Abstract
Pichia pastoris is an established protein expression host mainly applied for the production of biopharmaceuticals and industrial enzymes. This methylotrophic yeast is a distinguished production system for its growth to very high cell densities, for the available strong and tightly regulated promoters, and for the options to produce gram amounts of recombinant protein per litre of culture both intracellularly and in secretory fashion. However, not every protein of interest is produced in or secreted by P. pastoris to such high titres. Frequently, protein yields are clearly lower, particularly if complex proteins are expressed that are hetero-oligomers, membrane-attached or prone to proteolytic degradation. The last few years have been particularly fruitful because of numerous activities in improving the expression of such complex proteins with a focus on either protein engineering or on engineering the protein expression host P. pastoris. This review refers to established tools in protein expression in P. pastoris and highlights novel developments in the areas of expression vector design, host strain engineering and screening for high-level expression strains. Breakthroughs in membrane protein expression are discussed alongside numerous commercial applications of P. pastoris derived proteins.
Keywords: Yeast, Pichia pastoris, Protein expression, Protein secretion, Protease-deficient strains, Chaperone
Introduction
The methylotrophic yeast Pichia pastoris, currently reclassified as Komagataella pastoris, has become a substantial workhorse for biotechnology, especially for heterologous protein production (Kurtzman 2009). It was introduced more than 40 years ago by Phillips Petroleum for commercial production of single cell protein (SCP) as animal feed additive based on a high cell density fermentation process utilizing methanol as carbon source. However, the oil crisis in 1973 increased the price for methanol drastically and made SCP production uneconomical. In the 1980s, P. pastoris was developed as a heterologous protein expression system using the strong and tightly regulated AOX1 promoter (Cregg et al. 1985). In combination with the already developed fermentation process for SCP production, the AOX1 promoter provided exceptionally high levels of heterologous proteins. One of the first large-scale industrial production processes established in the 1990s was the production of the plant-derived enzyme hydroxynitrile lyase at >20 g of recombinant protein per litre of culture volume (Hasslacher et al. 1997). This enzyme is used as biocatalyst for the production of enantiopure m-phenoxybenzaldehyde cyanohydrin — a building block of synthetic pyrethroids — on the multi-ton scale.
Through a far-sighted decision this expression system, initially patented by Phillips Petroleum, was made available to the scientific community for research purposes. A major breakthrough was the publication of detailed genome sequences of the original SCP production strain CBS7435 (Küberl et al. 2011), the first host strain developed for heterologous protein expression GS115 (De Schutter et al. 2009), as well as of the related P. pastoris DSMZ 70382 strain (Mattanovich et al. 2009b). Equally important breakthroughs for the commercial application of the P. pastoris cell factory were the Food and Drug Administration (FDA) GRAS (generally recognized as safe) status for a protein used in animal feed, phospholipase C (Ciofalo et al. 2006), and the FDA approval of a recombinant biopharmaceutical product, Kalbitor®, a kallikrein inhibitor (Thompson 2010).
The classical P. pastoris expression system has been extensively reviewed over the years (Cereghino and Cregg 2000; Daly and Hearn 2005; Gasser et al. 2013; Jin et al. 2006; Macauley-Patrick et al. 2005). In this review, we focus on recent developments for heterologous protein production and describe examples for the commercial use of this expression system. In the first chapter, we refer to the established basic vector systems and elaborate on developments thereof with an emphasis on newly developed promoter systems. Herein, also some aspects of secretion will be summarized. The second part is devoted to the most recent developments regarding host strain development. As a specific novelty, a new platform based on the CBS7435 strain is described, for which patent protection has ceased and no specific material rights are pending. In the third chapter, we describe specific strategies for obtaining high-level expression strains and summarize important applications of P. pastoris for production of biopharmaceuticals, membrane proteins and industrial proteins. The last section provides an outlook on future perspectives covering recent progress in molecular and cell biology of P. pastoris and possibilities for implementing new strategies in expression strain development.
Basic systems for cloning and expression in P. pastoris
When devising strategies for cloning and expression of heterologous proteins in P. pastoris some points need to be considered from the start, that is, the choice of promoter–terminator combinations, suitable selection markers and application of vector systems for either intracellular or secreted expression including selection of proper secretion signals (Fig. 1). The choice of the proper expression vector and complementary host strain are a most important prerequisite for successful recombinant protein expression.
Fig. 1.
General considerations for heterologous gene expression in P. pastoris. Expression plasmids harbouring the gene(s) of interest (GOI) are linearized prior to transformation. Selectable markers (e.g., AmpR) and origin of replication (Ori) are required for plasmid propagation in E. coli. The expression level of the protein of interest may depend on (i) the chromosomal integration locus, which is targeted by the 5′ and 3′ homologous regions (5′HR and 3′HR), and (ii) on the gene copy number. A representative promoter (P) and transcription terminator (TT) pair are shown. Proper signal sequences will guide recombinant protein for intracellular or secretory expression, and will govern membrane integration or membrane anchoring
Promoters
The use of tightly regulated promoters such as the alcohol oxidase (AOX1) promoter holds advantages for overexpression of proteins. By uncoupling the growth from the production phase, biomass is accumulated prior to protein expression. Therefore, cells are not stressed by the accumulation of recombinant protein during growth phase, and even the production of proteins that are toxic to P. pastoris is possible. Furthermore, it may be desirable to co-express helper proteins like chaperones at defined time points, for example, before the actual target protein is formed. On the other hand, use of constitutive promoters may ease process handling. Constitutive promoters are usually also applied to express selection markers. Metabolic pathway engineering strategies might further take advantage of fine-tuned constitutive promoters to ensure a controlled flux of metabolites. An extensive summary of promoters used for heterologous expression in P. pastoris has recently been published by Vogl and Glieder (2013). An overview of broadly used and extensively studied as well as recently examined promoters is given in Table 1.
Table 1.
The most prominently used and very recently established promoters for heterologous expression in P. pastoris
| Inducible | Corresponding gene | Regulation | Reference |
| AOX1 | Alcohol oxidase 1 | Inducible with MeOH | (Tschopp et al. 1987a) |
| DAS | Dihydroxyacetone synthase | Inducible with MeOH | (Ellis et al. 1985; Tschopp et al. 1987a) |
| FLD1 | Formaldehyde dehydrogenase 1 | Inducible with MeOH or methylamine | (Shen et al. 1998) |
| ICL1 | Isocitrate lyase | Repressed by glucose, induction in absence of glucose/by addition of ethanol | (Menendez et al. 2003) |
| PHO89 | Putative Na+/phosphate symporter | Induction upon phosphate starvation | (Ahn et al. 2009) |
| THI11 | Thiamine biosynthesis gene | Repressed by thiamin | (Stadlmayr et al. 2010) |
| ADH1 | Alcohol dehydrogenase | Repressed on glucose and methanol, induced on glycerol and ethanol | (Cregg and Tolstorukov 2012) |
| ENO1 | Enolase | Repressed on glucose, methanol and ethanol, induced on glycerol | (Cregg and Tolstorukov 2012) |
| GUT1 | Glycerol kinase | Repressed on methanol, induced on glucose, glycerol and ethanol | (Cregg and Tolstorukov 2012) |
| Constitutive | Corresponding gene | Regulation | Reference |
| GAP | Glyceraldehyde-3-P dehydrogenase | Constitutive expression on glucose, to a lesser extent on glycerol and methanol | (Waterham et al. 1997) |
| TEF1 | Translation elongation factor 1 | Constitutive expression on glycerol and glucose | (Ahn et al. 2007) |
| PGK1 | 3-Phosphoglycerate kinase | Constitutive expression on glucose, to a lesser extent on glycerol and methanol | (de Almeida et al. 2005) |
| GCW14 | Potential glycosyl phosphatidyl inositol (GPI)-anchored protein | Constitutive expression on glycerol, glucose and methanol | (Liang et al. 2013b) |
| G1 | High affinity glucose transporter | Repressed on glycerol, induced upon glucose limitation | (Prielhofer et al. 2013) |
| G6 | Putative aldehyde dehydrogenase | Repressed on glycerol, induced upon glucose limitation | (Prielhofer et al. 2013) |
Inducible promoters
The tightly regulated AOX1 promoter (P AOX1), which was first employed for heterologous gene expression by Tschopp et al. (1987a), is still the most commonly used promoter (Lünsdorf et al. 2011; Sigoillot et al. 2012; Yu et al. 2013). P AOX1 is strongly repressed when P. pastoris is grown on glucose, glycerol or ethanol (Inan and Meagher 2001). Upon depletion of these carbon sources, the promoter is de-repressed, but is fully induced only upon addition of methanol. Several studies have identified multiple regulatory elements in the P AOX1 sequence (Hartner et al. 2008; Kranthi et al. 2006, 2009; Ohi et al. 1994; Parua et al. 2012; Staley et al. 2012; Xuan et al. 2009). Positively and negatively acting elements have been described (Kumar and Rangarajan 2012; Lin-Cereghino et al. 2006; Polupanov et al. 2012), but the molecular details of P AOX1 regulation are still not completely elucidated.
Methanol is a highly flammable and hazardous substance and, therefore, undesirable for large-scale fermentations. Alternative inducible promoters or P AOX1 variants, which can be induced without methanol but still reach high expression levels, are desired. A recently published patent application describes such a method, wherein expression is controlled by methanol-inducible promoters, such as AOX1, methanol oxidase (MOX) or formate dehydrogenase (FMDH), without the addition of methanol (Takagi et al. 2008). This was achieved by constitutively co-expressing the positively acting transcription factor Prm1p from either of the GAP, TEF or PGK promoters. The relative activity of a phytase reporter protein was 3-fold increased without addition of methanol as compared to a control strain with PRM1 under its native promoter. However, phytase expression levels were not compared for standard methanol induction and constitutive Prm1p expression conditions. Hartner et al. have constructed a synthetic AOX1 promoter library by deleting or duplicating transcription factor binding sites for fine-tuned expression in P. pastoris (Hartner et al. 2008). Using EGFP as reporter, some promoter variants were found to confer even higher expression levels than the native P AOX1 spanning a range between 6 % and 160 % of the native promoter activity. These P AOX1 variants have also proven to behave similarly when industrially relevant enzymes such as horseradish peroxidase and hydroxynitrile lyases were expressed.
Numerous further controllable promoters are currently being investigated for their ability to promote high-level expression (Table 1). For example, a recently published patent application describes the use of three novel inducible promoters from P. pastoris, ADH1 (alcohol dehydrogenase), GUT1 (glycerol kinase) and ENO1 (enolase), showing interesting regulatory features (Cregg and Tolstorukov 2012). However, due to a lack of absolute expression values the performance of these novel promoters cannot be compared to the widely used AOX1 and GAP promoters.
