E-17 | 0.053 | 1.00E+00 | −0.226 | 8.25E-05 | 0.047 | 1.00E+00 | |
| 0.296 | 4.63E-09 | 0.330 | 1.32E-11 | ||||||||||
| FA2G2 | −0.646 | 5.71E-57 | −0.609 | 1.06E-48 | −0.638 | 4.02E-55 | −0.617 | 2.15E-50 | −0.634 | 2.92E-54 | −0.626 | 1.77E-52 | |
| FA2BG2 | −0.393 | 2.89E-17 | −0.333 | 6.46E-12 | −0.507 | 4.34E-31 | −0.422 | 2.34E-20 | −0.520 | 5.09E-33 | −0.361 | 3.22E-14 | |
| FA2G1S1 | 0.062 | 1.00E+00 | −0.026 | 1.00E+00 | −0.494 | 2.92E-29 | 0.176 | 1.61E-02 | −0.331 | 1.01E-11 | 0.003 | 1.00E+00 | |
| FA2G2S1 | −0.588 | 1.35E-44 | −0.519 | 6.58E-33 | −0.589 | 6.72E-45 | −0.585 | 5.75E-44 | −0.619 | 7.82E-51 | −0.584 | 8.00E-44 | |
| Total IgG glycans-derived traits | FGS/(FG+FGS) | −0.259 | 1.08E-06 | −0.081 | 1.00E+00 | −0.446 | 5.31E-23 | −0.436 | 7.63E-22 | −0.372 | 2.79E-15 | −0.457 | 2.17E-24 |
| FGS/(F+FG+FGS) | −0.593 | 1.30E-45 | −0.336 | 4.04E-12 | −0.577 | 1.55E-42 | −0.543 | 1.25E-36 | −0.586 | 2.99E-44 | −0.557 | 6.38E-39 | |
| FG1S1/(FG1+FG1S1) | 0.194 | 2.84E-03 | 0.113 | 1.00E+00 | −0.072 | 1.00E+00 | 0.252 | 2.85E-06 | −0.074 | 1.00E+00 | 0.015 | 1.00E+00 | |
| FG2S1/(FG2+FG2S1+FG2S2) | 0.203 | 1.06E-03 | 0.070 | 1.00E+00 | −0.186 | 6.41E-03 | −0.063 | 1.00E+00 | −0.120 | 1.00E+00 | 0.087 | 1.00E+00 | |
| Neutral IgG glycans-derived traits | G0n | 0.626 | 1.68E-52 | 0.585 | 4.42E-44 | 0.639 | 3.36E-55 | 0.586 | 2.70E-44 | 0.638 | 4.71E-55 | 0.598 | 1.38E-46 |
| G1n | −0.473 | 2.05E-26 | −0.495 | 2.13E-29 | −0.605 | 5.94E-48 | −0.373 | 2.33E-15 | −0.580 | 4.40E-43 | −0.406 | 1.31E-18 | |
| G2n | −0.638 | 5.51E-55 | −0.605 | 6.26E-48 | −0.637 | 8.22E-55 | −0.618 | 1.15E-50 | −0.648 | 2.48E-57 | −0.622 | 1.24E-51 | |
a These glycan structures are measured as two isomers with UPLC-FLR and xCGE-LIF (with galactose on 6- and 3-arm), but as single masses in the MS methods.
An important observation is that both MS-based methods and chromatography/electrophoresis revealed some associations that were undetectable by other methods. For example, the association between monogalactosylated glycans and age was restricted to IgG glycans with galactose on the 6-arm (FA2[6]G1; GP8 measured by UPLC-FLR and P19 measured by xCGE-LIF in supplemental Table S2). This branch-specificity could not be observed with the MS-based methods because they generally do not provide linkage information. On the other hand, glycopeptide-based glycosylation profiling methods readily reveal subclass-specific glycosylation profiles of IgG1, IgG2, IgG3, and IgG4, which was also reflected in much stronger association between galactosylation and age for IgG2 and 3, than for IgG1.
