Abstract
Aberrant von Hippel Lindau (VHL) protein function is the underlying driver of VHL-related diseases, including both sporadic and inherited clear cell renal cell carcinoma (ccRCC). About one third of VHL mutations are missense point mutations, with R167Q being the most common VHL point mutation in hereditary VHL disease. Although it has been studied extensively, the ability of VHL-R167Q to downregulate hypoxia inducible factor 2α (HIF2α) is still controversial. In addition, the manner in which the mutation contributes to tumorigenesis is not fully understood. No therapeutic approach is available to target VHL-R167Q and similar missense point mutations. We analyzed VHL-R167Q proteostasis and function at normoxia, at hypoxia with different oxygen pressure, and in a xenograft mouse model. We showed that the protein levels of VHL-R167Q dictate its ability to downregulate HIF2α and suppress tumor growth. Strikingly, the proteasome inhibitors bortezomib and carfilzomib, which are currently in clinical use, stabilize VHL-R167Q and increase its ability to downregulate HIF2α. VHL-R167Q binds elongin C and elongin B with considerably less avidity than wild-type VHL does but retains residual capacity to generate a VHL-elongin C-elongin B complex, downregulate HIF2α, and suppress tumorigenesis, which could be rescued by increase VHL-R167Q levels. Finally, we used in silico approaches and identified other missense VHL mutants in addition to VHL-R167Q that might be rescued by similar strategies. Thus, our studies revealed detailed information describing how VHL-R167Q contributes to tumorigenesis and identified a potential targeted therapy for ccRCC and other VHL-related disease in patients carrying VHL-R167Q or similar missense mutations.
Keywords: VHL, ccRCC, Proteasome Inhibition, Point Mutations, Functional Restoration
Introduction
von Hippel Lindau (VHL) disease is an autosomal dominant disorder that predisposes individuals to benign and malignant tumors. VHL loss of function is the underlying driver of VHL-related diseases, including both sporadic and inherited clear cell renal cell carcinoma (ccRCC) (1). The establishment of VHL as the substrate recognizing component of an E3 ligase for hypoxia inducible factor (HIF) has led to an understanding that loss of VHL function and stabilization of HIF play a central role in tumor angiogenesis in ccRCC (2–4). Mutations in VHL that disrupt binding with HIF occur at high frequency in ccRCC (5). These early findings and the recognition that angiogenesis is a key driver of the ccRCC phenotype ushered in a new era of targeted therapies for ccRCC with the new drugs including sorafenib, sunitinib, pazopanib, axitinib and bevacizumab (6–10).
Approximately one quarter to one third of all VHL mutations are missense point mutations, generating a full-length protein (5, 11), though less stable than the wild-type protein. These point mutations, in some cases, maintain residual functionality (5, 12). The most common mutation in hereditary VHL disease, R167Q, is a representative type 2B mutation that predispose to a high risk of ccRCC (13). The R167Q mutation disrupts VHL binding with elongin C and therefore disrupts the functional VHL-elongin B-elongin C (VBC) E3 ligase complex (12, 14). However, the ability of VHL-R167Q to downregulate HIF2α has been debated. Some studies have shown that VHL R167Q is deficient or partially deficient in downregulating HIF2α (15), whereas other studies have shown that it efficiently downregulates HIF2α (16, 17). The manner in which the R167Q mutation of VHL contributes to HIF2α downregulation has not been systematically elucidated. No therapeutic approach has been proposed to target R167Q and similar point mutations in patients.
To address these issues, we characterized VHL-R167Q in greater detail and gathered functional information regarding its ability to downregulate HIF2α and suppress tumor formation. In addition, we explored whether stabilization of VHL protein harboring missense mutations could serve as a novel therapeutic approach in ccRCC. We demonstrated that the protein levels of VHL-R167Q dictate its functional capacity to downregulate HIF2α and suppress tumorigenesis and that proteasome inhibition increases the levels and function of VHL-R167Q. Our study provides a potential innovative targeted therapy to treat kidney cancer patients by stabilizing R167Q and other related mutations.
Materials and Methods
Cell culture and plasmids
786-0 and RCC4 human kidney cancer cells were obtained from ATCC (Manassas, VA). Cells were cultured in Dulbecco’s modified Eagle’s medium from Invitrogen (Carlsbad, CA) supplemented with 10% fetal calf serum from Gibco (Carlsbad, CA). Venus is an eYFP derivative with high fluorescence intensity (18). Retroviral vectors expressing VHL-wild-type (wt)-Venus and VHL-W117A-Venus were described previously (19). We created VHL-R167Q-Venus, VHL-L118P-Venus, and VHL-F148A-Venus mutations from VHL-wt-Venus with a Quikchange mutagenesis kit from Stratagene (La Jolla, CA) and confirmed them by sequencing. We also made VHL constructs without the Venus tag (VHL-wt and VHL-R167Q) by introducing a stop codon downstream of VHL. Cells were transfected using FuGENE 6 transfection reagent from Roche (Indianapolis, IN) following the manufacturer’s protocol. Retrovirus preparation and infection were performed as previously described (20).
Reagents and antibodies
Bortezomib was obtained from Selleck Chemicals (Houston, TX), and carfilzomib from Onyx Pharmaceuticals (San Francisco, CA). Antibodies against VHL (#2738) and tubulin (#2125) were from Cell Signaling Technology (Beverly, MA). Antibodies against elongin B (#11447), elongin C (#1559), and Cullin-2 (Cul2) (#10781) were from Santa Cruz (Dallas, TX). Anti-HIF2α antibody (#NB100-122) was from Novus Biologicals (Littleton, CO), anti-GLUT1 antibody (#GT12-A) from Alpha Diagnostic International (San Antonio, TX), anti-GAPDH antibody (#AM4300) from Ambion (Austin, TX). LC3B antibody was prepared in Dr. Eissa laboratory (Baylor College of Medicine, Houston, TX) as described previously (21).