Constitutive promoters
Constitutive expression eases process handling, omits the use of potentially hazardous inducers and provides continuous transcription of the gene of interest. For this purpose, the glyceraldehyde-3-phosphate promoter (P GAP) is commonly used, which — on glucose — reaches almost the same expression levels as methanol-induced P AOX1 (Waterham et al. 1997). Expression levels from P GAP drop to about one half on glycerol and to one third when cells are grown on methanol (Cereghino and Cregg 2000). Alternative constitutive promoters and promoter variants have been described recently (Table 1). The constitutive P GCW14 promoter, for example, was described to be a stronger promoter than the GAP and TEF1 promoters, which was assessed by secretory expression of EGFP (Liang et al. 2013b). It was found that EGFP expression from P GCW14 yielded in a 10-fold increase compared to P GAP driven expression when cells were cultivated on glycerol or methanol, and a 5-fold increase on glucose.
A recent DNA microarray study identified novel promoters that are repressed on glycerol, but are being induced upon shift to glucose-limited media (Prielhofer et al. 2013). Supposedly, the most interesting promoters discovered by this approach control expression of a high-affinity glucose transporter, HGT1, and of a putative aldehyde dehydrogenase. The former promoter was reported to drive EGFP expression to even higher levels than could be reached with P GAP. In glycerol fed-batch fermenter cultures, human serum album was expressed from the novel promoter to a 230 % increase in specific product yield as compared to P GAP driven expression.
In some cases, it is desired that expression levels can be fine-tuned in order to (1) co-express accessory proteins facilitating recombinant protein expression and secretion or (2) provide protein post-translational modifications as well as to (3) engineer whole metabolic pathways consisting of a cascade of different enzymatic steps. For such applications, a library of GAP promoter variants with relative strengths ranging from 0.6 % to 16.9-fold of the wild type promoter activity was developed and tested using three different reporter proteins, yEGFP, β-galactosidase and methionine acetyltransferase (Qin et al. 2011).
Vectors
The standard setup of vectors is a bi-functional system enabling replication in E. coli and maintenance in P. pastoris using as selection markers either auxotrophy markers (e.g., HIS4, MET2, ADE1, ARG4, URA3, URA5, GUT1) or genes conferring resistance to drugs such as Zeocin™, geneticin (G418) and blasticidin S. Although there are some reports of using episomal plasmids for heterologous protein expression or for the screening of mutant libraries in P. pastoris (Lee et al. 2005; Uchima and Arioka 2012), stable integration into the host genome is the most preferred method. Unlike in Saccharomyces cerevisiae, where homologous recombination (HR) predominates, non-homologous end-joining (NHEJ) is a frequent process in P. pastoris. The ratio of NHEJ and HR can be shifted towards HR by elongating the length of the homologous regions flanking the actual expression cassettes and by suppressing NHEJ efficiency (Näätsaari et al. 2012).
The standard vector systems for intracellular and secretory expression provided by Life Technologies (Carlsbad, CA, USA) include constitutive (P GAP) and inducible promoters triggered by methanol or methylamine (P AOX1, P FLD). The recently introduced PichiaPink™ expression kit for intracellular or secreted expression enables easy selection of multicopy integration clones by differences in colour formation based on ade2 knockout strains and truncated ADE2 promoters of varying strengths in front of the ADE2 marker gene (Du et al. 2012; Nett 2010).
Additionally, BioGrammatics (Carlsbad, CA, USA) holds licences for selling standard P. pastoris expression vectors and strains and also provides GlycoSwitch® vectors for humanized glycosylation of target proteins (Table 2). Several vectors for disruption of OCH1 and expression of different glycosidases or glycosyltransferases are available to achieve mammalian-type N-glycan structures in P. pastoris. These vectors harbour, for example, the human GlcNAc transferase I, the mannosidase II from rat, or the human galactosyl transferase I. A detailed protocol for humanizing the glycosylation pattern using the GlycoSwitch® vectors is provided (Jacobs et al. 2009).
Table 2.
Commercial vector systems
| Supplier | Promoter | Signal sequences | Selection in yeast | Selection in bacteria | Comments |
|---|---|---|---|---|---|
| Life Technologies™ | AOX1, FLD1,GAP | S. cerevisiae α-MF; P. pastoris PHO1 | Blasticidin, G418, Zeocin™, HIS4 | Zeocin™, Ampicillin, Blasticidin | c-myc epitope, V5 epitope, C-terminal 6× His-tag available for detection/purification |
| Life Technologies –PichiaPink™ | AOX1 | α-MF; set of eight different signal sequences – not ready to usea | ADE2 | Ampicillin | Low- and high-copy vectors available, TRP2 sequence for targeting |
| BioGrammatics | AOX1 | α-MF | Zeocin™, G418, Nourseothricin | Ampicillin | Intracellular or secreted expression |
| BioGrammatics – GlycoSwitch® | GAP | – | Zeocin™, G418, Hygromycin, HIS4, Nourseothricin | Zeocin™, Ampicillin, Kanamycin, Nurseothricin | Human GlcNAc transferase I, rat Mannosidase II, human Gal transferase I |
| DNA2.0 | AOX1 | Ten different signal sequences – ready to useb | Zeocin™, G418 | Zeocin™, Ampicillin | Intracellular or secreted |
aThe different secretion signals have to be cloned into the vector by a three-way ligation step
bThe α-MF secretion signal is provided once with Kex2p (KR) and Ste13p cleavage sites (EAEA), once lacking EA repeats, and once as truncated version (pre-region only)
James Cregg’s laboratory at the Keck Graduate Institute, Claremont, CA, USA, has developed a set of plasmids for protein secretion and intracellular expression in P. pastoris containing the strong AOX1 promoter. These vectors are based on different auxotrophy markers, such as ARG4, ADE1, URA3 and HIS4, for selection necessitating the use of the appropriate host strains (see section “Host strain development”). The vectors contain restriction sites for linearization within the marker genes to target the expression cassettes to the desired locus as well as for multicopy integration (Lin-Cereghino et al. 2001). Moreover, a set of integration vectors for sequential disruption of ARG1, ARG2, ARG3, HIS1, HIS2, HIS5 and HIS6 in P. pastoris was applied to provide the host strains for engineering the protein glycosylation pathway (Nett et al. 2005).
The Institute of Molecular Biotechnology, Graz University of Technology, Austria, provides vectors and strains to the P. pastoris community through the so-called ‘Pichia Pool’. The pPp plasmids described by Näätsaari et al. (2012) comprise vectors containing the GAP or AOX1 promoters and, for secretory expression, the S. cerevisiae α-mating factor (α-MF) secretion signal. The antibiotic selection marker cassettes were placed under the control of ADH1 or ILV5 promoters in the pPpB1 and pPpT4 vectors, respectively. It is described that the pPpT4-based vectors usually lead to lower gene copies in the cell as compared to the pPpB1-based vectors.
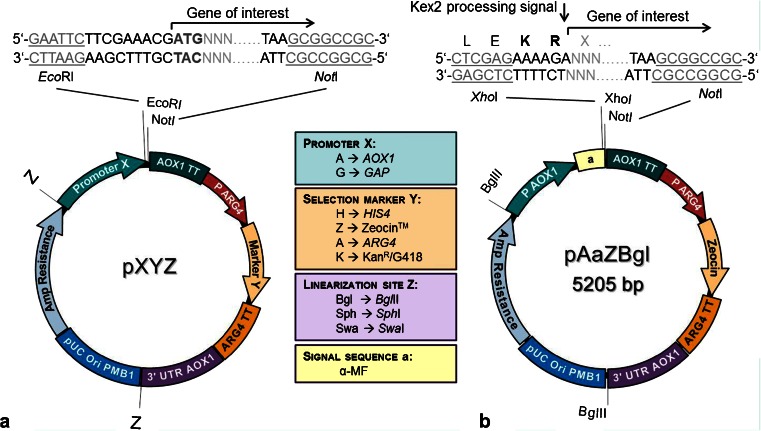
Further vectors based on either the GAP or the AOX1 promoter and a series of strains have recently been added to this pool, both for intracellular and secretory protein expression (M. Ahmad, unpublished results). For intracellular expression, cloning of the target genes is accomplished by using EcoRI and NotI, whereby the Kozak consensus sequence has to be restored for efficient translation initiation (Fig. 2a). A special characteristic of these vectors is that the EcoRI site has been introduced by a single point mutation directly into the AOX1 promoter sequence without changing the promoter activity. Thereby, the gene of interest may be fused to the promoter without having additional nucleotides between the promoter and the start codon. Another advantage is the use of the short ARG4 promoter for the expression of the selection markers. The weaker ARG4 promoter used for selection marker cassettes enables selection at lower concentrations of Zeocin™ (i.e., 25 instead of 100 μg/ml) without obtaining false-positive clones. For secretory expression governed by the S. cerevisiae α-MF signal sequence, XhoI and/or NotI sites are used for cloning the genes of interest (Fig. 2b).
Fig. 2.
Novel ‘Pichia Pool’ plasmid sets for intracellular and secretory expression. a General features of pXYZ vector for intracellular expression. Letters refer to the choice of promoters (X), selection markers (Y), and restriction enzymes (Z) for linearization. Available elements are shown in boxes. The vector backbone harbours an ampicillin resistance marker and origin of replication for maintenance of the plasmid in E. coli. The GOI is EcoRI–NotI cloned directly after the promoter of choice. The Kozak consensus sequence for yeast (i.e., CGAAACG), should be restored between the EcoRI cloning site and the start codon of the GOI in order to achieve optimal translation. In addition, sequence variation within this region will allow fine-tuning translation initiation efficiency. Expression in P. pastoris is driven either by the methanol inducible AOX1 or the constitutive GAP promoter. Positive clones can be selected for by antibiotic resistance (i.e., to Zeocin™ or geneticin sulphate) or by selection for His or Arg prototrophy. Selection marker expression is uniformly driven by the ARG4 promoter–terminator pair. b Plasmid pAaZBgl from ‘Pichia Pool’ is shown as an example of a vector made for secretory expression encoding S. cerevisiae α-MF signal sequence in front of the GOI cloning site. The Kex2 processing site AAAAGA should be restored between the XhoI cloning site and the fusion point of the GOI
Aspects of secretory expression
One of the main advantages of using P. pastoris as a protein production host is its ability to secrete high titres of properly folded, post-translationally processed and active recombinant proteins into the culture media. As a rule of thumb, proteins secreted in their native hosts will also be secreted in P. pastoris. However, there are also some reports of successful secretion of typically intracellular proteins such as GFP or human catalase (Eiden-Plach et al. 2004; Shi et al. 2007). The most commonly employed secretion signals in P. pastoris are derived from S. cerevisiae α-MF, S. cerevisiae invertase (SUC2) and the P. pastoris endogenous acid phosphatase (PHO1) (Daly and Hearn 2005). As listed in Table 2, commercial kits also provide vectors with different secretion signals, which allows for screening of the best-suited signal sequence.
