Abstract.
Presently, global efforts are being made to control and eradicate the deadliest tropical diseases through the improvement of adequate interventions. A critical point for programs to succeed is the prompt and accurate diagnosis in endemic regions. Rapid diagnostic tests (RDTs) are being massively deployed and used to improve diagnosis in tropical countries. In the present report, we evaluated the hypothesis of, after use for diagnosis, the reuse of the Leishmania RDT kit as a DNA source, which can be used downstream as a molecular surveillance and/or quality control tool. As a proof of principle, a polymerase chain reaction-based method was used to detect Leishmania spp. minicircle kinetoplast DNA from leishmaniasis RDT kits. Our results show that Leishmania spp. DNA can be extracted from used RDTs and may constitute an important, reliable, and affordable tool to assist in future leishmaniasis molecular surveillance methods.
Introduction
Leishmania is a protozoan parasite transmitted by phlebotomine sand flies that causes the neglected tropical disease commonly known as leishmaniasis. Within the Leishmania spp., parasites of the L. donovani complex are the causative agent of visceral leishmaniasis (VL) or kala-azar, the most severe form of leishmaniasis. VL is lethal if left untreated, and its mortality burden is estimated to be more than 50,000 deaths each year worldwide.1 In the Indian subcontinent, about 200 million people are estimated to be at risk of developing VL, accounting for 67% of the global disease burden.2
The classical microscopy diagnosis of VL from spleen or bone marrow fluids is an extremely invasive method. Recently, this method has been replaced by rapid diagnostic tests (RDTs) based on antibody detection using Leishmania antigens.2,3 The use of RDTs for tropical diseases has major advantages because of its high territory coverage, ease of performance, simple interpretation, and rapidity. Additionally, such methods do not require major laboratory facilities and constitute the only access to parasitological diagnostics for most of the people in developing countries.
Detection of Leishmania DNA in the peripheral blood has been previously reported4–10; herein, we report the capacity to extract DNA from VL RDTs and use it as a molecular tool for leishmaniasis molecular epidemiology studies. We show that the proposed approach can be used as an RDT quality control tool and a tool to evaluate the geographic prevalence of VL subspecies, which is important information for clinical and public health use for monitoring Leishmania parasites prevalence, fluctuation, and transmission routes in time and space in a scaled-up manner.
Materials and Methods
Sample collection.
In total, 147 samples were included in this study. The samples were obtained from suspected VL patients having irregular fever for more than 2 weeks combined with splenomegaly, hepatomegaly, weight loss, and signs of anemia who visited the Sukraraj Tropical and Infectious Disease Hospital (STIDH), Kathmandu, Nepal or Janakpur Zonal Hospital (JZH), Janakpur, Nepal for proper diagnosis and treatment. The included patients originated from different parts of Nepal, including areas so far believed to be endemic and non-endemic. The study was conducted from November of 2011 to June of 2012. Peripheral blood samples (1 mL) were collected in (ethylenedinitrilo) tetraacetic acid (EDTA) tubes from the suspected patients by a medical technologist for serological diagnosis. Eight microliters blood sample were used for the rK39 dipstick test (DiaMed-IT LEISH; Bio-Rad, Hercules, CA). This kit was used for all experiments throughout this study (we use RDT to refer to this specific test).
Paired samples were taken by spotting the same blood amount (8 μL) used for RDT in a Whatman–FTA classic card (GE Healthcare Ltd., Princeton, NJ) filter paper (FP) in four different spots. In all cases, FP in this work refers to this specific filter paper brand and model. Blood on FP was dried at room temperature. Both RDT kits and FPs were coded and kept in small plastic bags at room temperature in dry places away from direct sunlight.
The results of the RDTs were used by the physician for the management and treatment of patients. After confirmed leishmaniasis, patients were treated with either Amphotericin B at a dose of 1 mg/kg per day for 14 or 21 days intravenously (i.v.) or 50 mg miltefosine given orally two times per day for 28 days. Patients were discharged from the hospital after improvement based on their clinical symptoms.
Ethics statement.