DISCUSSION
In this study we have compared four different methods (UPLC-FLR, xCGE-LIF, MALDI-TOF-MS, and LC-ESI-MS) for the quantitative analysis of IgG N-glycosylation by analyzing the same 1201 IgG samples using all four methods. These four analytical methods, together with direct infusion MSn and LC-MS/MS, have been commonly used for glycosylation analysis in the past years, but there is currently no “gold standard” analytical method for the evaluation of other methods. Therefore, we have decided to use an innovative approach to determine the relative accuracy of the four most widely used methods by comparing association analysis of IgG glycans with genetic polymorphisms and correlations of glycans with age of studied individuals.
GWAS are routinely being used to identify genetic loci associated with specific traits. We have also successfully applied this approach in previous studies to identify genetic loci that are associated with the regulation of protein glycosylation (21, 22, 24, 45). For this study we decided to use GWAS in a different way. We analyzed IgG N-glycosylation with four different methods in the same individuals for whom genetic data was also available. Genetic association analysis was performed separately on glycan data generated by the four methods under the assumption that any imprecision in measurement will decrease power to detect the biological association between genetic polymorphisms and measured glycans. Therefore the analytical method that is the most precise is expected to show the strongest association with genetic loci relevant for IgG glycosylation.
The results presented in Table II (and supplemental Table S3) clearly show that all four methods generate glycan data of sufficiently high quality to be used to detect associations with genetic polymorphisms. The chromatography-based methods, UPLC-FLR and LC-ESI-MS, appear to be somewhat more precise because the measured glycome generally shows stronger associations with genetic polymorphisms, but MALDI-TOF-MS and xCGE-LIF offer the advantage of higher throughput (which could compensate in some circumstances for somewhat lower precision). In addition to GWAS of the minimal shared data set, we also performed the analysis of all glycans measured by all four methods (supplemental Table S3). The number of successfully analyzed samples and glycan traits was different for each method, thus direct comparison of methods is not possible, but the results presented in supplemental Table S3 generally support the conclusion that chromatography-based methods (UPLC-FLR and LC-ECI-MS) yield somewhat better associations with genetic polymorphisms. The same conclusion can also be derived from the analysis of correlation between IgG glycans and age (Table III). In this study we did not detect all genetic associations which were previously reported (24), but this is not unexpected because the number of studied individuals in this study is much lower. Actually, for a study on only 1100 individual, the number of genetic associations is very large indicating that glycans are under strong genetic regulation.
It is frequently argued that methods based on mass spectrometry are not quantitative, but this study clearly demonstrated that the relative quantification by both MALDI-TOF-MS and LC-ESI-MS is very reliable, and that very good associations with genetic polymorphisms and age can be obtained with glycans measured by both methods. Numeric values generated by mass spectrometers for different glycans or glycopeptides are not directly comparable because each molecular specie has its own response factors in mass spectrometry (44), but this difference is not of much relevance for comparisons of the same glycan (or glycopeptide) between different individuals within a studied population. This is evident from good associations with genetic polymorphisms and correlations with age observed in this study. However, if derived traits (like fucosylation, galactosylation, sialylation, etc.) are calculated from MS data, their numerical values may not correspond to real biological situation because they would be distorted by different response factors for individual glycans/glycopeptides, and this is something that needs to be considered when interpreting MS-based data. Furthermore, there are several potential complications, such as variations in allotype, incomplete digestion, chemical modifications (deamidation, oxidation), and alkylation side reactions occurring during cysteine alkylation, which might introduce a bias in glycoprofiling if they occur more frequently in association with certain types of glycopeptides.
In addition to providing important analytical characteristics of different methods for glycomics, this study also clarified one unresolved issue about IgG glycosylation. Previous studies reported irreconcilable differences in the amount of IgG sialylation measured by HPLC/UPLC or by MS. Although MS studies estimated IgG sialylation to be below 5% (33), HPLC/UPLC studies reported much higher levels, even including values of over 20% of IgGs sialylated (46–49). This difference was most often attributed to inclusion of Fab glycans in UPLC and CE analysis, but in the current study we also observed significant IgG Fc sialylation when quantified by LC-ESI-MS (Table I). Therefore the lower values of IgG Fc sialylation reported using MALDI-TOF-MS analysis appear to be caused by an experimental artifact most probably caused by loss of sialic acid during MALDI-TOF-MS analysis. This finding is very important in the context of further development of therapeutic intravenous immunoglobulins, because some studies indicate that IgG with sialylated Fc glycans is an anti-inflammatory agent (50).