The α-MF signal sequence is composed of a pre- and pro-region and has proven to be most effective in directing protein through the secretory pathway in P. pastoris. The pre-region is responsible for directing the nascent protein post-translationally into the endoplasmic reticulum (ER) and is cleaved off subsequently by signal peptidase (Waters et al. 1988). The pro-region is thought to play a role in transferring the protein from ER to Golgi compartment and is finally cleaved at the dibasic KR site by the endo-protease Kex2p (Julius et al. 1984). The two EA repeats are subsequently trimmed by the STE13 gene product (Brake et al. 1984). One of the common problems encountered while using the α-MF secretion signal is non-homogeneity of the N-termini of the recombinant proteins due to incomplete STE13 processing. Constructs without the EA repeats may enhance homogeneity at the N termini of recombinant proteins. However, the removal of these sequences may affect protein yield. While no reports on enhanced co-expression of STE13 are available, co-overexpression of HAC1, a transcription factor in the unfolded protein response (UPR) pathway, with the membrane protein adenosine A2 receptor had a positive effect on proper processing of the α-MF signal sequence (Guerfal et al. 2010). Recently, Yang et al. (2013) reported enhanced secretory protein production by optimizing the amino acid residues at the Kex2 P1′ site.
Multiple strategies have been followed to enhance the secretory potential of the α-MF signal sequence including codon optimization (Kjeldsen et al. 1998), directed evolution (Rakestraw et al. 2009), insertion of spacers and deletion mutagenesis (Lin-Cereghino et al. 2013). Directed evolution of the α-MF signal sequence in S. cerevisiae resulted in up to 16-fold enhanced full-length IgG1 secretion as compared to the wild type. Furthermore, when this improved leader sequence was combined with strain engineering strategies comprising PDI overexpression and elimination of proteins involved in vacuolar targeting, up to 180-fold enhanced secretion of the reporter protein was observed (Rakestraw et al. 2009). Deletion mutagenesis based on a predicted structure model of α-MF signal peptide resulted in 50 % increased secretion of horseradish peroxidase and C. antarctica lipase B (CALB) in P. pastoris (Lin-Cereghino et al. 2013). It appears that decreasing the hydrophobicity of the leader sequence by deleting hydrophobic residues or substituting them with more polar or charged residues increased the flexibility of the α-MF signal sequence structure, which enhanced the overall secretory capacity of the pro-region. Alternative signal sequences used to direct protein secretion and their features and applications are summarized in Table 3.
Table 3.
Signal sequences used to secrete the protein into the extracellular space
| Secretion signal | Source | Target protein(s) | Length | Reference |
|---|---|---|---|---|
| α-MF | S.c. α-mating factor | Most commonly used secretion signal in P. pastoris | 85 aa, with or without EA repeats | (Brake et al. 1984) |
| PHO1 | P.p. acid phosphatase | Mouse 5-HT5A, porcine pepsinogen, | 15 aa | (Payne et al. 1995; Weiss et al. 1995; Yoshimasu et al. 2002) |
| SUC2 | S.c. Invertase | Human interferon, α-amylase, α-1-antitrypsin | 19 aa | (Moir and Dumais 1987; Paifer et al. 1994; Tschopp et al. 1987b) |
| PHA-E | Phytohemagglutinin | GNA, GFP and native protein | 21 aa | (Raemaekers et al. 1999) |
| KILM1 | Kl toxin | CM cellulase | 44 aa | (Skipper et al. 1985) |
| pGKL | pGKL killer protein | Mouse α-amylase | 20 aa | (Kato et al. 2001) |
| CLY and CLY-L8 | C-lysozyme and syn. leucin-rich peptide | Human lysozyme | 18 and 16 aa | (Oka et al. 1999) |
| K28 pre-pro-toxin | K28 virus toxin | Green fluorescent protein | 36 aa | (Eiden-Plach et al. 2004) |
| Scw, Dse and Exg | P.p. Endogenous signal peptides | CALB and EGFP | 19, 20 and 23 aa | (Liang et al. 2013a) |
| Pp Pir1 | P.p. Pir1p | EGFP and Human α1-antitrypsin | 61 aa | (Khasa et al. 2011) |
| HBFI and HBFII | Hydrophobins of Trichoderma reesei | EGFP | 16 and 15 aa | (Kottmeier et al. 2011) |
Beyond the choice of the secretion signals there are several other factors that govern efficient protein secretion. The newly synthesized proteins are translocated co- or post-translationally into the ER lumen through the Sec61p translocon. Then, proteins may undergo one or several posttranslational modifications, folding into the native state, disulphide-bond formation, glycosylation and membrane-anchoring. When the recombinant protein fails to fold into its native state or protein expression exceeds the folding capacity of the ER (Sha et al. 2013), unfolded proteins may start to aggregate, triggering the UPR pathway. UPR is responsible for induction of genes that are involved in protein folding. In parallel to UPR pathway, ER-associated degradation (ERAD) by the proteasome may relieve blocks in protein secretion (recently reviewed by Idiris et al. 2010 and Damasceno et al. 2012). Inappropriate mRNA structure and gene copy numbers, limits in transcription, translation and protein translocation into the ER, incomplete protein folding and inefficient protein targeting to the exterior of the cell are major bottlenecks encountered in secretory expression of heterologous proteins. Commonly used strategies to overcome such secretory bottlenecks comprise the overexpression of folding helper proteins like BiP/Kar2p, DnaJ, PDI, PPIs and Ero1p or, alternatively, overexpression of HAC1, a transcriptional regulator of the UPR pathway genes. Unlike in S. cerevisiae, Guerfal et al. (2010) reported that HAC1 is constitutively expressed and spliced in P. pastoris under normal growth conditions, which may explain the higher titers of secreted proteins obtainable with this organism. A contradictory observation was reported by Whyteside et al. (2011). Un-spliced HAC1 mRNA was detected under normal growth conditions and splicing of HAC1 mRNA was only detected when cells were grown in presence of dithiothreitol (DTT) to activate the UPR. It should be mentioned, though, that sometimes overexpression of folding helpers actually reduced protein secretion or did not have any effect (van der Heide et al. 2002).
Host strain development
Elucidation of full genome sequences and gene annotation were great steps toward rational strain engineering, identifying new promoters and progressing in the (systems) biology of P. pastoris (Küberl et al. 2011; Mattanovich et al. 2009a; De Schutter et al. 2009). Two online databases (http://bioinformatics.psb.ugent.be/orcae/overview/Picpa and http://www.pichiagenome.org) provide convenient access to genome sequences and annotations. Frequently used commercially available strains are the his4 strain GS115, the reconstituted prototrophic strain X-33, the aox1 knockout strains KM71 and KM71H as well as protease-deficient strains SMD1168 and SMD1168H and the ade2 auxotrophic PichiaPink™ strain. Use of these strains for commercial applications, however, is restricted by patent protection and/or materials ownership policy. Strains derived from P. pastoris CBS7435, in contrast, are not covered by patent protection and, therefore represent an alternative for production purposes. Furthermore, the CBS7435 MutS strain provided by the Graz Pichia Pool has the advantage of being marker-free as it was constructed using the Flp/FRT recombinase system for marker removal (Näätsaari et al. 2012). Using the same strategy, ade1 and his4 knockout strains were created along with the CBS7435 ku70 strain (CBS 12694), which is impaired in the NHEJ mechanism, thereby enhancing the efficiency of HR. A selection of most relevant strains is compiled in Table 4.
Table 4.