This study was reviewed and approved by the Ethical Committee of Nepal Health Research Council and the Ethical Committee at the Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan. Blood samples were collected after obtaining written informed consent from patients and the concerned hospital. In the case of minors, consent was given by parents or guardians.
DNA extraction.
Blood from RDT kits and FPs was used for DNA extraction. The RDT strips and FPs were cut into small pieces by sterile scissors, and the portions containing the sample blotting site were used for DNA extraction. DNA was isolated from both RDTs and FPs using the QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol, and DNA was eluted to a volume of 50 μL. All samples were quantified using the ND-1000 spectrophotometer (Nano-Drop Technologies, Inc., Wilmington, DE).
Parasite culture.
The L. major (MHOM/SU/73-5-ASKH) strain was used as a source of control DNA, because the primer sites for polymerase chain reaction (PCR) detection are identical with the primer sites for L. donovani (described later). The promastigotes were cultured at 26°C in Medium 199 (GIBCO) supplemented with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), NaHCO3, 100 μg/mL streptomycin, 100 U/mL penicillin G potassium, and 10% heat-inactivated fetal calf serum (Nichirei Biosciences Inc, Tokyo, Japan).
Preparation of parasite DNA.
L. major promastigotes were mixed in 500 μL peripheral whole blood from a healthy volunteer. Limiting dilutions were performed sequentially in 1/10 (v/v) dilutions. After dilution, blood smears was prepared for each parasite solution, and parasite density was calculated using a hemocytometer against 100 white blood cells in a triplicate manner. Each parasite solution was spotted into Leishmania RDT kits (following the manufacturer's instructions) and FPs. DNA extraction was performed as described above.
PCR conditions.
We used a previously reported nested PCR method with primers targeting the parasite's minicircle kinetoplast DNA (kDNA; 6–7). For the first PCR amplification, 1 μL extracted DNA solution was used as a template in a final volume of 10 μL. The reaction mixture (10 μL) contains 1 × KOD-Plus-Neo buffer (Toyobo, Japan), 1.5 mM MgSO4, 0.2 mM each deoxynucleotide triphosphate (dNTPs), 1 U KOD-Plus-Neo-DNA polymerase, and 0.3 μM LinF4 (5′-GGGGTTGGTGTAAAATAGGG-3′) and LinR17 (5′-TTTGAACGGGATTTCTG-3′) primers. The PCR program was run for 35 cycles that consisted of denaturation at 94°C for 30 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 30 seconds.
Before the nested PCR, the amplified products from the first PCR were diluted at 1:20 with sterile distilled water. One microliter PCR product was subjected to the nested PCR with a set of inner primers KP1 (5′-TGTAAAATAGGGCCGGG-3′) and Lin19 (5′-CAGAACGCCCCTACCCG-3′). Ampdirect Plus reagent (Shimadzu Biotech, Tsukuba, Japan) was used to obtain maximum sensitivity of PCR amplification against the possible carryover of tissue-derived inhibitors for the enzymatic reaction in each sample of the nested PCR. For the nested PCR, 38 cycles were used, consisting of denaturation at 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 1 minute. Initial activation of Taq DNA polymerase was performed at 95°C for 10 minutes, and a final extension at 72°C for 10 minutes was included. Amplified PCR products were separated by electrophoresis on a 2% agarose gel with a 100-bp DNA ladder as a molecular size marker (SibEnzyme, Russia) and stained with ethidium bromide. Stained gels were visualized and photographed under ultraviolet (UV) light emission with a UV transilluminator (AE-6932 GXCFl; ATTO, Japan). Positive samples yielded a PCR product of 720 bp. In every run, sterile distilled water and DNA extracted from healthy human were used as negative controls, and DNA from cultured promastigotes served as a positive control. As a control, human DNA was PCR-amplified for specific human cytochrome b (cytb) as previously described.11
DNA sequencing.
The PCR products were extracted from agarose gels, purified using the Nucleospin Extraction II kit (Clontech Laboratories Inc., Mountain View, CA), and ligated into the pGEM-T-easy vector (Promega, Madison, WI) following the manufacturer's instructions, and Escherichia coli DH5α cells were transformed. Purified clones were checked for inserts by PCR analysis and sequenced for both strands using SP6 and T7 primers with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
Sequences were then used as queries to search for homologous sequences in the nucleotide database (nr) using the Basic Local Alignment Search Tool (BLAST).
