Abstract
This study describes our investigation on the prevalence and molecular identification of bartonellae from Rattus diardii and R. norvegicus in the urban areas of Malaysia. Of 95 rats investigated, Bartonella tribocorum, B. rattimassiliensis, B. coopersplainsensis, B. elizabethae, and B. queenslandensis were isolated from kidney and spleen homogenates of four rats. Bartonellae DNA was amplified from the rat organ tissues by using primers specific for the bartonellae RNA polymerase beta subunit (rpoB) gene in nine other rats. Sequence analysis of the rpoB gene fragments shows the identification of B. queenslandensis in five rats, B. elizabethae in three rats, and B. tribocorum in one rat. Combining the results of isolation and molecular detection of bartonellae, we found that the prevalence of Bartonella infection in the Rattus spp. investigated in this study was 13.7%. Implementation of effective rat control program in the urban areas is necessary to prevent the spillover of bartonellosis from rats to humans.
Bartonellae are bacterial parasites that infect mammalian erythrocytes and endothelial cells.1 These small gram-negative and fastidious bacteria are transmitted usually through the bites of hematophagous arthropods, such as fleas, lice, flies, and ticks. Bartonellosis is an emerging and reemerging zoonotic infection responsible for a variety of clinical syndromes in humans and animals.2 Members of the genus Bartonella exhibit high degrees of genetic diversity and ecologic plasticity.3 Since the first description of Bartonella bacilliformis, the type species of the genus, 29 Bartonella species have been reported (http://www.bacterio.net/b/bartonella.html). The increasing reports of new Bartonella species potentially causing human infections have spurred extensive investigations to identify the reservoir mammalian hosts and the arthropod vectors.4,5
The occurrence of bartonellae in rodents and other small mammals has been reported in several countries in Asia, including Bangladesh, China, Indonesia, Japan, Laos, Cambodia, Taiwan, Nepal, and Thailand.6–17 Bartonella tribocorum, B. rattimassiliensis, B. coopersplainsensis, B. elizabethae, and B. queenslandensis are among those that have been identified from the wild rats (Rattus species). Arthropod vectors, particularly fleas (Ctenocephalides felis) and ticks (Ixodes, Haemaphysalis), are often implicated in the natural maintenance of various species of bartonellae.16 A recent study in our laboratory reported the detection of B. henselae and B. clarridgeiae in C. felis fleas.17 However, no information is available on the presence of bartonellae in rodents in the urban areas of Malaysia. Thus, the main objective of this study was to determine the occurrence and type of bartonellae circulating in the urban wild rat population in Malaysia.
A total of 95 rodents were captured as part of rodent management program conducted by the pest control sections of the municipal councils, i.e., Kuala Lumpur (n = 59) and Pulau Pinang (n = 36), Malaysia during January 2008–December 2011. Rats were trapped by using live traps with tapioca and dried fish as baits. Rats were identified as Rattus diardii (n = 58) and R. norvegicus (n = 37) (Table 1), brought to our laboratory, and anesthetized by using an ether-charged chamber. Postmortem examination was conducted and organs (kidney, liver, and spleen) were harvested aseptically. A total of 295 tissue samples were obtained and kept at −80°C before processing.
Table 1.
Source and details of Rattus spp. investigated for bartonellae, Malaysia
| Rat species | Location | Sex | No. positive rats* |
|---|---|---|---|
| R. norvegicus (n = 37) | Kuala Lumpur (n = 12) | Male (n = 5) | 1 |
| Female (n = 7) | 1 | ||
| Pulau Pinang (n = 25) | Male (n = 19) | 3 | |
| Female (n = 6) | 0 | ||
| R. diardii (n = 58) | Kuala Lumpur (n = 47) | Male (n = 28) | 5 |
| Female (n = 19) | 3 | ||
| Pulau Pinang (n = 11) | Male (n = 6) | 0 | |
| Female (n = 5) | 0 | ||
| Total (n = 95) | Kuala Lumpur (n = 59) and Pulau Pinang (n = 36) | Male (n = 58) Female (n = 37) | 13 |
Combining results of isolation and direct amplification from rat tissues.