P. pastoris host strains
| Strain | Genotype | Phenotype | Source |
|---|---|---|---|
| Wild-type strains | |||
| CBS7435 (NRRL Y-11430) | WT | WT | Centraalbureau voor Schimmelcultures, the Netherlands |
| CBS704 (DSMZ 70382) | WT | WT | Centraalbureau voor Schimmelcultures, the Netherlands |
| X-33 | WT | WT | Life Technologies™ |
| Auxotrophic strains | |||
| GS115 | his4 | His− | Life Technologies™ |
| PichiaPink™ 1 | ade2 | Ade− | Life Technologies™ |
| KM71 | his4, aox1::ARG4, arg4 | His−, MutS | Life Technologies™ |
| KM71H | aox1::ARG4, arg4 | MutS | Life Technologies™ |
| BG09 | arg4::nourseo R Δlys2::hyg R | Lys−, Arg−, NourseothricinR, HygromycinR | BioGrammatics |
| GS190 | arg4 | Arg− | (Cregg et al. 1998) |
| GS200 | arg4 his4 | His−, Arg− | (Waterham et al. 1996) |
| JC220 | ade1 | Ade− | (Cregg et al. 1998) |
| JC254 | ura3 | Ura− | (Cregg et al. 1998) |
| JC227 | ade1 arg4 | Ade− Arg− | (Lin-Cereghino et al. 2001) |
| JC300-JC308 | Combinations of ade1 arg4 his4 ura3 | Combinations of Ade−, Arg−, His−, Ura− | (Lin-Cereghino et al. 2001) |
| YJN165 | ura5 | Ura− | (Nett and Gerngross 2003) |
| CBS7435 his4 a | his4 | His− | (Näätsaari et al. 2012) |
| CBS7435 MutS his4 a | aox1, his4 | MutS, His− | (Näätsaari et al. 2012) |
| CBS7435 MutS arg4 a | aox1, arg4 | MutS, Arg− | (Näätsaari et al. 2012) |
| CBS7435 met2 a | met2 | Met− | (Pp7030)b |
| CBS7435 met2 arg4 a | met2 arg4 | Met− Arg− | (Pp7031)b |
| CBS7435 met2 his4 a | met2 his4 | Met− His− | (Pp7032)b |
| CBS7435 lys2 a | lys2 | Lys− | (Pp7033)b |
| CBS7435 lys2 arg4 a | lys2 arg4 | Lys− Arg− | (Pp7034)b |
| CBS7435 lys2 his4 a | lys2 his4 | Lys− His− | (Pp7035)b |
| CBS7435 pro3 a | pro3 | Pro− | (Pp7036)b |
| CBS7435 tyr1 a | tyr1 | Tyr− | (Pp7037)b |
| Protease-deficient strains | |||
| SMD1163 | his4 pep4 prb1 | His− | (Gleeson et al. 1998) |
| SMD1165 | his4 prb1 | His− | (Gleeson et al. 1998) |
| SMD1168 | his4 pep4::URA3 ura3 | His− | Life Technologies™ |
| SMD1168H | pep4 | Life Technologies™ | |
| SMD1168 kex1::SUC2 | pep4::URA3 kex1::SUC2 his4 ura3 | His− | (Boehm et al. 1999) |
| PichiaPink 2-4 | Combinations of prb1/pep4 | Ade− | Life Technologies™ |
| BG21 | sub2 | BioGrammatics | |
| CBS7435 prc1 a | prc1 | (Pp6676)b | |
| CBS7435 sub2 a | sub2 | (Pp6668)b | |
| CBS7435 sub2 a | his4 pep4 | His− | (Pp6911)b |
| CBS7435 prb1 a | prb1 | (Pp6912)b | |
| CBS7435 his4 pep4 prb1 | his4 pep4 prb1 | His− | (Pp7013)b |
| Glyco-engineered strains | |||
| SuperMan5 | his4 och1::pGAPTrα1,2-mannosidase | His−, BlasticidinR | BioGrammatics |
| och1::pGAPTrα1,2-mannosidase | BlasticidinR | BioGrammatics | |
| pep4 och1::pGAPTrα1,2-mannosidase | BlasticidinR | BioGrammatics | |
| Other strains | |||
| GS241 | fld1 | Growth defect on methanol as sole C-source or methylamine as sole N-source | (Shen et al. 1998) |
| MS105 | his4 fld1 | See GS241; His− | (Shen et al. 1998) |
| MC100-3 | his4 arg4 aox1::ScARG4 aox2::PpHIS4 | Mut− | (Cregg et al. 1989) |
| CBS7435 ku70 a | ku70 | WT | (Näätsaari et al. 2012) |
| CBS7435 ku70 his4 a | ku70, his4 | His− | (Näätsaari et al. 2012) |
| CBS7435 ku70 gut1 | ku70, gut1 | Growth defect on glycerol; ZeocinR | (Näätsaari et al. 2012) |
| CBS7435 ku70 ade1 | ku70, ade1 | Ade−, ZeocinR | (Näätsaari et al. 2012) |
aThese P. pastoris CBS7435 derived strains are marker-free knockouts
bStrains from ‘Pichia Pool’ of TU Graz (M. Ahmad, unpublished results)
Auxotrophic strains
Several auxotrophic strains (e.g., ade1, arg4, his4, ura3, met2), and combinations thereof are available together with vectors harbouring the respective genes as selectable markers (Lin-Cereghino et al. 2001; Thor et al. 2005, Graz Pichia Pool). Auxotrophic strains have been useful for in vivo labelling of proteins, for example in the global fluorination of Candida antarctica lipase B (CALB) in a P. pastoris X-33 aro1 strain deficient in tryptophan, tyrosine, and phenylalanine biosynthesis (Budisa et al. 2010). Fluorinated analogues of these amino acids were supplemented and incorporated into the heterologous protein, thereby, for example, prolonging CALB shelf-life but lowering its lipase activity. The proteolytic pattern of CALB was retained, though. Another example is the use of a lys2 arg4 double knockout strain for stable isotope labelling by amino acids in cell culture (SILAC) (Austin et al. 2011).
Protease-deficient strains
Undesired proteolysis of heterologous proteins expressed in P. pastoris does not only lower the product yield or biological activity, but also complicates downstream processing of the intact product as the degradation products will have similar physicochemical and affinity properties. Proteolysis may occur either during vesicular transport of recombinant protein by secretory pathway-resident proteases (Werten and de Wolf 2005; Ni et al. 2008) or in the extracellular space by proteases being secreted, cell wall-associated (Kang et al. 2000) or released into the culture medium as a result of cell disruption during high cell density cultivation (Sinha et al. 2005). Different strategies have been employed to address the proteolysis problem, namely, modifying fermentation parameters (pH, temperature and specific growth rate), changing the media composition (rich medium, addition of casamino acids or peptone as competing substrates), lowering the salt concentration and addition of soytone (Zhao et al. 2008), applying protein engineering strategies (Gustavsson et al. 2001) and engineering of the expression host to obtain protease-deficient strains (reviewed by Idiris et al. 2010 and Macauley-Patrick et al. 2005). However, in some cases, optimization of the fermentation media and protein engineering strategies failed to alleviate the proteolysis problem and tuning the expression host itself was the only viable option (Li et al. 2010). The use of protease-deficient strains such as SMD1163 (Δhis4 Δpep4 Δprb1), SMD1165 (Δhis4 Δprb1) and SMD1168 (Δhis4 Δpep4) has been well documented for the expression of protease-sensitive proteins (Gleeson et al. 1998). PEP4 encodes a major vacuolar aspartyl protease which is able to activate itself as well as further proteases such as carboxypeptidase Y (PRC1) and proteinase B (PRB1). The use of protease-deficient strains other than the above mentioned (e.g., yps1, kex1, kex2) was reported with variable success (Ni et al. 2008; Werten and de Wolf 2005; Wu et al. 2013; Yao et al. 2009). A general conclusion from these studies is that in many cases several proteases are involved in degradation events and, therefore, it is not an easy task to optimize protein expression by knocking out just a single one. However, the pep4 and prb1 knockout strains are still the most effective ones in preventing recombinant protein degradation, and, hence, also the most widely applied. Although it has been reported that protease-deficient strains show typically slower growth rates, lower transformation efficiencies and reduced viability (Lin-Cereghino and Lin-Cereghino 2007), experiments in our laboratory showed robust growth behaviour of 28 protease-deficient strains that were recently created (M. Ahmad, unpublished results).
Glyco-engineered strains
When yeasts such as P. pastoris are chosen for production of therapeutic proteins, N- and O-linked glycosylation are of tremendous relevance. Although the assembly of the core glycans, that is, (Man)8-(GlcNAc)2, in the ER is highly conserved in mammals and yeasts, mammals provide a much higher diversity in the ultimate glycan structure assembled in the Golgi cisternae. Yeasts, in contrast, produce high mannose glycan structures, which may lead to decreased serum half-life and may trigger allergic reactions in the human body (Ballou 1990). While in P. pastoris the hyper-mannosylation is not as prominent as in S. cerevisiae, it is still a problem that needs to be tackled, and is therefore a target for intensive strain engineering. A very detailed summary of the glycosylation machinery and the targets for glyco-engineering in different yeast species, including P. pastoris, has been given recently (De Pourcq et al. 2010). To sum up briefly, engineering strategies included the introduction of a Trichoderma reesei α-1,2-mannosidase (Callewaert et al. 2001), the knockout of the highly conserved yeast Golgi protein α-1,6-mannosyltransferase encoded by OCH1, which is responsible for hyperglycosylation (Choi et al. 2003; Vervecken et al. 2004), as well as co-overexpression of several glycosyltransferases and glycosidases carrying proper targeting signals (Hamilton et al. 2003). Terminally sialylated glycoproteins produced for the first in P. pastoris were obtained by introducing a complex sialic acid pathway (Hamilton et al. 2006). Key to success was the correct localization of the heterologous glycosyltransferases and glycosidases in the ER and Golgi networks. Combinatorial genetic libraries and high throughput screening methods were successfully applied to find the best targeting signal/enzyme combinations for N-linked glycoengineering (Nett et al. 2011). Furthermore, a useful guide to glyco-engineering in P. pastoris by using the GlycoSwitch® technology was described by Jacobs et al. (2009). These strategies, altogether, enable the production of valuable biopharmaceuticals with a more homogeneous, ‘humanized’ N-glycosylation pattern.
However, as yeasts also carry out O-glycosylation that differs structurally from the mammalian type (Strahl-Bolsinger et al. 1999), O-glycosylation has also been an interesting target for engineering. In P. pastoris, O-linked glycosylation is initiated with a mannose monosaccharide, which is further elongated by α-1,2-mannose residues and finally capped with β- or phospho-mannose residues. Until lately, the engineering strategies were limited to the use of an inhibitor of the major ER located protein-O-mannosyltransferases (PMTs) as the deletion of these genes did not yield robust and viable strains. The characterization of the P. pastoris PMT gene family was an important step forward in O-glycosylation engineering (Nett et al. 2013). In this study, the knockout of PMTs as well as the use of PMT inhibitors led to a reduced number of O-mannosylation events and, furthermore, to reduced chain lengths of the O-glycans. A follow-up study described the production of a TNFR2:Fc1 fusion protein carrying sialylated O-linked glycans in P. pastoris (Hamilton et al. 2013). Therein, an α-1,2-mannosidase as well as a protein-O-linked-mannose β-1,2-N-acetylglucosaminyl-transferase 1 (PomGnT1) were co-expressed in a P. pastoris strain, that was already engineered in its N-glycosylation pathway. Hence, the mannose residues were first trimmed to single O-linked mannose residues, which were then capped with N-acetylglucosamine. This structure was extended with sialic acid residues to achieve human-like O-glycan residues similar to the α-dystroglycan-type. However, there is still room for improvement, for example by engineering P. pastoris towards human mucin-type O-glycosylation.
Expression strategies and industrial applications
Screening for high level expression
Subsequent to the choice of suitable expression vectors and proper host strains, and transformation of the expression cassettes, it is important to select for transformants which show high expression levels of the desired protein. Single copy transformants can be easily generated by targeting the linear expression cassettes to the AOX1 locus resulting in gene replacement events. Ectopic integrations may simultaneously occur, however. Transformants resulting from gene replacement at the AOX1 locus have methanol utilization slow phenotype (MutS) and can be easily identified by replica-plating on minimal methanol plates. The most commonly applied strategy to screen for high-yielding P. pastoris transformants focusses on screening for clones having multicopy integrations of the expression cassette. A recent detailed review describes the methods applied to obtain strains containing multiple expression cassettes and provides a summary of published data showing correlations between copy number and expression levels of intracellular as well as secreted proteins. It also highlights the problem of genetic instability of the integration cassettes that might be encountered when cultivating multicopy strains. Due to the highly recombinogenic nature of P. pastoris, expression cassettes might be excised through loop-out recombination. This effect seems to be more pronounced the more copies are integrated (Aw and Polizzi 2013).