Very weak associations between sialylated glycans measured by MALDI-TOF-MS and genetic loci and age further support the hypothesis that MALDI is underperforming in quantitative analysis of sialylated glycans, and stabilization of sialic acids may be needed for a more robust quantitation of sialic acids by MALDI methods. Interestingly, xCGE-LIF also showed lower relative quantitative values for some of the sialylated glycans that resulted in weaker associations with both genetic polymorphisms and age. Each of the methods reveals some additional complementary information about the glycome, indicating that in some situations the combined analysis by different methods can yield additional useful information, which helps interpretation of complex biological systems.
CONCLUSIONS
It is increasingly recognized that variation in glycan structures is likely to play an essential and ubiquitous role in human physiology and pathophysiology. This recognition has led to glycomics being declared a research priority for the next decade (3), and it is expected that an increasing number of future large clinical and population studies will include glycan analysis (1). However, methods for high-throughput glycan analysis have been developed only recently, and thorough evaluation and standardization of the analytical methods is needed before a significant amount of time and other resources should be invested in large-scale studies. In this study we have used association with genetic polymorphisms and age as the evaluation criterion to compare four methods (UPLC-FLR, xCGE-LIF, MALDI-TOF-MS, and LC-ESI-MS) that are currently being used to study protein glycosylation. All four methods delivered reliable quantitative data. In this study we identify a number of specific advantages and disadvantages of each method (Table IV) in order to guide selection of the most appropriate and cost-effective approach for any given research study.
Table IV. Comparison of four methods for high-throughput glycomic and glycoproteomic analysis.
| UPLC-FLR | xCGE-LIF | MALDI-TOF-MS | LC-ESI-MS | |
|---|---|---|---|---|
| Acceptance/usage for glycomics | Widely used | Rarely used | Widely used | Moderately used |
| Throughput | Medium, approximately 50 samples per instrument per day | (Very) high, multiplexing with up to 96 capillaries enables analysis of thousands of samples | (Very) high, as measurement of a sample can be performed at a sub-minute time scale | Medium, approximately 100 samples per day per instrument |
| Required expertise | Medium | Medium | High | Very high |
| Resolution | High | High | Very high | Very high |
| Isomer separation | Good | Very good | None | Some |
| Quantification | Very good | Good | Medium | Good |
| Costs of equipment | Ca. Euro 40–70,000 | Ca. Euro 100,000 for a 4-capillary instrument | Ca. Euro 100–500,000 | Euro 200–500,000 |
| Costs per sample in high throughput mode | Rather high costs, mainly due to low throughput and costs of consumables | Low costs per sample, due to low running costs and parallelization by multiplexing | Low costs per sample due to high throughput per instrument | Very high costs, mainly due to expensive equipment and low throughput per instrument |
| Main advantages for genetic and epidemiological studies | Reliable quantification, robustness | Less demanding in sample preparation, low costs, high robustness and high throughput, no sample carry over; reliable relative quantification, very sensitive (low LOD) | Low cost and high throughput, site specific glycosylation analysis, sensitive, enables structural elucidation via fragmentation experiments | Reliable quantification, site specific glycosylation analysis, sensitive, enables structural elucidation via fragmentation experiments |
| Main disadvantages for genetic and epidemiological studies | Inability to perform site specific glycosylation analysis, relatively low throughput and high cost | Inability to perform site specific glycosylation analysis, comparatively small database (to be enlarged) | Less reliable quantification, loss of sialic acids | Relatively high costs |
| Specific advantages for IgG glycosylation analysis | Differentiation of galactosylation on 3- and 6-arms, accurate quantification of IgG sialylation | Differentiation of galactosylation on 3- and 6-arms, accurate quantification of IgG sialylation | Differentiation of glycans on different IgG subclasses, analysis of only Fc glycans | Differentiation of glycans on different IgG subclasses, analysis of only Fc glycans, accurate quantification of IgG sialylation |
Supplementary Material
Acknowledgments
We thank Carolien A. M. Koeleman for expert technical assistance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The CROATIA-Vis and CROATIA-Korčula studies would like to acknowledge the invaluable contribution of the recruitment teams (including those from the Institute of Anthropological Research in Zagreb) in Vis and Korčula, the administrative teams in Croatia and Edinburgh and the people of Vis and Korčula.