Western blotting and GFP-Trap
To prepare cell lysates for western blotting, cells were lysed in RIPA buffer (Tris-HCl, pH 7.4, 50 mM; NaCl, 150 mM; NP-40, 1%; sodium deoxycholate, 0.5%; SDS, 0.1%) supplemented with protease inhibitor and phosphatase inhibitor cocktail (Pierce, Rockford, IL). Western blotting was performed as previously described (20). Samples were resolved on 10% SDS polyacrylamide gels. Scanning densitometric values were obtained using ImageJ software (version 1.46r; National Institutes of Health, Bethesda, MD). To prepare cell lysates for the GFP-Trap, cells were lysed in 0.5% NP-40 buffer (Tris-HCl, pH 7.4, 50 mM; NaCl, 150 mM; NP-40, 0.5%; sodium deoxycholate, 0.5%) supplemented with protease inhibitor and phosphatase inhibitor cocktail (Pierce, Rockford, IL). The GFP-Trap was performed according to the manufacturer’s protocol (Allele Biotechnology, San Diego, CA). Proteins pulled down by GFP-Trap beads were analyzed by western blotting using SDS-polyacrylamide denaturing gels.
Real-time PCR
Total RNA was extracted and purified using RNeasy kits and treated with DNase according to the manufacturer’s protocol (Qiagen, Venlo, Netherlands). cDNA was synthesized using the SuperScript First Strand Synthesis System (#18080-051, Invitrogen) with 1 μg RNA as the template. Real-time PCR was performed with ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) in 25μl volume (Power SYBR® Green PCR Master Mix, #4367659, Applied Biosystems) using 1μl of cDNA and 0.3 μM forward and reverse primers (HIF2α: forward, 5′-TGGCCGCTCAGCCTATGAAT-3′; reverse, 5′-TGGGTCTCCAGCCACACGTA-3′ (22); GFP: forward, 5′-GAGCGCACCATCTTCTTCAAG-3′; reverse, 5′-TGTCGCCCTCGAACTTCAC-3′ (23); GAPDH: forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse, 5′-GAAGATGGTGATGGGATTTC-3′ (24)). Experiments were performed in triplicates and the relative quantification results were calculated using the 2−ΔΔCt method. VHL-wt-Venus, VHL-R167Q-Venus, and HIF2α mRNA expressions were normalized to GAPDH. A dissociation curve was generated for each PCR amplicon to ensure single-product amplification.
Xenografts
All procedures were approved by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center. Kidney cancer 786-0 cells expressing VHL proteins or GFP control were cultured in DMEM medium supplemented with 10% fetal bovine serum. Log-phase cells (1 × 107 cells/mouse) were harvested, mixed with Matrigel (100 μL), and injected subcutaneously into the flanks of 6- to 8-week-old athymic nu/nu mice (NCI). Seven female mice were used in each group. Tumor growth was measured twice a week by calipers. Mice were euthanized and tumors were harvested. Half of each tumor was snap frozen in liquid nitrogen for molecular analysis, and the other half was fixed in formalin and embedded in paraffin for histological analysis.
In silico analysis of VHL mutations
We analyzed 18 VHL crystal structures available in the Protein Data Bank (PDB) (25) and chose the recently solved VBC-HIF1α peptide complex (PDB id: 4AJY) at a high resolution (1.73 Å) (26) as the reference VHL structure. Point mutations were classified as buried or exposed. Buried mutations are likely capable of inducing protein destabilization, whereas exposed mutations probably alter interactions with substrates and other binding partners. The impact of missense mutations on VHL protein function can be estimated by measuring the changes in the shape and physicochemical properties of the interaction interfaces (27).
The Prediction of Protein Mutant Stability Changes (PoPMuSiC) program (28) was used to compute the variation in the folding free energy (ΔΔG) of the VHL complex. A positive ΔΔG value represents destabilizing mutations, whereas a negative ΔΔG value represents stabilizing mutations. We chose a value of 3.0 kcal/mol, which corresponds to the loss of two or three hydrogen bonds, as the cutoff. Thus, a ΔΔG value <3.0 kcal/mol indicates that destabilization caused by a mutation could be compensated by increasing the VHL protein level and thus that the mutation can be rescued (i.e., that the wild-type phenotype can be restored). On the other hand, a ΔΔG value of >3.0 kcal/mol suggests the mutation cannot be rescued.
Results
Protein level of VHL-R167Q dictates functional capacity of VHL in downregulating HIF2α and suppressing tumorigenesis
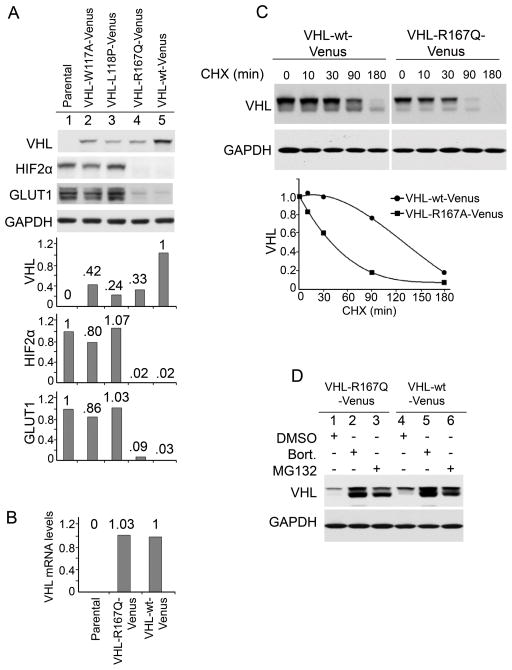
We first examined the effect of the R167Q point mutation on VHL protein stability using Venus tagged proteins. VHL-null 786-0 kidney cancer cells were infected with retroviral vectors to stably express VHL-R167Q-Venus and VHL-wt-Venus. VHL-R167Q-Venus protein levels were significantly lower than VHL-wt-Venus levels (Fig. 1A), which is consistent with a previous study that demonstrated that the R167Q mutation destabilizes VHL (12). Two other point mutations, W117A and L118P, which disrupt TRiC binding and destabilize VHL (29), also had decreased protein levels compared with VHL-wt-Venus (Fig. 1A). mRNA levels of VHL-R167Q-Venus and VHL-wt-Venus were comparable, indicating that the different protein levels were not due to differences in transcription level (Fig. 1B). The half-life of VHL-R167Q-Venus (~ 40 mins) was significantly shorter than that of VHL-wt-Venus (~130 mins) determined in the cells treated with the translation inhibitor cycloheximide (Fig. 1C). The proteasome inhibitor MG132 significantly increased VHL-R167Q levels, which became comparable to VHL-wt levels after MG132 treatment (Fig. 1D). All of the above data suggest that elevated proteasomal degradation of VHL-R167Q-Venus was responsible for the lower protein level.