Regarding the correlation between copy number and expression level, a number of recent studies have shown a direct correlation especially for intracellular expression (Marx et al. 2009; Vassileva et al. 2001). The direct correlation of expression level and gene copy number is, however, not necessarily valid when the protein is directed to the secretory pathway. The most commonly employed method of generating multicopy expression strains in P. pastoris is based on plating the transformation mixture directly on selection plates containing increasing concentrations of antibiotics (e.g., 100 to 2,000 μg/ml of Zeocin™). The majority of transformants will have a single copy of the expression vector integrated into the genome, and numerous clones will have to be screened to find high-copy transformants (Lin-Cereghino and Lin-Cereghino 2007). Therefore, several high-throughput methods have been established to screen a large number of clones based on small-scale cultivation in deep well plates (Mellitzer et al. 2012; Weinhandl et al. 2012; Weis et al. 2004). The selected clones, however, might not perform as well in fermenter cultivations due to different cultivation conditions. A further pronounced problem of resistance marker based screening is a high prevalence of false-positive colonies. This so-called high transformation background is supposedly caused by cell stress and cell rupture. Depending on the mechanism of antibiotic resistance conferred by the resistance marker, un-transformed cells may survive in the vicinity of ruptured transformants. This problem was addressed by constructing expression vectors based on marker gene expression driven by the weak ARG4 promoter (Pichia Pool, Fig. 2). This ensures basal levels of expression, thereby allowing handlers to select single copy to multicopy strains by plating the transformants directly on low concentrations of Zeocin™ (i.e., 25 μg/ml for single copy and up to 400 μg/ml for multi-copy transformants). Thus, transformants having 1 to 20 (±5) copies can be selected. To reduce the chances of having single copy transformants, regeneration time should be kept short and transformants should be plated directly on increased concentrations of antibiotic. By employing this method, only few transformants survive on high concentrations of antibiotic, but will most likely contain multiple copies, which can be determined by quantitative (qPCR) or Southern blot analysis (M. Ahmad, unpublished results). Performance can then be tested directly under production conditions in bioreactor cultivations instead of small-scale cultivations in deep well plates or shake flasks.
Membrane protein expression
P. pastoris has been shown to produce 15+ g of soluble recombinant protein per litre of culture intracellularly (Hasslacher et al. 1997) or in secretory mode (Werten et al. 1999). Key to such high titres is the ability of P. pastoris to grow to very high cell densities reaching up to 150 g cell dry weight per litre of fermentation broth in fed-batch bioreactor cultivations (Jahic et al. 2006). At very high cell densities, even proteins that are present in limited entities per single cell can be produced with reasonable volumetric yields in P. pastoris. Typical examples of non-abundant proteins with high scientific and commercial relevance are integral membrane proteins. Being the targets of >50 % of drugs applied on humans (Arinaminpathy et al. 2009), only very few membrane proteins have been characterized on the molecular level regarding structure–function relationships. The simple reason is that it is difficult to obtain sufficient purified membrane protein for structural and biochemical studies, unless affinity-tagged membrane proteins are obtained at reasonable yield. Actually, P. pastoris has been applied routinely to produce affinity-tagged membrane proteins for protein purification and subsequent biochemical studies (Cohen et al. 2005; Haviv et al. 2007; Lifshitz et al. 2007). Furthermore, P. pastoris has been the expression host of choice for elucidating the crystal structures of membrane proteins from diverse origins, even from higher eukaryotes (Brohawn et al. 2012; Hino et al. 2012; Ho et al. 2009).
Evolutionary proximity of a heterologous expression host and the origin of an expressed membrane protein are beneficial for successful recombinant expression (Grisshammer and Tateu 2009). In addition to the intramolecular forces and bonds, ions, cofactors and interacting proteins that stabilize soluble proteins, membrane proteins are usually interacting with and are partially also stabilized by the lipids of the surrounding bilayers (Adamian et al. 2011). As P. pastoris and other yeast expression hosts do significantly differ in their membrane compositions from bacterial, plant or animal cells (Wriessnegger et al. 2007, 2009; Zinser and Daum 1995), heterologous membrane proteins may face stability issues upon expression in distantly related hosts. Thus, multiple approaches have been undertaken to improve P. pastoris host strains and expression conditions for membrane protein production. Applying similar tools as for the optimisation of soluble protein expression — that is, manipulation of expression conditions, addition of chemical chaperones, co-expression of chaperones or of proteins activating UPR, use of protease deficient strains, etc. — has been showing some, however often target-specific success in membrane protein expression. A novel approach is the engineering of P. pastoris cellular membranes for improved accommodation of heterologous membrane proteins. In the first reported example, a cholesterol-producing P. pastoris strain was shown to stably express an enhanced level of ligand-binding human Na,K-ATPase moieties on the cell surface (Hirz et al. 2013).
Products on — or on the way to — the market
The P. pastoris expression system has gained importance for industrial application as highlighted by the number of patents published on heterologous expression in and cell engineering of P. pastoris (Bollok et al. 2009). Products obtained by heterologous expression in P. pastoris have already found their way to the market, as FDA approved biopharmaceuticals or industrial enzymes have shown. The www.pichia.com web page provides a list of proteins produced in P. pastoris with the commercial expression system licensed by Research Corporation Technologies (RCT) and their applications: Phytase (Phytex, Sheridan, IN, USA) is applied as animal feed additive to cleave plant derived phytate, thereby providing a source of phosphate. Trypsin (Roche Applied Science, Germany) is used, for example, as protease in proteomics research to obtain peptide patterns for MS analysis. Further examples listed are nitrate reductase (The Nitrate Elimination Co., Lake Linden, MI, USA), used for water testing and treatment, phospholipase C (Verenium, San Diego, CA, USA/DSM, The Netherlands), used for degumming of vegetable oils, and Collagen (Fibrogen, San Francisco, CA, USA), used in medical research and as dermal filler. Thermo Scientific (Waltham, MA, USA) sells recombinant Tritirachium album Proteinase K produced in P. pastoris. Concerning biopharmaceuticals, a famous example is Kalbitor® (ecallantide), produced in P. pastoris by Dyax (Cambridge, MA, USA). Kalbitor® is a plasma kallikrein inhibitor indicated against hereditary angioedema. This product was the first biopharmaceutical to be approved by the FDA for market release in 2009 (Walsh 2010). As can be found on the web page of RCT (www.rctech.com), Pichia-manufactured Jetrea®, a drug used for treatment of symptomatic vitreomacular adhesion, was recently approved by the FDA and the European Commission. Other Pichia-derived products provided by the Indian company Biocon are recombinant human insulin and analogues thereof (Insulin, Glargine). Products under development, such as Elastase inhibitor against Cystic fibrosis or Nanobody® ALX antibody fragments developed by Ablynx (Belgium), are also listed by Gerngross (2004) and on www.pichia.com. In 2008, Novozymes (Denmark), which found a highly active antimicrobial agent, the plectasin peptide derivative NZ2114 (Andes et al. 2009; Mygind et al. 2005), granted Sanofi-Aventis (France) an exclusive licence for the production and commercialisation of this compound in P. pastoris. This might be the first antimicrobial peptide approved for the market in the future.
Although not yet approved for medical use, many products can be found on the market for research purposes. GenScript (Piscataway, NJ, USA) provides recombinant cytokines and growth factors, such as human HSA-IFN-Alpha 2b, human Stem Cell Factor SCF, murine TNF-α and ovine IFN-τ, to name just a few examples. Recombinant human angiostatin can be found for instance in the reagents offered by Sigma-Aldrich (St. Louis, MO, USA).
Future perspectives — outlook
Successful expression of many industrial enzymes as well as pharmaceutically relevant proteins has rendered the methylotrophic yeast P. pastoris one of the most suitable and powerful protein production host systems. It is also an emerging host for the expression of membrane proteins (Hirz et al. 2013) and of small bioactive and antimicrobial peptides, which could be a forthcoming alternative to chemical synthesis (Zhang et al. 2014). Although many basic elements of this expression system are now well developed and one can make use of a broad variety of vectors and host strains, there is still space for further optimization of protein expression and secretion, which, in many cases, will be highly dependent on the desired product. One general interest is to find effective alternatives for induction to replace methanol for industrial scale fermentations (Delic et al. 2013; Prielhofer et al. 2013; Stadlmayr et al. 2010).
Improving protein secretion performance is one of the first and foremost goals for engineering P. pastoris. There is still potential to increase yields, for example, by employing different secretion signals (Vadhana et al. 2013) or mutating S. cerevisiae α-MF (Lin-Cereghino et al. 2013). In contrast to the well-studied secretory pathway of S. cerevisiae, P. pastoris still is a black box regarding factors influencing secretion efficiency. Current studies try to identify these factors by mutagenesis approaches and screening for enhanced secretion of reporter proteins (Larsen et al. 2013; C. Winkler and H. Pichler, unpublished results). The well-developed tools for strain engineering, including marker-free integration and deletion of desired genes, will provide a powerful set of engineered designer host strains in the near future. These will provide optimized cell factories by fine-tuned co-expression of important homologous or heterologous protein functions needed for efficient and accurate functional expression, secretion and post-translational modification of proteins. Moreover, knockout or knockdown of undesired functions such as proteolytic decay will increase product quality and process performance. Considering the scope of this review on heterologous protein expression, it was not feasible to address all possible applications for P. pastoris as production organism, such as metabolic engineering for production of small molecules and metabolites, or for whole-cell biocatalysis. However, developments in these fields may also be relevant for constructing improved host strains dedicated for protein production. There are several recent reviews and research articles describing advances in these fields in detail (Abad et al. 2010; Araya-Garay et al. 2012; Wriessnegger and Pichler 2013).
Footnotes
Ectodomain of tumor necrosis factor 2 with crystallizable fragment of IgG1 (Fc)
Mudassar Ahmad and Melanie Hirz contributed equally to this work.