Footnotes
Author contributions: Y.S.A., E.R., M.W., and G.L. designed research; M.P., R.H., M.H.S., M.N., J.K., M.B., and T.M. performed research; O.G., A.F.W., I.R., C.H., H.C., A.M.D., and U.R. contributed new reagents or analytic tools; J.E.H., L.K., F.V., O.P., G.R., E.T., and Y.S.A. analyzed data; J.E.H., M.P., L.K., R.H., M.H.S., R.H.P., I.R., Y.S.A., E.R., M.W., and G.L. wrote the paper.
* The CROATIA-Vis and CROATIA-Korčula studies in the Croatian islands of Vis and Korčula were supported by grants from the Medical Research Council (UK), the Ministry of Science, Education and Sport of the Republic of Croatia (grant number 108–1080315-0302) and the European Union framework program 6 European Special Populations Research Network project (contract LSHG-CT-2006–018947). SNP genotyping for CROATIA-Vis was performed at the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh, UK and CROATIA-Korcula at Helmholz Zentrum München, GmbH, Neuherberg, Germany. Glycome analysis was supported by the Croatian Ministry of Science, Education and Sport (grant number 309-0061194-2023), the European Commission GlycoBioM (contract #259869), HighGlycan (contract #278535), MIMOmics (contract #305280), HTP-GlycoMet (contract #324400) and IntegraLife (contract# 315997) grants and a Zenith grant from The Netherlands Organization for Scientific Research (#93511033). The work of YSA was supported by Russian Foundation for Basic Research grant 12-04-33182. MHJ Selman thanks Hoffmann la Roche for financial support.
 This article contains supplemental Tables S1 to S5.
This article contains supplemental Tables S1 to S5.
Conflict of interests: GL declares that he is a founder and owner, and LK, FV, MPB, JK and MN declare that they are employees of Genos Ltd, which offers commercial service of glycomic analysis and has several patents in this field. ER and RH declare that they are founders and owners of glyXera GmbH, which offers commercial service of glycomic analysis. MB declares that he was employee and RH and TM declare that they are part time employees of glyXera. ER and MW have several patents in the field of glycosylation analysis. YSA declares that he is a founder and owner of “Yurii Aulchenko” consulting.
1 The abbreviations used are:
- IgG
- immunoglobulin G
- GWAS
- genome-wide association studies
- UPLC-FLR
- ultraperformance liquid chromatography with fluorescence detection
- CGE-LIF
- multiplex capillary gel electrophoresis with laser induced fluorescence detection
- MALDI-TOF-MS
- matrix assisted laser desorption/ionization time of flight MS
- LC-ESI-MS
- liquid chromatography electrospray MS.