Figure 1. R167Q mutation decreases protein stability of VHL in 786-0 cells.
(A) R167Q mutation decreased VHL protein levels. VHL-null 786-0 cells were infected to stably express the indicated proteins. Cells were cultured in complete medium and were lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Samples were resolved on 10% SDS-polyacrylamide gels. Scanning densitometric values were obtained using ImageJ software. Protein levels were normalized to the loading control (GAPDH) and presented as relative conversion to values in VHL-wt-Venus cells (for VHL) or in parental cells (for HIF2α and GLUT1). (B) Comparable mRNA levels of VHL-R167Q-Venus and VHL-wt-Venus. Parental 786-0 cells, cells expressing VHL-R167Q-Venus, or cells expressing VHL-wt-Venus were cultured in complete medium. mRNA levels were measured by real-time PCR. Data are presented as relative conversion to values in VHL-wt-Venus cells. (C) Half-lives of VHL-R167Q-Venus and VHL-wt-Venus. Cells were cultured in complete medium and treated with cycloheximide (CHX) (50 μg/mL) for the indicated times. Scanning densitometric values were obtained using ImageJ software and were used in the trendline chart. (D) Proteasome inhibitor MG132 stabilizes VHL-R167Q-Venus and VHL-wt-Venus. Cells were cultured in complete medium and treated with MG132 (5 μg/mL) for 6 hours.
We then examined the functional capacity of VHL-R167Q-Venus to downregulate HIF2α, the most studied VHL target (2–4). Surprisingly, VHL-R167Q-Venus mediated complete degradation of HIF2α, comparable to the effect of VHL-wt-Venus (Fig. 1A). A similar result was shown in a previous study (16). Glucose transporter 1 (GLUT1), a target of HIF2α, was also comparably downregulated by VHL-R167Q-Venus. In contrast, as we expected, two mutant proteins that disrupt interactions between VHL and HIF2α, VHL-W117A-Venus and VHL-L118P-Venus, could not downregulate HIF2α or GLUT1 (Fig. 1A). These results indicated that VHL-R167Q-Venus was fully capable of downregulating HIF2α in 786-0 cells. Although the results are consistent with some published data (16), they are inconsistent with the fact that the R167Q mutation is the most common disease causing mutation in hereditary VHL disease and with the widely accepted notion that the R167Q mutation diminishes VHL binding to elongin B and elongin C and subsequently its ability to downregulate HIF (12, 13).
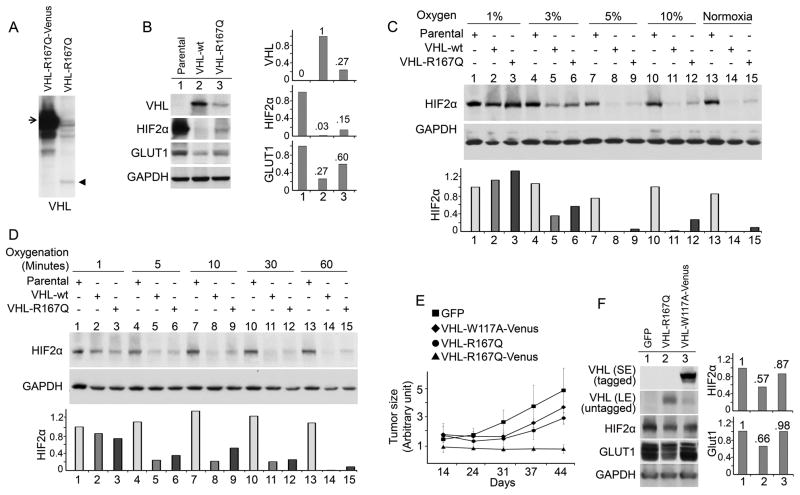
To explore the tumorigenesis mechanism of the R167Q mutation, we sought to characterize it in more depth. We observed that VHL-R167Q-Venus was expressed at a much higher level than VHL-R167Q without any tag in 786-0 cells (Fig. 2A), likely due to a stabilization effect of the Venus tag. We then examined whether the protein levels of VHL-R167Q affected its function. We stably expressed untagged VHL-R167Q and VHL-wt in 786-0 cells (Fig. 2B), which were expressed at much lower levels than Venus-tagged VHL. Although it was expressed at lower levels than VHL-wt-Venus, VHL-wt still mediated complete HIF2α degradation. In contrast, VHL-R167Q could only mediate partial HIF2α degradation, suggesting that at lower expression levels, VHL-R167Q was not sufficient for complete HIF2α downregulation. GLUT1 levels were higher in VHL-R167Q cells than in VHL-wt cells, indicating that HIF2α was only partially functioning in VHL-R167Q cells (Fig. 2B), possibly due to a stoichiometric effect.
Figure 2. R167Q mutation decreases the ability of VHL to downregulate HIF2α in 786-0 cells and suppress tumor growths.