A 20% homogenized tissue was prepared by grinding approximately 0.4 grams of rat tissue in 2 mL of Schneider's liquid medium (Sigma, St. Louis, MO) by using a mortar and pestle. Two hundred microliters of the tissue homogenate were then inoculated on a commercially available Columbia agar plate (Isolabs Sdn. Bhd, Selangor, Malaysia) supplemented with 5% sheep blood, and incubated at 35°C in 5% CO2 incubator for at least one month. Bacterial growth was monitored at least once per week after initial plating. Suspected colonies were streaked on a fresh agar plate and subjected for amplification targeting citrate synthase (gltA)18 and RNA polymerase beta subunit (rpoB)19 genes. Sequence determination of the amplified fragments was performed in an ABI PRISM 377 DNA Sequencer (Applied Biosystems, Foster City, CA) with primers for gltA (BhCS.781p and BhCS.1137n) and rpoB (1400D and 2300R). Resulting sequences were compared with known Bartonella sequences deposited in GenBank by using the Basic Local Alignment Search Tool (BLAST) program (National Center for Biotechnology Information, Bethesda, MD). Based on rpoB sequences, a dendrogram was constructed by using the neighbor-joining method in MEGA software and bootstrap analysis with 1,000 resamplings.20
The bartonellae isolation rate from the wild rats was low. Only five Bartonella isolates were successfully recovered from four rats caught in Kuala Lumpur after an incubation period of 4–8 days. One isolate was obtained from the kidney homogenate of a R. norvegicus rat, and the remaining four isolates were obtained from kidney, liver, and spleen homogenates of three R. diardii rats (Table 2). Mixed infection of Bartonella spp. was detected in an R. diardii rat (DKK4) by isolation of different Bartonella species in kidney and spleen homogenates. Based on the BLAST analysis of the gltA (276 nuleotides) and rpoB sequences (750 bp), the isolates obtained in this study were identified as B. tribocorum, B. rattimassiliensis, B. coopersplainsensis, B. elizabethae, and B. queenslandensis. All isolates had ≥ 97% sequence similarities with their respective type strains. Thus, they are regarded as members of the same species, in accordance with a proposal by La Scola and others,21 that bartonellae should be considered as the same species if the sequence similarities for their gltA and rpoB genes are > 95.4% and 96.0%, respectively.
Table 2.
Sequence analysis of bartonellae identified from Rattus spp., Malaysia*
| Rat | Tissue, rat species, location | Gene | Closest relative (gene accession no., % similarity) |
|---|---|---|---|
| Isolation (n = 4 rats) | |||
| DKK5 | Kidney, R. norvegicus, KL | rpoB | B. tribocorum strain IBS 506 (AF165996, 96.9) |
| gltA | B. tribocorum strain IBS 506 (AJ005494, 99.6) | ||
| DKK4-(1) | Kidney, R. diardii, KL | rpoB | B. rattimassiliensis strain 15908 (AY515130, 98.2) |
| gltA | B. rattimassiliensis strain 15908 (AY515124, 97.8) | ||
| -(2) | Spleen, R. diardii, KL | rpoB | B. coopersplainsensis AUST/NH20 (EU111792, 100.0) |
| gltA | B. coopersplainsensis AUST/NH20 (EU111803, 99.6) | ||
| KL16 | Kidney, spleen and liver, R. diardii, KL | rpoB | B. queenslandensis AUST/NH12 (EU111787, 97.6) |
| gltA | B. queenslandensis AUST/NH12 (EU111798, 97.8) | ||
| KL20 | Kidney, spleen and liver, R. diardii, KL | rpoB | B. elizabethae F9251 (AF165992, 100.0) |
| gltA | B. elizabethae F9251 (Z70009, 100.0) | ||
| Direct amplification from rat organ tissue (n = 9 rats) | |||
| KL4, KL7, KL8 | Spleen, R. diardii, KL | rpoB | B. queenslandensis AUST/NH12 (EU111787, 97.1) |
| KL12 | Spleen, R. diardii, KL | rpoB | B. queenslandensis strain AUST/NH12 (EU111787, 99.8) |
| KL5 | Spleen, R. diardii, KL | rpoB | B. elizabethae (AF165992, 100.0) |
| KL11 | Spleen, kidney, R. norvegicus, KL | rpoB | B. queenslandensis AUST/NH12 (EU111787, 97.1) |
| RP4 | Spleen, kidney, R. norvegicus, PP | rpoB | B. tribocorum (AF165996, 97.6) |
| RP31, RP33 | Spleen, kidney, R. norvegicus, PP | rpoB | B. elizabethae (AF165992, 100.0) |
KL = Kuala Lumpur; rpoB = RNA polymerase beta subunit; gltA = citrate synthase; PP = Pulau Pinang.