Data analysis.
All statistical tests were performed using GraphPad InStat (GraphPad Software Inc., La Jolla, CA). DNA sequences were aligned with MUSCLE,12 and the phylogenetic tree was constructed by the neighbor-joining method implemented in PHYLIP v3.68. Geographic location and a map were obtained through Google Maps (http://maps.google.com).
Results
Patient data.
In total, 147 samples were collected from patients clinically diagnosed as VL: 47 samples at STIDH and 100 samples at JZH. The most common clinical features were fever, headache, splenomegaly, and hepatomegaly (Table 1). We found a significant difference in the sex of people seeking health assistance: 62% were male, whereas 38% were female. The age of the patients ranged from 4 months to 70 years (median = 28 years), with splenomegaly ranging from just palpable to 17 cm from the costal margin along the midclavicular line. Patients were subjected to routine laboratory checks, including white blood cells (WBCs), platelets, and hemoglobin (Hb) levels.
Table 1.
Clinical data of patients included in the present study
| CI95 | ||
|---|---|---|
| Age, years (N = 74) | ||
| Median | 28 | 24–32 |
| Minimum | 0.3 | |
| Maximum | 70 | |
| Hb, g/dL (N = 74) | ||
| Median | 9.0 | 8.5–10.0 |
| Minimum | 3.6 | |
| Maximum | 14.2 | |
| WBC, /μL (N = 73) | ||
| Median | 5,400 | 4,200–6,500 |
| Minimum | 1,100 | |
| Maximum | 21,800 | |
| Platelets, /μL (N = 44) | ||
| Median | 146,000 | 134,000–161,000 |
| Minimum | 69,000 | |
| Maximum | 526,000 | |
| Mean | ||
| Sex (N = 146) | ||
| Male | 62% | 53.6–69.1% |
| Female | 38% | 30.9–46.5% |
| Fever (N = 143) | ||
| Yes | 99% | 95.8–> 99.99% |
| No | 1% | < 0.01–4.3% |
| Headache (N = 140) | ||
| Yes | 89% | 82.1–92.9% |
| No | 11% | 7.1–17.9% |
| Splenomegaly (N = 145) | ||
| Yes | 90% | 84.3–94.3% |
| No | 10% | 5.7–15.7% |
| Hepatomegaly (N = 120) | ||
| Yes | 81% | 72.8–86.9% |
| No | 19% | 13.1–27.2% |
| rK39 RDT (N = 147) | ||
| Positive | 44% | 35.8–51.6% |
| Negative | 56% | 48.4–64.2% |
Of 147 patients suspicious of being Leishmania-infected included in our study, 64 (44%) patients had a positive RDT result; conversely, 83 (56%) patients were diagnosed negative for Leishmania using the RDT kit.
PCR amplification of kDNA from RDTs.
We then optimized a PCR-based method (6) using DNA extracted from the RDT kit to detect Leishmania parasites by targeting the minicircle kDNA, which is defined in this work as the RDT-PCR method. RDT-PCR was compared with DNA extracted from the FP classical dried blood spot method, which is defined in this work as the FP-PCR method.