REFERENCES
- 1. Hart G. W., Copeland R. J. (2010) Glycomics hits the big time. Cell 143, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zoldos V., Novokmet M., Beceheli I., Lauc G. (2013) Genomics and epigenomics of the human glycome. Glycoconj. J. 30, 41–50 [DOI] [PubMed] [Google Scholar]
- 3. Walt D., Aoki-Kinoshita K. F., Bendiak B., Bertozzi C. R., Boons G. J., Darvill A., Hart G., Kiessling L. L., Lowe J., Moon R., Paulson J., Sasisekharan R., Varki A., Wong C. H. (2012) Transforming Glycoscience: A Roadmap for the Future, Nacional Academies Press, Washington: [PubMed] [Google Scholar]
- 4. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 5. Knežević A., Polašek O., Gornik O., Rudan I., Campbell H., Hayward C., Wright A., Kolčić I., O'Donoghue N., Bones J., Rudd P. M., Lauc G. (2009) Variability, heritability and environmental determinants of human plasma N-glycome. J. Proteome Res. 8, 694–701 [DOI] [PubMed] [Google Scholar]
- 6. Pučić M., Pinto S., Novokmet M., Knežević A., Gornik O., Polašek O., Vlahoviček K., Wei W., Rudd P. M., Wright A. F., Campbell H., Rudan I., Lauc G. (2010) Common aberrations from normal human N-glycan plasma profile. Glycobiology 20, 970–975 [DOI] [PubMed] [Google Scholar]
- 7. Gornik O., Pavic T., Lauc G. (2012) Alternative glycosylation modulates function of IgG and other proteins - implications on evolution and disease. Biochim. Biophys. Acta 1820, 1318–1326 [DOI] [PubMed] [Google Scholar]
- 8. Pucic M., Knezevic A., Vidic J., Adamczyk B., Novokmet M., Polasek O., Gornik O., Supraha-Goreta S., Wormald M. R., Redzic I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josic D., Lauc G. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteomics 10, M111 010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mechref Y., Hu Y., Garcia A., Hussein A. (2012) Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis 33, 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adamczyk B., Tharmalingam T., Rudd P. M. (2012) Glycans as cancer biomarkers. Biochim. Biophys. Acta 1820, 1347–1353 [DOI] [PubMed] [Google Scholar]
- 11. Theodoratou E., Campbell H., Ventham N. T., McGovern D. P., Satsangi J., Lauc G., and IBD-BIOM (2014) The role of glycosylation in inflammatory bowel disease. Nat. Rev. Gastroentero. revised version submitted [DOI] [PubMed] [Google Scholar]
- 12. Theodoratou E., Montazeri Z., Hawken S., Allum G. C., Gong J., Tait V., Kirac I., Tazari M., Farrington S. M., Demarsh A., Zgaga L., Landry D., Benson H. E., Read S. H., Rudan I., Tenesa A., Dunlop M. G., Campbell H., Little J. (2012) Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J. Natl. Cancer Inst. 104, 1433–1457 [DOI] [PubMed] [Google Scholar]
- 13. Siontis K. C., Patsopoulos N. A., Ioannidis J. P. (2010) Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur. J. Hum. Genet. 18, 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visscher P. M., Brown M. A., McCarthy M. I., Yang J. (2012) Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Callewaert N., Van Vlierberghe H., Van Hecke A., Laroy W., Delanghe J., Contreras R. (2004) Noninvasive diagnosis of liver cirrhosis using DNA sequencer-based total serum protein glycomics. Nat. Med. 10, 429–434 [DOI] [PubMed] [Google Scholar]
- 16. Miura Y., Kato K., Takegawa Y., Kurogochi M., Furukawa J., Shinohara Y., Nagahori N., Amano M., Hinou H., Nishimura S. (2010) Glycoblotting-assisted O-glycomics: ammonium carbamate allows for highly efficient o-glycan release from glycoproteins. Anal. Chem. 82, 10021–10029 [DOI] [PubMed] [Google Scholar]
- 17. Winnik W. M., Dekroon R. M., Jeong J. S., Mocanu M., Robinette J. B., Osorio C., Dicheva N. N., Hamlett E., Alzate O. (2012) Analysis of proteins using DIGE and MALDI mass spectrometry. Methods Mol. Biol. 854, 47–66 [DOI] [PubMed] [Google Scholar]
- 18. Reusch D., Haberger M., Kailich T., Heidenreich A. K., Kampe M., Bulau P., Wuhrer M. (2013) High-throughput glycosylation analysis of therapeutic immunoglobulin G by capillary gel electrophoresis using a DNA analyzer. mAbs 6, published online [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Royle L., Campbell M. P., Radcliffe C. M., White D. M., Harvey D. J., Abrahams J. L., Kim Y. G., Henry G. W., Shadick N. A., Weinblatt M. E., Lee D. M., Rudd P. M., Dwek R. A. (2008) HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376, 1–12 [DOI] [PubMed] [Google Scholar]