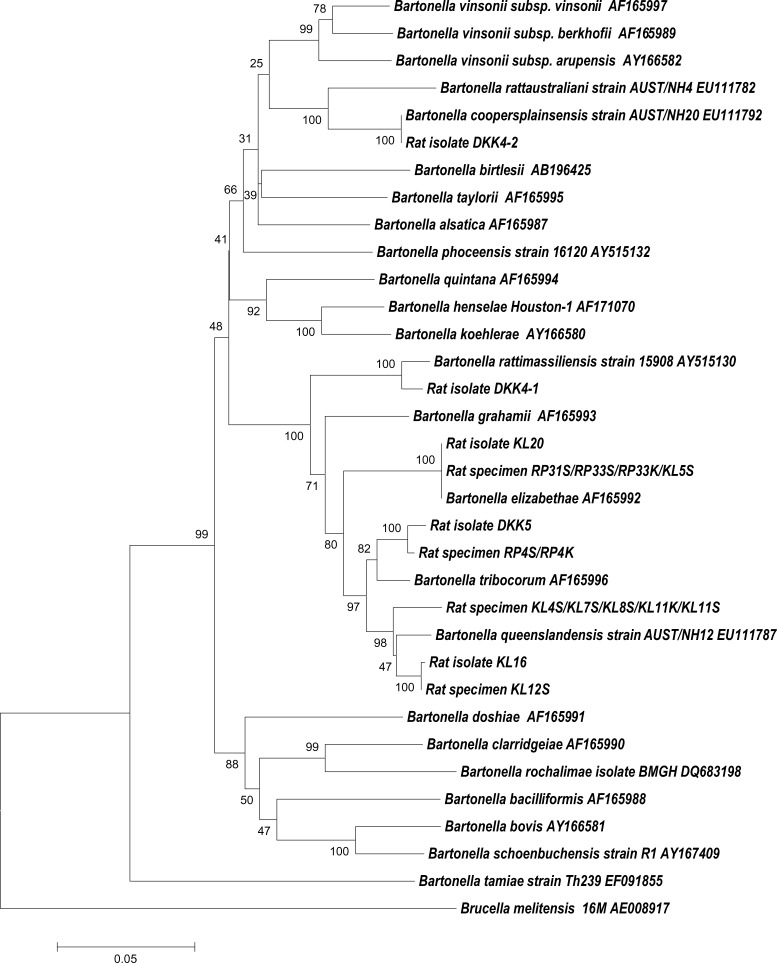
Direct amplification of bartonellae DNA from rat tissues was performed to improve the detection of bartonellae from the rat tissue homogenates in this study. DNA was extracted from 200 μL of each rat tissue homogenate by using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Polymerase chain reactions (PCRs) specific for gltA18 and rpoB19 genes were performed as described above. Amplification of the bartonellae gltA gene did not show a positive result. However, bartonellae DNA was detected from spleen and kidney samples of nine rats (four R. norvegicus and five R. diardii) by using rpoB PCRs (Table 2). Sequence analysis of the amplified rpoB genes identified B. queenslandensis in five rats, B. elizabethae in three rats, and B. tribocorum in one rat (Table 2). The species status of bartonellae in this study was confirmed when isolates clustered with known Bartonella species (Figure 1).