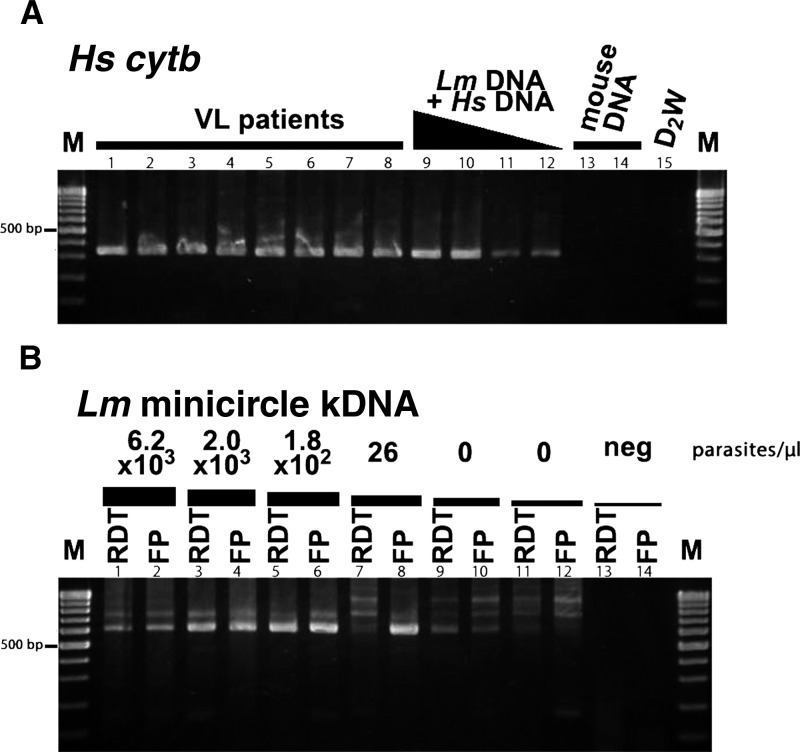
DNA extraction efficiency from RDT kit and FP was assessed by spectrophotometry and through PCR. Human DNA (cytb) was used as a positive control and detected in all samples analyzed (Figure 1). The median DNA concentration obtained for the RDT group was 5.120 ng/μL (SD = 3.0231), which was significantly lower than the median for the FP group (6.580 ng/μL; SD = 9.1597; P value = 0.0309 by Mann–Whitney test).
Figure 1.
Sensitivity of the PCR and extraction methods to detect Leishmania parasite in RDT or FP. In A (Hs = Homo sapiens), a human-specific cytb PCR product (330-bp band) is shown to detect human DNA in eight patients (lanes 1–8), L. major(Lm) limiting diluted samples (lanes 9–12), mouse blood (lanes 13 and 14), and distilled water (D2W, lane 15). In B, detection of L. major kDNA (620-bp band) in limiting dilution solutions of L. major plotted in the RDT kits or FP with a parasite density (parasites per microliter) of 6.2 × 103 (lane 1, RDT; lane 2, FP), 2 × 103 (lane 3, RDT; lane 4, FP), 1.8 × 102 (lane 5, RDT; lane 6, FP), 26 (lane 7, RDT; lane 8, FP), 0 (lane 9, RDT; lane 10, FP), and 0 (lane 11, RDT; lane 12, FP). Lanes 13 and 14 correspond for RDT- and FP-negative controls, respectively. Note that 0 parasites/μL means that parasites were not detected under Giemsa staining microscopy. The M lanes stand for 100-bp molecular marker.
To determine the PCR detection threshold of parasite DNA extracted from RDT or FP, we prepared a series of DNA solutions by serially diluting an L. major solution (up to 0 parasites/μL) and spotting it onto the RDT kit or FP. Detection threshold by PCR was shown to be below the microscopic detection limit (Figure 1) and not significantly altered, even when kept for a long period (3 months) at room temperature (data not shown).
Then, we randomly selected 12 samples that were VL-positive by both RDT and PCR and 3 samples that were VL-negative by RDT diagnostics but PCR-positive to determine the identity of the PCR product. We cloned and evaluate 44 sequences obtained from 15 RDTs (sequences deposited to DNA Data Base in Japan/European Molecular Biology Laboratory/GenBank with accession numbers AB786560–AB786603); 10 of 15 samples were shown to possess heterogeneous kDNA, which ranged from one to six different kDNA sequences.
Using the BLASTN database search through the National Center for Biotechnology Information (NCBI), we found that all kDNA sequences possessed high homology with the L. donovani complex species, the causative agent of VL. Highest hits unveiled high homology with three L. donovani subspecies: L. donovani donovani, L. donovani infantum, and L. donovani chagasi.