(A) VHL-R167Q protein levels were lower than VHL-R167Q-Venus protein levels. VHL-null 786-0 cells were infected to stably express VHL-R167Q or VHL-R167Q-Venus, cultured in complete medium, and lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Samples were resolved in 10% SDS-polyacrylamide gels. The arrow designates VHL-R167Q-Venus. The arrowhead designates VHL-R167Q. (B) R167Q mutation decreased VHL function in HIF2α downregulation. VHL-null 786-0 cells were infected to stably express the indicated proteins. Cells were cultured in complete medium, and lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Scanning densitometric values were obtained using ImageJ software. Protein levels were normalized to the loading control (GAPDH) and presented as relative conversion to values in VHL-wt cells (for VHL) and in parental cells (for HIF2α and GLUT1). (C) R167Q mutation decreased VHL function in HIF2α downregulation under hypoxia. Parental, VHL-R167Q, and VHL-wt 786-0 cells were cultured in complete medium with the indicated oxygen levels for 72 hours. Cells were lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Normoxia: 21% oxygen. Scanning densitometric values were obtained using ImageJ software. HIF2α protein levels are presented as relative conversion to values in Lane 1. (D) R167Q mutation decreased VHL function in HIF2α downregulation after reoxygenation. Parental, VHL-R167Q, and VHL-wt 786-0 cells were cultured in complete medium under hypoxia (1% oxygen) for 72 hours, moved to normoxic conditions (21% oxygen) to reoxygenate the cells, and immediately lysed after reoxygenation in RIPA buffer for the indicated times. (E) VHL-R167Q-Venus but not VHL-R167Q completely suppressed tumor growth. Mice were injected with 786-0 cells expressing green fluorescent protein (GFP), VHL-W117A-Venus, VHL-R167Q, or VHL-R167Q-Venus. Tumors were measured on the indicated days after injection. (F) VHL-R167Q partially downregulated HIF2α and GLUT1 in xenografts. Lysates were prepared from snap frozen xenograft tissue. SE, short exposure; LE, long exposure.
We then compared the ability of VHL-wt and VHL-R167Q in 786-0 cells to downregulate HIF2α under different partial pressure of oxygen, which likely resembles conditions that tumor cells encounter in vivo (30). Under commonly used hypoxia conditions with 1% oxygen, HIF2α was stabilized in both VHL-wt and VHL-R167Q cells (Fig. 2C). As the percentage of oxygen increased, VHL-R167Q was less able to downregulate HIF2α than VHL-wt (Fig. 2C). We also compared the ability of VHL-wt and VHL-R167Q to downregulate HIF2α when cells were reoxygenated after hypoxia; this scenario may better reflect tissue oxygen fluctuation in vivo. VHL-R167Q was less able to downregulate HIF2α than VHL-wt (Fig. 2D).
We further examined the tumor suppressive function of VHL-R167Q in a mouse xenograft model. As expected, control 786-0 cells (which expressed GFP only) underwent rapid tumor growth (Fig. 2E). Consistent with its ability to downregulate HIF2α in tissue culture, VHL-R167Q-Venus completely suppressed tumor growth, whereas VHL-R167Q only partially suppressed tumor growth. VHL-W117A-Venus, a HIF2α-binding deficient mutation (4), did not efficiently suppress tumor growth (Fig. 2E) despite being expressed at levels comparable to that of VHL-R167Q-Venus (Fig. 1A). Our data indicated that, at high levels, VHL-R167Q-Venus completely suppresses tumor growth but VHL-R167Q at low levels only partially suppresses it.
We verified the protein levels of VHL, HIF2α, and the HIF target GLUT1 in xenografts (Fig. 2F). Consistent with the data for cultured cells, the VHL-R167Q level was significantly lower than that of VHL-W117A-Venus in xenografts. The levels of HIF2α and GLUT1 were lower in VHL-R167Q xenografts than in GFP or VHL-W117A-Venus xenografts; this finding indicated that VHL-R167Q retains partial ability to downregulate HIF and subsequently HIF targets in vivo.
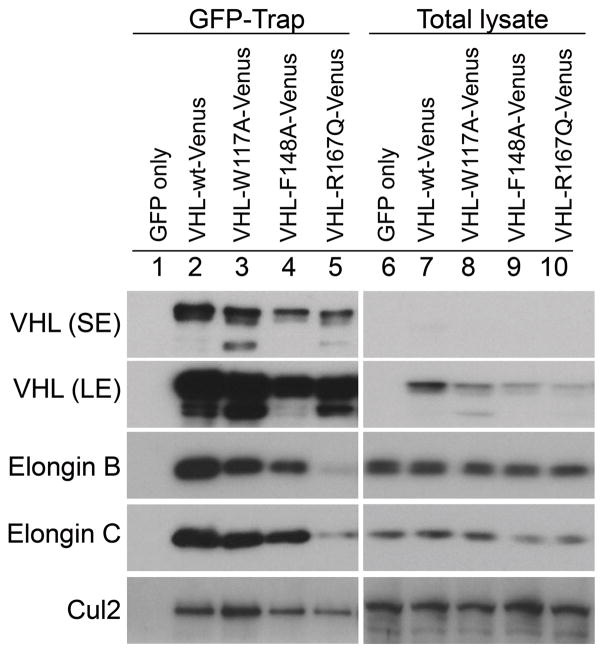
VHL-R167Q-Venus forms significantly less VBC complex compared with VHL-wt-Venus but is still sufficient for HIF2α downregulation
VHL serves as a substrate-binding component of an E3 ubiquitin ligase complex for HIF2α (2, 3). The mutated protein VHL-R167Q retains the ability to bind to the HIF2α substrate but is less efficient in binding elongin C and is therefore deficient in forming a functional VBC complex (12). We examined whether VHL-R167Q-Venus can form a functioning VBC complex for the observed HIF2α downregulation. We used a GFP-Trap assay that pulls down GFP fusion proteins and binding partners with high efficiency and specificity. Results showed that compared with VHL-wt-Venus, VHL-R167Q-Venus bound to substantially less elongin B, elongin C, and Cul2 (Fig. 3), consistent with results from a previous study (17). However, because VHL-R167Q-Venus was expressed at high levels, the residual VBC complex was likely still sufficient for complete HIF2α downregulation. VHL-W117A-Venus and VHL-F148A-Venus both bound to elongin B, elongin C, and Cul2 efficiently, though with modest changes compared with VHL-wt-Venus (Fig. 3).