Figure 1.
Identification of Bartonellae by comparing the sequences of RNA polymerase beta subunit gene fragments of known Bartonella species. The dendrogram was constructed using the neighbor-joining method in MEGA software and bootstrap analysis with 1,000 resamplings. Source of the isolates and specimens is indicated in Table 1. S = spleen; K = kidney. Scale bar indicates nucleotide substitutions per site.
The prevalence of bartonellae in rodents varied in different geographic regions, ranging from 8.7% in Thailand15 to 64.2%, in United Kingdom.22 Combining the results of isolation and direct amplification from the rat organ tissues, we found that the overall prevalence of Bartonella infection in wild rats in this study was 13.7% (13 of 95 rats were positive). This finding suggests that R. rattus (13.5% positive) and R. norvegicus (13.8% positive) in cities may serve as a main reservoir for several Bartonella species in Malaysia.
Three Bartonella species (B. rattimassiliensis, B. tribocorum, and B. elizabethae) identified in this study are of public health significance in Southeast Asia because these species have been isolated from febrile patients in Thailand.5 Bartonella elizabethae has been identified as a causative agent of human endocarditis and neuroretinitis in Indonesia.8 These species have also been reported from small mammals in different regions e.g., France,23,24 the United States, and Portugal.25
Bartonella queenslandensis was the predominant Bartonella species identified in 6 of 13 (5 R. diardii and 1 R. norvegicus) rats in this study. The Bartonella species was originally isolated from R. fuscipes rats in Australia,26 and has been isolated from small mammals in Bangladesh,6 Nepal13 and three countries in southeastern Asia (Cambodia, Laos, and Thailand).11 Bartonella coopersplainsensis was first isolated from the blood of R. leucopus in Coopers Plains, Queensland, Australia,26 and later, from Rattus spp. and Bandicota spp. in Thailand.14,16 Nevertheless, the zoonotic potential of B. queenslandensis and B. coopersplainsensis has not been reported.
This study demonstrated the prevalence and genetic heterogeneity of Bartonella organisms in the urban wild rat population in Malaysia. Because identification of Bartonella species by using conventional microbiologic tests is difficult, PCR, followed by sequence analysis of specific target genes (gltA and rpoB), assisted our investigation. A confirmed case of bartonellosis has not been documented in Malaysia. However; in view of the identification of several bartonellae of medical importance in this study, implementation of effective rat control program in the urban areas is necessary to prevent the spilling over of bartonellosis from rats to human population.
ACKNOWLEDGMENTS
We thank B. Douadi, S. Samulong, N. Sahimin, J. Nagappan, Chai KS, all staff of the pest control unit of the Kuala Lumpur City Hall, and the Municipal Council of Penang Island for technical assistance.
Footnotes
Financial support: This study was supported by grants HIR/E000013-20001 (subprogramme 4) and RP013A/2012 from University of Malaya, Kuala Lumpur, Malaysia.
Authors' addresses: Sun Tee Tay and Aida Syafinaz Mokhtar, Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, E-mails: tayst@um.edu.my and aidasyafinaz87@gmail.com. Siti Nursheena Mohd Zain, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia, E-mail: nsheena@um.edu.my. Kiat Cheong Low, Laboratory Animal Resource Unit, Universiti Kebangsaan Malaysia, Kuala Lumpur, E-mail: lkc_mike@yahoo.com.