- 20. Ruhaak L. R., Hennig R., Huhn C., Borowiak M., Dolhain R. J., Deelder A. M., Rapp E., Wuhrer M. (2010) Optimized workflow for preparation of APTS-labeled N-glycans allowing high-throughput analysis of human plasma glycomes using 48-channel multiplexed CGE-LIF. J. Proteome Res. 9, 6655–6664 [DOI] [PubMed] [Google Scholar]
- 21. Huffman J. E., Knezevic A., Vitart V., Kattla J., Adamczyk B., Novokmet M., Igl W., Pucic M., Zgaga L., Johannson A., Redzic I., Gornik O., Zemunik T., Polasek O., Kolcic I., Pehlic M., Koeleman C. A., Campbell S., Wild S. H., Hastie N. D., Campbell H., Gyllensten U., Wuhrer M., Wilson J. F., Hayward C., Rudan I., Rudd P. M., Wright A. F., Lauc G. (2011) Polymorphisms in B3GAT1, SLC9A9, and MGAT5 are associated with variation within the human plasma N-glycome of 3533 European adults. Hum. Mol. Genet. 20, 5000–5011 [DOI] [PubMed] [Google Scholar]
- 22. Lauc G., Essafi A., Huffman J. E., Hayward C., Knežević A., Kattla J. J., Polašek O., Gornik O., Vitart V., Abrahams J. L., Pučić M., Novokmet M., Redžić I., Campbell S., Wild S. H., Borovečki F., Wang W., Kolčić I., Zgaga L., Gyllensten U., Wilson J. F., Wright A. F., Hastie N. D., Campbell H., Rudd P. M., Rudan I. (2010) Genomics meets glycomics - The first GWAS study of human N-glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet. 6, e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kutalik Z., Benyamin B., Bergmann S., Mooser V., Waeber G., Montgomery G. W., Martin N. G., Madden P. A., Heath A. C., Beckmann J. S., Vollenweider P., Marques-Vidal P., Whitfield J. B. (2011) Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. Hum. Mol. Genet. 20, 3710–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lauc G., Huffman J. E., Pucic M., Zgaga L., Adamczyk B., Muzinic A., Novokmet M., Polasek O., Gornik O., Kristic J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolcic I., Lukic I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., Rudan I. (2013) Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet 9, e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudan I., Marusic A., Jankovic S., Rotim K., Boban M., Lauc G., Grkovic I., Dogas Z., Zemunik T., Vatavuk Z., Bencic G., Rudan D., Mulic R., Krzelj V., Terzic J., Stojanovic D., Puntaric D., Bilic E., Ropac D., Vorko-Jovic A., Znaor A., Stevanovic R., Biloglav Z., Polasek O. (2009) “10001 Dalmatians:” Croatia launches its national biobank. Croat. Med. J. 50, 4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Selman M. H., Hoffmann M., Zauner G., McDonnell L. A., Balog C. I., Rapp E., Deelder A. M., Wuhrer M. (2012) MALDI-TOF-MS analysis of sialylated glycans and glycopeptides using 4-chloro-alpha-cyanocinnamic acid matrix. Proteomics 12, 1337–1348 [DOI] [PubMed] [Google Scholar]
- 27. Parekh R. B., Dwek R. A., Sutton B. J., Fernandes D. L., Leung A., Stanworth D., Rademacher T. W., Mizuochi T., Taniguchi T., Matsuta K., et al. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 316, 452–457 [DOI] [PubMed] [Google Scholar]
- 28. Selman M. H., McDonnell L. A., Palmblad M., Ruhaak L. R., Deelder A. M., Wuhrer M. (2010) Immunoglobulin G glycopeptide profiling by matrix-assisted laser desorption ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 82, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 29. Shikata K., Yasuda T., Takeuchi F., Konishi T., Nakata M., Mizuochi T. (1998) Structural changes in the oligosaccharide moiety of human IgG with aging. Glycoconj. J. 15, 683–689 [DOI] [PubMed] [Google Scholar]
- 30. Stadlmann J., Pabst M., Kolarich D., Kunert R., Altmann F. (2008) Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8, 2858–2871 [DOI] [PubMed] [Google Scholar]
- 31. Wuhrer M., Stam J. C., van de Geijn F. E., Koeleman C. A., Verrips C. T., Dolhain R. J., Hokke C. H., Deelder A. M. (2007) Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 7, 4070–4081 [DOI] [PubMed] [Google Scholar]