References
- Abad S, Nahalka J, Bergler G, Arnold SA, Speight R, Fotheringham I, Nidetzky B, Glieder A. Stepwise engineering of a Pichia pastorisd-amino acid oxidase whole cell catalyst. Microb Cell Factories. 2010;9:24. doi: 10.1186/1475-2859-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamian L, Naveed H, Liang J. Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim Biophys Acta. 2011;1808:1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Hong J, Lee H, Park M, Lee E, Kim C, Choi E, Jung J, Lee H. Translation elongation factor 1-alpha gene from Pichia pastoris: molecular cloning, sequence, and use of its promoter. Appl Microbiol Biotechnol. 2007;74:601–608. doi: 10.1007/s00253-006-0698-6. [DOI] [PubMed] [Google Scholar]
- Ahn J, Hong J, Park M, Lee H, Lee E, Kim C, Lee J, Choi E, Jung J, Lee H. Phosphate-responsive promoter of a Pichia pastoris sodium phosphate symporter. Appl Environ Microbiol. 2009;75:3528–3534. doi: 10.1128/AEM.02913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes D, Craig W, Nielsen LA, Kristensen HH, In vivo pharmacodynamic characterization of a novel plectasin antibiotic, NZ2114, in a murine infection model. Antimicrob Agents Chemother. 2009;53:3003–3009. doi: 10.1128/AAC.01584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya-Garay JM, Feijoo-Siota L, Rosa-dos-Santos F, Veiga-Crespo P, Villa TG. Construction of new Pichia pastoris X-33 strains for production of lycopene and β-carotene. Appl Microbiol Biotechnol. 2012;93:2483–2492. doi: 10.1007/s00253-011-3764-7. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today. 2009;14:1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RJ, Kuestner RE, Chang DK, Madden KR, Martin DB. SILAC compatible strain of Pichia pastoris for expression of isotopically labeled protein standards and quantitative proteomics. J Proteome Res. 2011;10:5251–5259. doi: 10.1021/pr200551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw R, Polizzi KM. Can too many copies spoil the broth? Microb Cell Factories. 2013;12:128. doi: 10.1186/1475-2859-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou CE. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- Boehm T, Pirie-Shepherd S, Trinh LB, Shiloach J, Folkman J. Disruption of the KEX1 gene in Pichia pastoris allows expression of full-length murine and human endostatin. Yeast. 1999;15:563–572. doi: 10.1002/(SICI)1097-0061(199905)15:7<563::AID-YEA398>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Bollok M, Resina D, Valero F, Ferrer P. Recent patents on the Pichia pastoris expression system: expanding the toolbox for recombinant protein production. Recent Pat Biotechnol. 2009;3:192–201. doi: 10.2174/187220809789389126. [DOI] [PubMed] [Google Scholar]
- Brake AJ, Merryweather JP, Coit DG, Heberlein UA, Masiarz FR, Mullenbach GT, Urdea MS, Valenzuela P, Barr PJ. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984;81:4642–4646. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, del Mármol J, MacKinnon R. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science. 2012;335:436–441. doi: 10.1126/science.1213808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa N, Wenger W, Wiltschi B. Residue-specific global fluorination of Candida antarctica lipase B in Pichia pastoris. Mol BioSyst. 2010;6:1630–1639. doi: 10.1039/c002256j. [DOI] [PubMed] [Google Scholar]
- Callewaert N, Laroy W, Cadirgi H, Geysens S, Saelens X, Min Jou W, Contreras R. Use of HDEL-tagged Trichoderma reesei mannosyl oligosaccharide 1,2-alpha-d-mannosidase for N-glycan engineering in Pichia pastoris. FEBS Lett. 2001;503:173–178. doi: 10.1016/s0014-5793(01)02676-x. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Choi B-K, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li H, Miele RG, Nett JH, Wildt S, Gerngross TU. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc Natl Acad Sci U S A. 2003;100:5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofalo V, Barton N, Kreps J, Coats I, Shanahan D. Safety evaluation of a lipase enzyme preparation, expressed in Pichia pastoris, intended for use in the degumming of edible vegetable oil. Regul Toxicol Pharmacol. 2006;45:1–8. doi: 10.1016/j.yrtph.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Cohen E, Goldshleger R, Shainskaya A, Tal DM, Ebel C, le Maire M, Karlish SJD (2005) Purification of Na+, K+-ATPase expressed in Pichia pastoris reveals an essential role of phospholipid-protein interactions. J Biol Chem 280:16610–16618. doi:10.1074/jbc.M414290200 [DOI] [PubMed]
- Cregg J, Tolstorukov I (2012) P. pastoris ADH promoter and use thereof to direct expression of proteins. United States patent US 8,222,386
- Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Madden KR, Barringer KJ, Thill GP, Stillman CA. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol Cell Biol. 1989;9:1316–1323. doi: 10.1128/mcb.9.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Shen S, Johnson M, Waterham HR. Classical genetic manipulation. Methods Mol Biol. 1998;103:17–26. doi: 10.1385/0-89603-421-6:17. [DOI] [PubMed] [Google Scholar]
- Daly R, Hearn MTW. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- Damasceno LM, Huang C-J, Batt CA. Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol. 2012;93:31–39. doi: 10.1007/s00253-011-3654-z. [DOI] [PubMed] [Google Scholar]
- de Almeida JRM, de Moraes LMP, Torres FAG. Molecular characterization of the 3-phosphoglycerate kinase gene (PGK1) from the methylotrophic yeast Pichia pastoris. Yeast. 2005;22:725–737. doi: 10.1002/yea.1243. [DOI] [PubMed] [Google Scholar]
- De Pourcq K, De Schutter K, Callewaert N. Engineering of glycosylation in yeast and other fungi: current state and perspectives. Appl Microbiol Biotechnol. 2010;87:1617–1631. doi: 10.1007/s00253-010-2721-1. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Lin Y-C, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Delic M, Mattanovich D, Gasser B. Repressible promoters — a novel tool to generate conditional mutants in Pichia pastoris. Microb Cell Factories. 2013;12:6. doi: 10.1186/1475-2859-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Battles MB, Nett JH. A color-based stable multi-copy integrant selection system for Pichia pastoris using the attenuated ADE1 and ADE2 genes as auxotrophic markers. Bioeng Bugs. 2012;3:32–37. doi: 10.4161/bbug.3.1.17936. [DOI] [PubMed] [Google Scholar]
- Eiden-Plach A, Zagorc T, Heintel T, Carius Y, Breinig F, Schmitt MJ. Viral preprotoxin signal sequence allows efficient secretion of green fluorescent protein by Candida glabrata, Pichia pastoris, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. Appl Environ Microbiol. 2004;70:961–966. doi: 10.1128/AEM.70.2.961-966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SB, Brust PF, Koutz PJ, Waters AF, Harpold MM, Gingeras TR. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol Cell Biol. 1985;5:1111–1121. doi: 10.1128/mcb.5.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol. 2013;8:191–208. doi: 10.2217/fmb.12.133. [DOI] [PubMed] [Google Scholar]
- Gerngross TU. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- Gleeson MA, White CE, Meininger DP, Komives EA. Generation of protease-deficient strains and their use in heterologous protein expression. Methods Mol Biol. 1998;103:81–94. doi: 10.1385/0-89603-421-6:81. [DOI] [PubMed] [Google Scholar]
- Grisshammer R, Tateu CG. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 2009;28:315. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- Guerfal M, Ryckaert S, Jacobs PP, Ameloot P, Van Craenenbroeck K, Derycke R, Callewaert N. The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb Cell Factories. 2010;9:49. doi: 10.1186/1475-2859-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Lehtio J, Denman S, Teeri TT, Hult K, Martinelle M. Stable linker peptides for a cellulose-binding domain–lipase fusion protein expressed in Pichia pastoris. Protein Eng Des Sel. 2001;14:711–715. doi: 10.1093/protein/14.9.711. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, Wildt S, Gerngross TU. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Davidson RC, Sethuraman N, Nett JH, Jiang Y, Rios S, Bobrowicz P, Stadheim TA, Li H, Choi B, Hopkins D, Wischnewski H, Roser J, Mitchell T, Strawbridge RR, Hoopes J, Wildt S, Gerngross TU. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Cook WJ, Gomathinayagam S, Burnina I, Bukowski J, Hopkins D, Schwartz S, Du M, Sharkey NJ, Bobrowicz P, Wildt S, Li H, Stadheim TA, Nett JH. Production of sialylated O-linked glycans in Pichia pastoris. Glycobiology. 2013;23:1192–1203. doi: 10.1093/glycob/cwt056. [DOI] [PubMed] [Google Scholar]
- Hartner FS, Ruth C, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder A. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008;36:e76. doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasslacher M, Schall M, Hayn M, Bona R, Rumbold K, Lückl J, Griengl H, Kohlwein SD, Schwab H. High-level intracellular expression of hydroxynitrile lyase from the tropical rubber tree Hevea brasiliensis in microbial hosts. Protein Expr Purif. 1997;11:61–71. doi: 10.1006/prep.1997.0765. [DOI] [PubMed] [Google Scholar]
- Haviv H, Cohen E, Lifshitz Y, Tal DM, Goldshleger R, Karlish SJD. Stabilization of Na(+), K(+)-ATPase purified from Pichia pastoris membranes by specific interactions with lipids. Biochemistry. 2007;46:12855–12867. doi: 10.1021/bi701248y. [DOI] [PubMed] [Google Scholar]
- Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N, Cameron AD, Kobayashi T, Hamakubo T, Iwata S, Murata T. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirz M, Richter G, Leitner E, Wriessnegger T, Pichler H (2013) A novel cholesterol-producing Pichia pastoris strain is an ideal host for functional expression of human Na, K-ATPase α3β1 isoform. Appl Microbiol Biotechnol 97:9465–9478. doi:10.1007/s00253-013-5156-7 [DOI] [PubMed]
- Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WEC, Robbins RA, Miercke LJW, Stroud RM. Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance. Proc Natl Acad Sci U S A. 2009;106:7437–7442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idiris A, Tohda H, Kumagai H, Takegawa K. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl Microbiol Biotechnol. 2010;86:403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- Inan M, Meagher MM. Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J Biosci Bioeng. 2001;92:585–589. doi: 10.1263/jbb.92.585. [DOI] [PubMed] [Google Scholar]
- Jacobs PP, Geysens S, Vervecken W, Contreras R, Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat Protoc. 2009;4:58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- Jahic M, Veide A, Charoenrat T, Teeri T, Enfors S-O. Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol Prog. 2006;22:1465–1473. doi: 10.1021/bp060171t. [DOI] [PubMed] [Google Scholar]