References
- 1.Breitschwerdt EB, Linder KL, Day MJ, Maggi RG, Chomel BB, Kempf VA. Koch's postulates and the pathogenesis of comparative infectious disease causation associated with Bartonella species. J Comp Pathol. 2013;148:115–125. doi: 10.1016/j.jcpa.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care. 2010;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 3.Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol. 2012;12:894–904. doi: 10.1016/j.meegid.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser PO, Riess T, O'Rourke F, Linke D, Kempf VA. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol. 2011;301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, Boonmar S, Bhengsri S, Dowell SF, Sitdhirasdr A, Lerdthusnee K, Richardson J, Peruski LF. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg. 2010;82:1140–1145. doi: 10.4269/ajtmh.2010.09-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y, Montgomery SP, Sheff KW, Chowdhury MA, Breiman RF, Kabeya H, Kosoy MY. Bartonella strains in small mammals from Dhaka, Bangladesh, related to Bartonella in America and Europe. Am J Trop Med Hyg. 2007;77:567–570. [PubMed] [Google Scholar]
- 7.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 8.Winoto IL, Goethert H, Ibrahim IN, Yuniherlina I, Stoops C, Susanti I, Kania W, Maguire JD, Bangs MJ, Telford SR, III, Wongsrichanalai C. Bartonella species in rodents and shrews in the greater Jakarta area. Southeast Asian J Trop Med Public Health. 2005;36:1523–1529. [PubMed] [Google Scholar]
- 9.Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, Yukawa M, Masuzawa T, Kawamori F, Kadosaka T, Takada N, Fujita H, Kawabata H. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–5092. doi: 10.1128/AEM.00071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelakis E, Khamphoukeo K, Grice D, Newton PN, Roux V, Aplin K, Raoult D, Rolain JM. Molecular detection of Bartonella species in rodents from the Lao PDR. Clin Microbiol Infect. 2009;15((Suppl 2)):95–97. doi: 10.1111/j.1469-0691.2008.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiyipong T, Jittapalapong S, Morand S, Raoult D, Rolain JM. Prevalence and genetic diversity of Bartonella spp. in small mammals from Southeastern Asia. Appl Environ Microbiol. 2012;78:8463–8466. doi: 10.1128/AEM.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol. 2008;57:1496–1501. doi: 10.1099/jmm.0.2008/004671-0. [DOI] [PubMed] [Google Scholar]
- 13.Gundi VA, Kosoy MY, Myint KS, Shrestha SK, Shrestha MP, Pavlin JA, Gibbons RV. Prevalence and genetic diversity of Bartonella species detected in different tissues of small mammals in Nepal. Appl Environ Microbiol. 2010;76:8247–8254. doi: 10.1128/AEM.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg. 2009;81:811–886. doi: 10.4269/ajtmh.2009.09-0294. [DOI] [PubMed] [Google Scholar]
- 15.Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Leepitakrat W, Monkanna T, Khlaimanee N, Chandranoi K, Jones JW, Coleman RE. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–433. [PubMed] [Google Scholar]
- 16.Saisongkorh W, Rolain JM, Suputtamongkol Y, Raoult D. Emerging Bartonella in humans and animals in Asia and Australia. J Med Assoc Thai. 2009;92:707–731. [PubMed] [Google Scholar]
- 17.Mokhtar AS, Tay ST. Molecular detection of Rickettsia felis, Bartonella henselae, and B. clarridgeiae in fleas from domestic dogs and cats in Malaysia. Am J Trop Med Hyg. 2011;85:931–933. doi: 10.4269/ajtmh.2011.10-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–321. doi: 10.1016/s0966-842x(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 22.Birtles RJ, Harrison TG, Molyneux DH. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann Trop Med Parasitol. 1994;88:317–327. doi: 10.1080/00034983.1994.11812872. [DOI] [PubMed] [Google Scholar]
- 23.Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piémont Y. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 25.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, Marston E, Ksiazek TG, Jones D, Childs JE. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 26.Gundi VA, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol. 2009;59:2956–2561. doi: 10.1099/ijs.0.002865-0. [DOI] [PubMed] [Google Scholar]