Figure 3. R167Q mutation decreases VHL binding to elongin c, elongin B and CulII in 786-0 cells.
VHL-null 786-0 cells were infected to stably express the indicated proteins. Cells were cultured in complete medium and were lysed in 0.5% NP-40 buffer supplemented with protease inhibitors and phosphatase inhibitors. GFP-Trap elutes and total lysates were resolved on 10% SDS-polyacrylamide denaturing gels and analyzed with the indicated antibodies. Long exposure (LE) blots show weak bands. SE, short exposure.
Proteasome inhibition restores the expression levels and function of VHL-R167Q
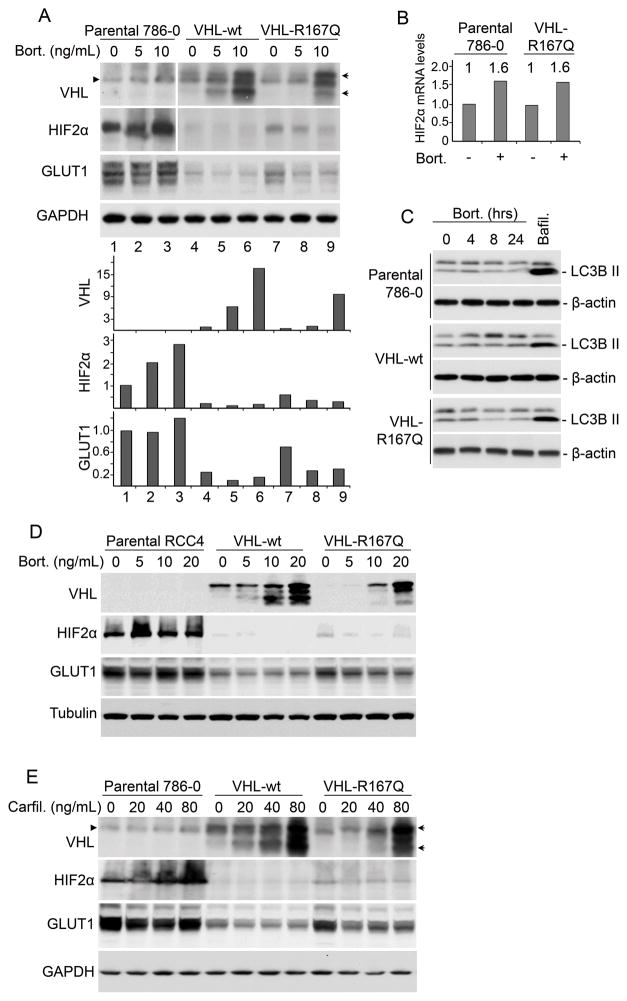
On the basis of our finding that the protein level of VHL-R167Q determines its ability to downregulate HIF2α and suppress tumors, we predicted that stabilization of this mutant protein would increase its functional ability. Treatment with bortezomib, a proteasome inhibitor currently used in cancer therapy, resulted in a significant increase in the protein levels of both VHL-wt and VHL-R167Q (Fig. 4A). In parental 786-0 cells with no VHL, bortezomib treatment resulted in an increase of HIF2α level which was likely due to blockade of VHL-independent HIF2α proteasomal degradation (Fig. 4A). In contrast, in cells expressing VHL-R167Q, bortezomib treatment significantly decreased HIF2α levels (Fig. 4A), which was consistent with stabilized VHL-R167Q facilitating HIF2α degradation. Bortezomib did not decrease HIF2α mRNA levels indicating the decrease in HIF2α protein levels was likely due to protein degradation (Fig. 4B). Indeed, bortezomib increased HIF2α mRNA levels to a certain extent, which may be responsible for elevated HIF2α protein levels when VHL is not present (Fig. 4A). Autophagy is a conserved process involved in lysosomal degradation of long lived proteins, protein aggregates, and damaged organelles (31). In order to explore the role of autophagy in bortezomib-induced HIF2α decrease, we checked the changes in type II LC3B (LC3B-II), an indicator of autophagy (31). Here we observed that bortezomib at the concentration we used didn’t increase LC3B-II, while autophagy-lysosome inhibition by bafilomycin A1 led to obvious accumulation of LC3B-II (Fig. 4C). These results suggest that autophagy was not responsible for the decrease of HIF2α levels in the presence of bortezomib. GLUT1 levels increased in parental cells but decreased in VHL-R167Q cells after bortezomib treatment, consistent with the changes in HIF2α levels (Fig. 4A). Moreover, in VHL-wt cells, both HIF2α and GLUT1 were at very low levels regardless of bortezomib treatment (Fig. 4A), indicating that HIF2α was efficiently degraded when VHL-wt was present.
Figure 4. Proteasome inhibitors stabilize VHL-R167Q protein and restore its function to downregulate HIF2α.