- Jin F, Xu X, Zhang W, Gu D. Expression and characterization of a housefly cecropin gene in the methylotrophic yeast, Pichia pastoris. Protein Expr Purif. 2006;49:39–46. doi: 10.1016/j.pep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine–arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Kang HA, Choi ES, Hong WK, Kim JY, Ko SM, Sohn JH, Rhee SK. Proteolytic stability of recombinant human serum albumin secreted in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2000;53:575–582. doi: 10.1007/s002530051659. [DOI] [PubMed] [Google Scholar]
- Kato S, Ishibashi M, Tatsuda D, Tokunaga H, Tokunaga M. Efficient expression, purification and characterization of mouse salivary alpha-amylase secreted from methylotrophic yeast, Pichia pastoris. Yeast. 2001;18:643–655. doi: 10.1002/yea.714. [DOI] [PubMed] [Google Scholar]
- Khasa YP, Conrad S, Sengul M, Plautz S, Meagher MM, Inan M. Isolation of Pichia pastoris PIR genes and their utilization for cell surface display and recombinant protein secretion. Yeast. 2011;28:213–226. doi: 10.1002/yea.1832. [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Hach M, Balschmidt P, Havelund S, Pettersson AF, Markussen J. Prepro-leaders lacking N-linked glycosylation for secretory expression in the yeast Saccharomyces cerevisiae. Protein Expr Purif. 1998;14:309–316. doi: 10.1006/prep.1998.0977. [DOI] [PubMed] [Google Scholar]
- Kottmeier K, Ostermann K, Bley T, Rödel G. Hydrophobin signal sequence mediates efficient secretion of recombinant proteins in Pichia pastoris. Appl Microbiol Biotechnol. 2011;91:133–141. doi: 10.1007/s00253-011-3246-y. [DOI] [PubMed] [Google Scholar]
- Kranthi BV, Balasubramanian N, Rangarajan PN. Isolation of a single-stranded DNA-binding protein from the methylotrophic yeast, Pichia pastoris and its identification as zeta crystallin. Nucleic Acids Res. 2006;34:4060–4068. doi: 10.1093/nar/gkl577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranthi BV, Kumar R, Kumar NV, Rao DN, Rangarajan PN. Identification of key DNA elements involved in promoter recognition by Mxr1p, a master regulator of methanol utilization pathway in Pichia pastoris. Biochim Biophys Acta. 2009;1789:460–468. doi: 10.1016/j.bbagrm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Küberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, Hajek T, Jaenicke S, Brinkrolf K, Goesmann A, Szczepanowski R, Pühler A, Schwab H, Glieder A, Pichler H. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 2011;154:312–320. doi: 10.1016/j.jbiotec.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kumar NV, Rangarajan PN. The zinc finger proteins Mxr1p and repressor of phosphoenolpyruvate carboxykinase (ROP) have the same DNA binding specificity but regulate methanol metabolism antagonistically in Pichia pastoris. J Biol Chem. 2012;287:34465–34473. doi: 10.1074/jbc.M112.365304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol. 2009;36:1435–1438. doi: 10.1007/s10295-009-0638-4. [DOI] [PubMed] [Google Scholar]
- Larsen S, Weaver J, de Sa CK, Bulahan R, Nguyen J, Grove H, Huang A, Low L, Tran N, Gomez S, Yau J, Ilustrisimo T, Kawilarang J, Lau J, Tranphung M, Chen I, Tran C, Fox M, Lin-Cereghino J, Lin-Cereghino GP. Mutant strains of Pichia pastoris with enhanced secretion of recombinant proteins. Biotechnol Lett. 2013 doi: 10.1007/s10529-013-1290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Williams TG, Wong DWS, Robertson GH. An episomal expression vector for screening mutant gene libraries in Pichia pastoris. Plasmid. 2005;54:80–85. doi: 10.1016/j.plasmid.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Li Z, Leung W, Yon A, Nguyen J, Perez VC, Vu J, Giang W, Luong LT, Phan T, Salazar KA, Gomez SR, Au C, Xiang F, Thomas DW, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Secretion and proteolysis of heterologous proteins fused to the Escherichia coli maltose binding protein in Pichia pastoris. Protein Expr Purif. 2010;72:113–124. doi: 10.1016/j.pep.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Li C, Ye Y, Lin Y. Endogenous signal peptides efficiently mediate the secretion of recombinant proteins in Pichia pastoris. Biotechnol Lett. 2013;35:97–105. doi: 10.1007/s10529-012-1055-8. [DOI] [PubMed] [Google Scholar]
- Liang S, Zou C, Lin Y, Zhang X, Ye Y (2013b) Identification and characterization of P GCW14 : a novel, strong constitutive promoter of Pichia pastoris. Biotechnol Lett 35:1865–1871. doi:10.1007/s10529-013-1265-8 [DOI] [PubMed]
- Lifshitz Y, Petrovich E, Haviv H, Goldshleger R, Tal DM, Garty H, Karlish SJD (2007) Purification of the human alpha2 Isoform of Na, K-ATPase expressed in Pichia pastoris. Stabilization by lipids and FXYD1. Biochemistry 46:14937–14950. doi:10.1021/bi701812c [DOI] [PubMed]
- Lin-Cereghino J, Lin-Cereghino GP. Vectors and strains for expression. Methods Mol Biol. 2007;389:11–26. doi: 10.1007/978-1-59745-456-8_2. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Lin-Cereghino J, Jay Sunga A, Johnson MA, Lim M, Gleeson MAG, Cregg JM. New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene. 2001;263:159–169. doi: 10.1016/s0378-1119(00)00576-x. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, de la Cruz BJ, De JS, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM (2006) Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol 26:883–897. doi:10.1128/MCB.26.3.883-897.2006 [DOI] [PMC free article] [PubMed]
- Lin-Cereghino GP, Stark CM, Kim D, Chang JWJ, Shaheen N, Poerwanto H, Agari K, Moua P, Low LK, Tran N, Huang AD, Nattestad M, Oshiro KT, Chavan A, Tsai JW, Lin-Cereghino J. The effect of α-mating factor secretion signal mutations on recombinant protein expression in Pichia pastoris. Gene. 2013;519:311–317. doi: 10.1016/j.gene.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lünsdorf H, Gurramkonda C, Adnan A, Khanna N, Rinas U. Virus-like particle production with yeast: ultrastructural and immunocytochemical insights into Pichia pastoris producing high levels of the hepatitis B surface antigen. Microb Cell Factories. 2011;10:48. doi: 10.1186/1475-2859-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- Marx H, Mecklenbräuker A, Gasser B, Sauer M, Mattanovich D. Directed gene copy number amplification in Pichia pastoris by vector integration into the ribosomal DNA locus. FEMS Yeast Res. 2009;9:1260–1270. doi: 10.1111/j.1567-1364.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Callewaert N, Rouzé P, Lin Y-C, Graf A, Redl A, Tiels P, Gasser B, De Schutter K. Open access to sequence: browsing the Pichia pastoris genome. Microb Cell Factories. 2009;8:53. doi: 10.1186/1475-2859-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich D, Graf A, Stadlmann J, Dragosits M, Redl A, Maurer M, Kleinheinz M, Sauer M, Altmann F, Gasser B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb Cell Factories. 2009;8:29. doi: 10.1186/1475-2859-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer A, Glieder A, Weis R, Reisinger C, Flicker K. Sensitive high-throughput screening for the detection of reducing sugars. Biotechnol J. 2012;7:155–162. doi: 10.1002/biot.201100001. [DOI] [PubMed] [Google Scholar]
- Menendez J, Valdes I, Cabrera N. The ICL1 gene of Pichia pastoris, transcriptional regulation and use of its promoter. Yeast. 2003;20:1097–1108. doi: 10.1002/yea.1028. [DOI] [PubMed] [Google Scholar]
- Moir DT, Dumais DR. Glycosylation and secretion of human alpha-1-antitrypsin by yeast. Gene. 1987;56:209–217. doi: 10.1016/0378-1119(87)90138-7. [DOI] [PubMed] [Google Scholar]
- Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sönksen CP, Ludvigsen S, Raventós D, Buskov S, Christensen B, De Maria L, Taboureau O, Yaver D, Elvig-Jørgensen SG, Sørensen MV, Christensen BE, Kjaerulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen H-H. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- Näätsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J (2010) Yeast strains for protein production. United States patent US 2010/0279348
- Nett JH, Gerngross TU. Cloning and disruption of the PpURA5 gene and construction of a set of integration vectors for the stable genetic modification of Pichia pastoris. Yeast. 2003;20:1279–1290. doi: 10.1002/yea.1049. [DOI] [PubMed] [Google Scholar]
- Nett JH, Hodel N, Rausch S, Wildt S. Cloning and disruption of the Pichia pastoris ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, HIS6 genes and their use as auxotrophic markers. Yeast. 2005;22:295–304. doi: 10.1002/yea.1202. [DOI] [PubMed] [Google Scholar]
- Nett JH, Stadheim TA, Li H, Bobrowicz P, Hamilton SR, Davidson RC, Choi B-K, Mitchell T, Bobrowicz B, Rittenhour A, Wildt S, Gerngross TU. A combinatorial genetic library approach to target heterologous glycosylation enzymes to the endoplasmic reticulum or the Golgi apparatus of Pichia pastoris. Yeast. 2011;28:237–252. doi: 10.1002/yea.1835. [DOI] [PubMed] [Google Scholar]
- Nett JH, Cook WJ, Chen M-T, Davidson RC, Bobrowicz P, Kett W, Brevnova E, Potgieter TI, Mellon MT, Prinz B, Choi B-K, Zha D, Burnina I, Bukowski JT, Du M, Wildt S, Hamilton SR. Characterization of the Pichia pastoris protein-O-mannosyltransferase gene family. PLoS ONE. 2013;8:e68325. doi: 10.1371/journal.pone.0068325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Zhou X, Sun X, Wang Y, Zhang Y. Decrease of hirudin degradation by deleting the KEX1 gene in recombinant Pichia pastoris. Yeast. 2008;25:1–8. doi: 10.1002/yea.1542. [DOI] [PubMed] [Google Scholar]
- Ohi H, Miura M, Hiramatsu R, Ohmura T. The positive and negative cis-acting elements for methanol regulation in the Pichia pastoris AOX2 gene. Mol Gen Genet. 1994;243:489–499. doi: 10.1007/BF00284196. [DOI] [PubMed] [Google Scholar]
- Oka C, Tanaka M, Muraki M, Harata K, Suzuki K, Jigami Y. Human lysozyme secretion increased by alpha-factor pro-sequence in Pichia pastoris. Biosci Biotechnol Biochem. 1999;63:1977–1983. doi: 10.1271/bbb.63.1977. [DOI] [PubMed] [Google Scholar]
- Paifer E, Margolles E, Cremata J, Montesino R, Herrera L, Delgado JM. Efficient expression and secretion of recombinant alpha amylase in Pichia pastoris using two different signal sequences. Yeast. 1994;10:1415–1419. doi: 10.1002/yea.320101104. [DOI] [PubMed] [Google Scholar]
- Parua PK, Ryan PM, Trang K, Young ET. Pichia pastoris 14-3-3 regulates transcriptional activity of the methanol inducible transcription factor Mxr1 by direct interaction. Mol Microbiol. 2012;85:282–298. doi: 10.1111/j.1365-2958.2012.08112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne WE, Gannon PM, Kaiser CA. An inducible acid phosphatase from the yeast Pichia pastoris: characterization of the gene and its product. Gene. 1995;163:19–26. doi: 10.1016/0378-1119(95)00379-k. [DOI] [PubMed] [Google Scholar]