- 32. Yamada E., Tsukamoto Y., Sasaki R., Yagyu K., Takahashi N. (1997) Structural changes of immunoglobulin G oligosaccharides with age in healthy human serum. Glycoconjugate J. 14, 401–405 [DOI] [PubMed] [Google Scholar]
- 33. Bakovic M. P., Selman M. H., Hoffmann M., Rudan I., Campbell H., Deelder A. M., Lauc G., Wuhrer M. (2013) High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J. Proteome Res. 12, 821–831 [DOI] [PubMed] [Google Scholar]
- 34. Balbin M., Grubb A., de Lange G. G., Grubb R. (1994) DNA sequences specific for Caucasian G3m(b) and (g) allotypes: allotyping at the genomic level. Immunogenetics 39, 187–193 [DOI] [PubMed] [Google Scholar]
- 35. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 36. Nevedomskaya E., Derks R., Deelder A. M., Mayboroda O. A., Palmblad M. (2009) Alignment of capillary electrophoresis-mass spectrometry datasets using accurate mass information. Anal. Bioanal. Chem. 395, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 37. Alfred V., Aho B. W. K. a. P. J. W. (1988) The AWK programming language, Addison-Wesley: Reading, Massachusetts [Google Scholar]
- 38. Strittmatter E. F., Ferguson P. L., Tang K., Smith R. D. (2003) Proteome analyses using accurate mass and elution time peptide tags with capillary LC time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 14, 980–991 [DOI] [PubMed] [Google Scholar]
- 39. Selman M. H., Derks R. J., Bondt A., Palmblad M., Schoenmaker B., Koeleman C. A., van de Geijn F. E., Dolhain R. J., Deelder A. M., Wuhrer M. (2012) Fc specific IgG glycosylation profiling by robust nano-reverse phase HPLC-MS using a sheath-flow ESI sprayer interface. J. Proteomics 75, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 40. Hennig R., Reichl U., Rapp E. (2011) A Software Tool for Automated High-Throughput Processing of CGE-LIF Based Glycoanalysis Data, Generated by a Multiplexing Capillary DNA Sequencer. Glycoconjugate J. 28, 331–332 [Google Scholar]
- 41. Behne A., Muth T., Borowiak M., Reichl U., Rapp E. (2013) glyXalign: high-throughput migration time alignment preprocessing of electrophoretic data retrieved via multiplexed capillary gel electrophoresis with laser-induced fluorescence detection-based glycoprofiling. Electrophoresis 34, 2311–2315 [DOI] [PubMed] [Google Scholar]
- 42. Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M. (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 [DOI] [PubMed] [Google Scholar]
- 43. R-Core-Team (2013) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 44. Stavenhagen K., Hinneburg H., Thaysen-Andersen M., Hartmann L., Varon Silva D., Fuchser J., Kaspar S., Rapp E., Seeberger P. H., Kolarich D. (2013) Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom.: JMS 48, 627–639 [DOI] [PubMed] [Google Scholar]
- 45. Zoldos V., Horvat T., Lauc G. (2013) Glycomics meets genomics, epigenomics and other high throughput omics for system biology studies. Curr. Opin. Chem. Biol. 17, 34–40 [DOI] [PubMed] [Google Scholar]
- 46. Anumula K. R. (2012) Quantitative glycan profiling of normal human plasma derived immunoglobulin and its fragments Fab and Fc. J. Immunol. Methods 382, 167–176 [DOI] [PubMed] [Google Scholar]
- 47. Kobata A. (2008) The N-linked sugar chains of human immunoglobulin G: their unique pattern, and their functional roles. Biochim. Biophys. Acta 1780, 472–478 [DOI] [PubMed] [Google Scholar]
- 48. Thobhani S., Yuen C. T., Bailey M. J., Jones C. (2009) Identification and quantification of N-linked oligosaccharides released from glycoproteins: an inter-laboratory study. Glycobiology 19, 201–211 [DOI] [PubMed] [Google Scholar]
- 49. Youings A., Chang S. C., Dwek R. A., Scragg I. G. (1996) Site-specific glycosylation of human immunoglobulin G is altered in four rheumatoid arthritis patients. Biochem. J. 314 (Pt 2), 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nimmerjahn F., Ravetch J. V. (2008) Anti-inflammatory actions of intravenous immunoglobulin. Annu. Rev. Immunol. 26, 513–533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.