(A) Bortezomib (Bort.) stabilized VHL-R167Q protein and increased its ability to downregulate HIF2α in 786-0 cells. Parental, VHL-wt, and VHL-R167Q 786-0 cells were cultured in complete medium and treated with bortezomib for 24 hours (or left untreated) at the indicated concentrations. Cells were lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Scanning densitometric values were obtained using ImageJ software. Protein levels were normalized to the loading control and presented as relative conversion to values in VHL-wt cells (for VHL) or in parental cells (for HIF2α and GLUT1) without bortezomib treatment. The arrowhead designates a nonspecific band. The arrows designate VHL. (B) Bortezomib increased HIF2α mRNA levels. Parental and VHL-R167Q-Venus 786-0 cells were treated with bortezomib (10ng/ml) for 24 hrs (or untreated). HIF2α mRNA levels were measured by real-time PCR. Data are presented as relative conversion to values in untreated parental cells. (C) Bortezomib did not induce autophagy. Parental, VHL-wt, and VHL-R167Q 786-0 cells were treated with bortezomib (10ng/ml) for indicated time. Bafilomycin (Bafil.) (200 nM) treatment for 4 hrs was used as a positive control to induce LC3B-II accumulation. Cell lysates were analyzed by Western blotting using antibodies against LC3B and β-actin. (D) Bortezomib stabilized VHL-R167Q and increased its function to downregulate HIF2α in RCC4 cells. Parental, VHL-R167Q, and VHL-wt RCC4 cells were cultured in complete medium and treated with bortezomib for 24 hours (or left untreated) at the indicated concentrations. Cells were lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. (E) Carfilzomib (Carfil.) stabilized VHL-R167Q protein in 786-0 cells. Parental, VHL-wt, and VHL-R167Q 786-0 cells were cultured in complete medium and treated with carfilzomib for 24 hours (or left untreated) at the indicated concentrations. The arrowhead designates a nonspecific band. The arrows designate VHL.
We further examined the effect of bortezomib in another kidney cancer cell line. Bortezomib treatment resulted in similar VHL-R167Q upregulation together with HIF2α and GLUT1 downregulation in RCC4 cells (Fig. 4D), indicating the effect of bortezomib was not cell line specific. Another clinically used proteasome inhibitor, carfilzomib, showed similar effects in stabilizing VHL-R167Q protein levels and restoring its function in downregulating HIF2α and GLUT1 (Fig. 4E), indicating the effect of proteasome inhibition on VHL was not compound specific.
In silico identification of VHL mutations for rescue
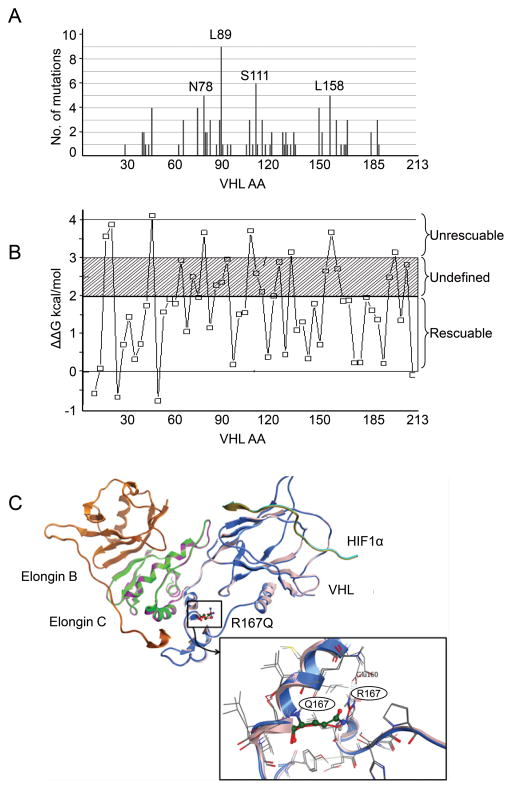
We showed that the protein levels and function of one type 2B VHL mutation, R167Q, could be rescued by increasing proteostasis. We then sought to develop in silico approaches to identify other VHL missense mutations that could be rescued and therefore to prioritize the mutations for future in vivo studies. We first compiled VHL point missense mutations in sporadic ccRCC from The Cancer Genome Atlas (TCGA) database (32–34) (Fig. 5A and Supplementary table 1). For sporadic ccRCC in the TCGA sample set, L89 was the most frequent missense mutation site; other prevalent missense mutations were on N78, S111, or L158 (Fig. 5A). We classified the sporadic missense mutations as buried or exposed (5) based on the recently solved VBC-HIF1α peptide complex (PDB id: 4AJY) (26). We used the PoPMuSiC program (28) to compute the variation in ΔΔG of the VHL complex with the introduction of a point mutation. Data from some experimentally tested mutations are shown in Fig. 5B. Using 3.0 kcal/mol as a cutoff, which corresponds to the loss of two or three hydrogen bonds, we would be able to rescue approximately 79% of missense point mutants (Fig. 5B and Supplementary table 2). If the cutoff value were more stringent, e.g., ΔΔG = 2.0 kcal/mol (corresponding to the loss of one or two hydrogen bonds), approximately 57% VHL missense mutations would be candidates for rescue (Supplementary table 2). Since missense point mutations make up approximately a third of all VHL mutations, we would theoretically be able to rescue between 14% and 25% of VHL mutations, which potentially affects a large number of VHL patients. Using the PoPMuSiC program, we predicted that the R167Q mutation would cause a 1.38 kcal/mol change in ΔΔG, which is consistent with an unstable but rescuable mutation. Structural analysis indicated that this destabilization was caused primarily by a change in the non-bonded intramolecular interactions of the residue Arg167 with Glu160, Asp187, and Asp190 (Fig. 5C). The R167W (ΔΔG = 1.24 kcal/mol) and L158P (ΔΔG = 1.86 kcal/mol) mutations were similarly characterized as rescuable mutations. We also calculated that the W117A (ΔΔG = 4.35 kcal/mol) mutation would greatly destabilize the complex structure and thus disrupt function; this prediction was consistent with our in vivo observation that the function of W117A mutation was not rescuable by increasing W117A expression (Fig. 1A). Using PoPMuSiC, we identified several frequent mutations, including L89H, L158P, and C162R, as potentially rescuable mutations in addition to R167Q. Our in silico results verified the experimental refunctionalization data of the VHL R167Q mutant and predicted more rescuable VHL mutants for future in vivo studies.