- Polupanov AS, Nazarko VY, Sibirny AA. Gss1 protein of the methylotrophic yeast Pichia pastoris is involved in glucose sensing, pexophagy and catabolite repression. Int J Biochem Cell Biol. 2012;44:1906–1918. doi: 10.1016/j.biocel.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Prielhofer R, Maurer M, Klein J, Wenger J, Kiziak C, Gasser B, Mattanovich D. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb Cell Factories. 2013;12:5. doi: 10.1186/1475-2859-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Qian J, Yao G, Zhuang Y, Zhang S, Chu J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol. 2011;77:3600–3608. doi: 10.1128/AEM.02843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raemaekers RJM, de Muro L, Gatehouse JA, Fordham-Skelton AP. Functional phytohemagglutinin (PHA) and Galanthus nivalis agglutinin (GNA) expressed in Pichia pastoris. Correct N-terminal processing and secretion of heterologous proteins expressed using the PHA-E signal peptide. Eur J Biochem. 1999;265:394–403. doi: 10.1046/j.1432-1327.1999.00749.x. [DOI] [PubMed] [Google Scholar]
- Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha C, Yu X-W, Li F, Xu Y. Impact of gene dosage on the production of lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol. 2013;169:1160–1172. doi: 10.1007/s12010-012-0050-9. [DOI] [PubMed] [Google Scholar]
- Shen S, Sulter G, Jeffries TW, Cregg JM. A strong nitrogen source-regulated promoter for controlled expression of foreign genes in the yeast Pichia pastoris. Gene. 1998;216:93–102. doi: 10.1016/s0378-1119(98)00315-1. [DOI] [PubMed] [Google Scholar]
- Shi X-L, Feng M-Q, Shi J, Shi Z-H, Zhong J, Zhou P. High-level expression and purification of recombinant human catalase in Pichia pastoris. Protein Expr Purif. 2007;54:24–29. doi: 10.1016/j.pep.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Sigoillot M, Brockhoff A, Lescop E, Poirier N, Meyerhof W, Briand L. Optimization of the production of gurmarin, a sweet-taste-suppressing protein, secreted by the methylotrophic yeast Pichia pastoris. Appl Microbiol Biotechnol. 2012;96:1253–1263. doi: 10.1007/s00253-012-3897-3. [DOI] [PubMed] [Google Scholar]
- Sinha J, Plantz BA, Inan M, Meagher MM. Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant ovine interferon-tau. Biotechnol Bioeng. 2005;89:102–112. doi: 10.1002/bit.20318. [DOI] [PubMed] [Google Scholar]
- Skipper N, Sutherland M, Davies RW, Kilburn D, Miller RC, Warren A, Wong R. Secretion of a bacterial cellulase by yeast. Science. 1985;230:958–960. doi: 10.1126/science.230.4728.958. [DOI] [PubMed] [Google Scholar]
- Stadlmayr G, Mecklenbräuker A, Rothmüller M, Maurer M, Sauer M, Mattanovich D, Gasser B. Identification and characterisation of novel Pichia pastoris promoters for heterologous protein production. J Biotechnol. 2010;150:519–529. doi: 10.1016/j.jbiotec.2010.09.957. [DOI] [PubMed] [Google Scholar]
- Staley CA, Huang A, Nattestad M, Oshiro KT, Ray LE, Mulye T, Li ZH, Le T, Stephens JJ, Gomez SR, Moy AD, Nguyen JC, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Analysis of the 5′ untranslated region (5′UTR) of the alcohol oxidase 1 (AOX1) gene in recombinant protein expression in Pichia pastoris. Gene. 2012;496:118–127. doi: 10.1016/j.gene.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta Gen Subj. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- Takagi S, Tsutsumi N, Terui Y, Kong XY (2008) Method for methanol independent induction from methanol inducible promoters in Pichia. United States patent US 8,143,023
- Thompson CA. FDA approves kallikrein inhibitor to treat hereditary angioedema. Am J Health Syst Pharm. 2010;67:93. doi: 10.2146/news100005. [DOI] [PubMed] [Google Scholar]
- Thor D, Xiong S, Orazem CC, Kwan A-C, Cregg JM, Lin-Cereghino J, Lin-Cereghino GP. Cloning and characterization of the Pichia pastoris MET2 gene as a selectable marker. FEMS Yeast Res. 2005;5:935–942. doi: 10.1016/j.femsyr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Tschopp JF, Brust PF, Cregg JM, Stillman CA, Gingeras TR. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. Nucleic Acids Res. 1987;15:3859–3876. doi: 10.1093/nar/15.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp JF, Sverlow G, Kosson R, Craig W, Grinna L. High-level secretion of glycosylated invertase in the methylotrophic yeast, Pichia pastoris. Nat Biotechnol. 1987;5:1305–1308. [Google Scholar]
- Uchima CA, Arioka M. Expression and one-step purification of recombinant proteins using an alternative episomal vector for the expression of N-tagged heterologous proteins in Pichia pastoris. Biosci Biotechnol Biochem. 2012;76:368–371. doi: 10.1271/bbb.110628. [DOI] [PubMed] [Google Scholar]
- Vadhana AKP, Samuel P, Berin RM, Krishna J, Kamatchi K, Meenakshisundaram S. Improved secretion of Candida antarctica lipase B with its native signal peptide in Pichia pastoris. Enzym Microb Technol. 2013;52:177–183. doi: 10.1016/j.enzmictec.2013.01.001. [DOI] [PubMed] [Google Scholar]
- van der Heide M, Hollenberg CP, van der Klei IJ, Veenhuis M. Overproduction of BiP negatively affects the secretion of Aspergillus niger glucose oxidase by the yeast Hansenula polymorpha. Appl Microbiol Biotechnol. 2002;58:487–494. doi: 10.1007/s00253-001-0907-2. [DOI] [PubMed] [Google Scholar]
- Vassileva A, Chugh DA, Swaminathan S, Khanna N. Effect of copy number on the expression levels of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris. Protein Expr Purif. 2001;21:71–80. doi: 10.1006/prep.2000.1335. [DOI] [PubMed] [Google Scholar]
- Vervecken W, Kaigorodov V, Callewaert N, Geysens S, De Vusser K, Contreras R (2004) In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. 70:2639–46. doi:10.1128/AEM.70.5.2639 [DOI] [PMC free article] [PubMed]
- Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. Nat Biotechnol. 2013;30:385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2010. Nat Biotechnol. 2010;28:917–924. doi: 10.1038/nbt0910-917. [DOI] [PubMed] [Google Scholar]
- Waterham HR, de Vries Y, Russel KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene. 1997;186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- Waters MG, Evans EA, Blobel G. Prepro-alpha-factor has a cleavable signal sequence. J Biol Chem. 1988;263:6209–6214. [PubMed] [Google Scholar]
- Weinhandl K, Winkler M, Glieder A, Camattari A. A novel multi-enzymatic high throughput assay for transaminase activity. Tetrahedron. 2012;68:7586–7590. [Google Scholar]
- Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;5:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Weiss HM, Haase W, Michel H, Reiländer H. Expression of functional mouse 5-HT5A serotonin receptor in the methylotrophic yeast Pichia pastoris: pharmacological characterization and localization. FEBS Lett. 1995;377:451–456. doi: 10.1016/0014-5793(95)01389-x. [DOI] [PubMed] [Google Scholar]
- Werten MWT, de Wolf FA (2005) Reduced proteolysis of secreted gelatin and Yps1-mediated alpha-factor leader processing in a Pichia pastoris kex2 disruptant. Appl Environ Microbiol 71:2310–2317. doi:10.1128/AEM.71.5.2310-2317.2005 [DOI] [PMC free article] [PubMed]
- Werten MWT, van den Bosch TJ, Wind RD, Mooibroek H, de Wolf FA. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast. 1999;15:1087–1096. doi: 10.1002/(SICI)1097-0061(199908)15:11<1087::AID-YEA436>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Whyteside G, Nor RM, Alcocer MJC, Archer DB. Activation of the unfolded protein response in Pichia pastoris requires splicing of a HAC1 mRNA intron and retention of the C-terminal tail of Hac1p. FEBS Lett. 2011;585:1037–1041. doi: 10.1016/j.febslet.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Wriessnegger T, Pichler H. Yeast metabolic engineering—targeting sterol metabolism and terpenoid formation. Prog Lipid Res. 2013;52:277–293. doi: 10.1016/j.plipres.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Wriessnegger T, Gübitz G, Leitner E, Ingolic E, Cregg J, de la Cruz BJ, Daum G. Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim Biophys Acta. 2007;1771:455–461. doi: 10.1016/j.bbalip.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Wriessnegger T, Leitner E, Belegratis MR, Ingolic E, Daum G. Lipid analysis of mitochondrial membranes from the yeast Pichia pastoris. Biochim Biophys Acta. 2009;1791:166–172. doi: 10.1016/j.bbalip.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Wu M, Shen Q, Yang Y, Zhang S, Qu W, Chen J, Sun H, Chen S. Disruption of YPS1 and PEP4 genes reduces proteolytic degradation of secreted HSA/PTH in Pichia pastoris GS115. J Ind Microbiol Biotechnol. 2013;40:589–599. doi: 10.1007/s10295-013-1264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Y, Zhou X, Zhang W, Zhang X, Song Z, Zhang Y. An upstream activation sequence controls the expression of AOX1 gene in Pichia pastoris. FEMS Yeast Res. 2009;9:1271–1282. doi: 10.1111/j.1567-1364.2009.00571.x. [DOI] [PubMed] [Google Scholar]
- Yang S, Kuang Y, Li H, Liu Y, Hui X, Li P, Jiang Z, Zhou Y, Wang Y, Xu A, Li S, Liu P, Wu D. Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1′ site. PLoS ONE. 2013;8:e75347. doi: 10.1371/journal.pone.0075347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XQ, Zhao HL, Xue C, Zhang W, Xiong XH, Wang ZW, Li XY, Liu ZM. Degradation of HSA-AX15(R13K) when expressed in Pichia pastoris can be reduced via the disruption of YPS1 gene in this yeast. J Biotechnol. 2009;139:131–136. doi: 10.1016/j.jbiotec.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Yoshimasu MA, Ahn J-K, Tanaka T, Yada RY. Soluble expression and purification of porcine pepsinogen from Pichia pastoris. Protein Expr Purif. 2002;25:229–236. doi: 10.1016/s1046-5928(02)00003-1. [DOI] [PubMed] [Google Scholar]
- Yu XW, Sha C, Guo YL, Xiao R, Xu Y. High-level expression and characterization of a chimeric lipase from Rhizopus oryzae for biodiesel production. Biotechnol Biofuels. 2013;6:29. doi: 10.1186/1754-6834-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Teng D, Mao R, Wang X, Xi D, Hu X, Wang J. High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl Microbiol Biotechnol. 2014;98:681–694. doi: 10.1007/s00253-013-4881-2. [DOI] [PubMed] [Google Scholar]
- Zhao HL, Xue C, Wang Y, Yao XQ, Liu ZM. Increasing the cell viability and heterologous protein expression of Pichia pastoris mutant deficient in PMR1 gene by culture condition optimization. Appl Microbiol Biotechnol. 2008;81:235–241. doi: 10.1007/s00253-008-1666-0. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]