Figure 5. In silico identification of VHL mutations for rescue.
(A) Distribution of VHL missense point mutations in sporadic ccRCC in the TCGA dataset. VHL mutation data of ccRCC samples were downloaded from cBioPortal. Missense point mutations were selected for the distribution chart. (B) Depiction of folding free energy (ΔΔG) values of the experimental mutations computed using PoPMuSiC. (C) Depiction of structural changes of the VHL-ElonginB-ElonginC-Hif1a complex (pdb id: 4AJY) with the R167Q mutation. The detailed interactions of arginine with the neighboring residues are magnified.
Discussion
In this study, we demonstrated that the protein level of VHL-R167Q determines its functional capacity to downregulate HIF2α and suppress tumorigenesis. VHL-R167Q expressed at a low level fails to completely downregulate HIF2α and suppress tumorigenesis in xenograft models, whereas VHL-R167Q-Venus expressed at a high level completely downregulates HIF2α and suppresses tumorigenesis. We systematically analyzed the function of VHL-R167Q under normoxic, hypoxic, and reoxygenation conditions and in a xenograft model and demonstrated that VHL-R167Q is less able to downregulate HIF2α than VHL-wt is but still maintains rescuable functionality. In addition, we demonstrated that proteasome inhibition increases the protein level of the VHL-R167Q mutant and, more importantly, its ability to downregulate HIF2α and a HIF target.
Our results indicate that proteasome inhibition increases the levels and function of a type 2B missense VHL mutant, which provides a potential innovative therapeutic approach to treat kidney cancer patients by stabilizing these missense VHL mutants. We observed a commensurate decrease in both HIF2α and the HIF target GLUT1 after stabilization of the R167Q-VHL mutant. An important finding was that although proteasome inhibition stabilized VHL, it did not counteract the rescue of VHL activity by stabilizing HIF2α but actually resulted in a lowering of HIF2α levels in a VHL level dependent fashion. Therefore it is likely that under the proteasome inhibition conditions we used, specific activities of the proteasome were inhibited, or the proteasome was globally inhibited to an extent that VHL, but not HIF2α, degradation was blocked. A high level of proteasome inhibitor can stabilize the HIF protein; however, stabilized HIF that is ubiquitinated but not degraded does not appear to be functional (35). Our studies using bortezomib and carfilzomib suggest that with newer proteasome inhibitors, there is a therapeutic window for upregulating VHL without a commensurate increase in HIF, thereby creating a favorable therapeutic index.
We previously proposed that modulation of the proteostasis of missense point-mutated VHL could serve as a novel approach to treating kidney cancer (19). Our data in this study support the idea that proteasome inhibitors can be used to stabilize missense VHL mutations. A clinical study testing bortezomib in patients with refractory kidney cancer showed that a subset of patients had a dramatic and prolonged response to this agent (36). Unfortunately, no molecular markers of benefit were assessed in that study and the toxicity profile of bortezomib precluded its further development for patients with kidney cancer. On the basis of our data, a clinical study testing carfilzomib for refractory kidney cancer was launched at The University of Texas MD Anderson Cancer Center (NCT01775930) and will assess the ability of this agent to control kidney cancer and whether VHL subtypes can predict the response to this agent.
Proteasome inhibitors represent only one of many kinds of compounds that may stabilize missense VHL mutations. VHL proteostasis is affected by multiple steps that could be targeted. For example, nascent VHL binds to Heat Shock Protein 70 and is shuttled to the eukaryotic type II chaperonin tailless complex polypeptide-1 ring complex (TRiC), where it is folded into a mature form to stabilize the VHL protein (29, 37). Mutations in VHL amino acids 114–119 and 148–155 interfere with TRiC binding, resulting in the failure of VHL to fold properly (29, 37). VHL binds to elongin C and elongin B to form the functional VBC complex which also stabilizes VHL, and mutations in VHL amino acids 155–181 decrease elongin C binding and stability (14). Destabilized VHL binds to heat shock proteins and is directed to proteasome degradation (38). Compounds that improve VHL shuttling or binding to the folding machinery TRiC, or that inhibit VHL ubiquitination or shuttling to the proteasome, may stabilize missense VHL mutants. We previously performed a drug screen and identified multiple compounds that stabilize mutant VHL (19). A recent study also showed that histone deacetylase inhibitors may stabilize mutant VHL (39). The in silico approach identified other missense VHL mutants that might be rescued. Thus, proteostasis rescue may be applicable to a significant number of VHL mutants and could affect a large number of VHL patients.
Our xenograft data showed that VHL-R167Q at low levels failed to completely suppress tumorigenesis. This finding is consistent with previous studies showing that VHL-R167Q knockin mice were more susceptible than wild-type mice to carcinogen-promoted renal neoplasia (12). Our data further showed that VHL-R167Q at high levels completely suppressed tumorigenesis, providing in vivo evidence that VHL-R167Q levels dictate how well it functions to suppress tumors. We also found that proteasome inhibitors stabilize missense VHL and rescue its ability to downregulate HIF2α in vitro. Further studies are needed to explore compounds that mediate VHL stabilization in vivo, which is important for developing and implementing modulation of the proteostasis of missense VHL mutants as a novel therapeutic approach for kidney cancer.
In conclusion, we characterized the VHL-R167Q mutation in detail and demonstrated that proteasome inhibition increases the levels and function of the type 2B missense VHL mutant R167Q. Our results support the innovative concept of modulating the proteostasis of missense point-mutated VHL as therapy for ccRCC. Considering that multiple proteasome inhibitors are currently in clinical use, our results are of significance and may further the development and implementation of proteasome inhibitors as therapy for kidney cancer.
Supplementary Material
Acknowledgments
We thank Darius Jonasch and Jeremy Pustilnik for technical assistance. The work is supported by a grant from the University of Texas MD Anderson Cancer Center Kidney Cancer Multidisciplinary Research Program (ZD), NIH 5 PN2 EY016525-09 (EJ), and NCI CCSG grant (P30 CA016672).
References
- 1.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. von Hippel-Lindau disease. Lancet. 2003;361:2059–67. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 2.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–41. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–33. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 5.Rechsteiner MP, von Teichman A, Nowicka A, Sulser T, Schraml P, Moch H. VHL gene mutations and their effects on hypoxia inducible factor HIFalpha: identification of potential driver and passenger mutations. Cancer Res. 2011;71:5500–11. doi: 10.1158/0008-5472.CAN-11-0757. [DOI] [PubMed] [Google Scholar]
- 6.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–34. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 7.Escudier B, Pluzanska A, Koralewski P, Ravaud A, Bracarda S, Szczylik C, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet. 2007;370:2103–11. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 8.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 9.Rini BI, Escudier B, Tomczak P, Kaprin A, Szczylik C, Hutson TE, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet. 2012;378:1931–9. doi: 10.1016/S0140-6736(11)61613-9. [DOI] [PubMed] [Google Scholar]
- 10.Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol. 2010;28:1061–8. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 11.Banks RE, Tirukonda P, Taylor C, Hornigold N, Astuti D, Cohen D, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–11. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 12.Lee CM, Hickey MM, Sanford CA, McGuire CG, Cowey CL, Simon MC, et al. VHL Type 2B gene mutation moderates HIF dosage in vitro and in vivo. Oncogene. 2009;28:1694–705. doi: 10.1038/onc.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum Mol Genet. 2001;10:1029–38. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- 14.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–6. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 15.Chung J, Roberts AM, Chow J, Coady-Osberg N, Ohh M. Homotypic association between tumour-associated VHL proteins leads to the restoration of HIF pathway. Oncogene. 2006;25:3079–83. doi: 10.1038/sj.onc.1209328. [DOI] [PubMed] [Google Scholar]
- 16.Bangiyeva V, Rosenbloom A, Alexander AE, Isanova B, Popko T, Schoenfeld AR. Differences in regulation of tight junctions and cell morphology between VHL mutations from disease subtypes. BMC Cancer. 2009;9:229. doi: 10.1186/1471-2407-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker KE, Lee CM, Rathmell WK. VHL type 2B mutations retain VBC complex form and function. PLoS One. 2008;3:e3801. doi: 10.1371/journal.pone.0003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–45. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Z, German P, Bai S, Feng Z, Gao M, Si W, et al. Agents That Stabilize Mutated von Hippel-Lindau (VHL) Protein: Results of a High-Throughput Screen to Identify Compounds That Modulate VHL Proteostasis. J Biomol Screen. 2012;17:572–80. doi: 10.1177/1087057112436557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Z, Liang J, Lu Y, Yu Q, Songyang Z, Lin SY, et al. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A. 2006;103:15014–9. doi: 10.1073/pnas.0606917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X-D, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, et al. Transient Aggregation of Ubiquitinated Proteins Is a Cytosolic Unfolded Protein Response to Inflammation and Endoplasmic Reticulum Stress. Journal of Biological Chemistry 2012. 2012;287:19687–98. doi: 10.1074/jbc.M112.350934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maranchie JK, Zhan Y. Nox4 is critical for hypoxia-inducible factor 2-alpha transcriptional activity in von Hippel-Lindau-deficient renal cell carcinoma. Cancer Res. 2005;65:9190–3. doi: 10.1158/0008-5472.CAN-05-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolacsek O, Krizsik V, Schamberger A, Erdei Z, Apati A, Varady G, et al. Reliable transgene-independent method for determining Sleeping Beauty transposon copy numbers. Mobile DNA. 2011;2:5. doi: 10.1186/1759-8753-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukasiak P, Antczak M, Ratajczak T, Bujnicki JM, Szachniuk M, Adamiak RW, et al. RNAlyzer—novel approach for quality analysis of RNA structural models. Nucleic Acids Research. 2013;41:5978–90. doi: 10.1093/nar/gkt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Molle I, Thomann A, Buckley DL, So EC, Lang S, Crews CM, et al. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein:hypoxia inducible factor 1alpha protein-protein interface. Chemistry & biology. 2012;19:1300–12. doi: 10.1016/j.chembiol.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy AS, Jalahalli V, Kumar S, Garg R, Zhang X, Sastry GN. Analysis of HIV protease binding pockets based on 3D shape and electrostatic potential descriptors. Chemical biology & drug design. 2011;77:137–51. doi: 10.1111/j.1747-0285.2010.01065.x. [DOI] [PubMed] [Google Scholar]
- 28.Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell. 2003;12:1213–24. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 30.Carreau A, Hafny-Rahbi BE, Matejuk A, Grillon C, Kieda C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. Journal of Cellular and Molecular Medicine. 2011;15:1239–53. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yoshimori T, Levine B. Methods in Mammalian Autophagy Research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin DH, Li SH, Chun YS, Huang LE, Kim MS, Park JW. CITED2 mediates the paradoxical responses of HIF-1[alpha] to proteasome inhibition. Oncogene. 2007;27:1939–44. doi: 10.1038/sj.onc.1210826. [DOI] [PubMed] [Google Scholar]
- 36.Kondagunta GV, Drucker B, Schwartz L, Bacik J, Marion S, Russo P, et al. Phase II trial of bortezomib for patients with advanced renal cell carcinoma. J Clin Oncol. 2004;22:3720–5. doi: 10.1200/JCO.2004.10.155. [DOI] [PubMed] [Google Scholar]
- 37.Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–61. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 38.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–48. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 39.Yang C, Huntoon K, Ksendzovsky A, Zhuang Z, Lonser RR. Proteostasis modulators prolong missense VHL protein activity and halt tumor progression. Cell reports. 2013;3:52–9. doi: 10.1016/j.celrep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.