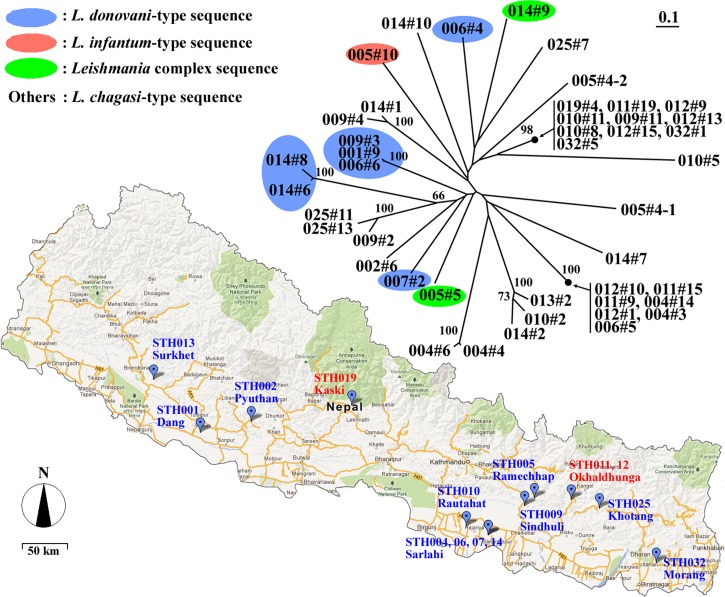
As a proof of concept, we sought to evaluate the genetic diversity of kDNA sequences and the origin of the infection in Nepal. For that purpose, we performed a multiple sequence alignment that was further analyzed in a dendrogram (Figure 2). The sequences were shown to be highly diverse, and no clear geographic association was found (Figure 2).
Figure 2.
kDNA diversity of Leishmania parasites in Nepal. Patient's origin and unrooted dendrogram of Leishmania sequences obtained in Nepal based on the minicircle kDNA nucleotide sequence are shown. Numbers on branches indicate bootstrap values based on 1,000 pseudoreplicates, and only values above 70 are shown.
RDT-PCR field application.
Application of the PCR method showed that 41 of 64 RDT-positive (64%; confidence interval95 [CI95] = 51.8–74.7%) results were confirmed by PCR performed in DNA extracted from RDTs. Similar results were obtained when using paired samples collected in FP (55%; CI95 = 42.6–66.3%). Comparison of PCRs performed from FP and RDTs samples can be found in Supplemental Table 1.
RDT-PCR for negative RDTs diagnosis was able to detect parasites in 26 of 86 negative RDTs (31%; CI95 = 22.3–42.0%), and FP-PCR for negative FP diagnosis was able to detect parasites in 12 of 86 negative FPs (14%; CI95 = 8.3–23.8%).
Evaluation of diagnostic capacity of RDT assessed by RDT-PCR showed a sensitivity of 61% (CI95 = 49.2–72.0%) and specificity of 71% (CI95 = 60.5–80.1%). With a poor/fair κ-coefficient of 0.326 ± 0.078, the diagnostic capacity of RDT for positive predictive value (PPV) was 64% (CI95 = 51.8–74.7%), and the negative predictive value (NPV) was 69% (CI95 = 58.0–77.7%) (Table 2).
Table 2.
Results of the rK39-based RDT compared with the results of the RDT-PCR for 147 clinically suspected VL patients
| RDT | RDT-PCR | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 41 | 23 | 64 |
| Negative | 26 | 57 | 83 |
| Total | 67 | 80 | 147 |
Discussion
We conducted a study to develop/evaluate new tools for VL diagnosis using RDTs and PCR-based methods. From all clinically suspicious leishmaniasis cases included in our study, we were just able to confirm approximately 50% of the cases by either immunologic or molecular tests. This result translates the difficulties in clinically diagnosing VL; it also shows its repercussions, leading to an overestimation at the diagnosis level. These observations support the urgent need for novel VL diagnostic tools.
The developed RDT-PCR method was shown to be compatible with relatively long storage periods of the sample, maintaining sensitivity below the microscopic detection limit. This feature makes the proposed method comparable with the conditions found in field settings. Despite the fact that higher DNA concentrations were observed in the FP group compared with RDT group, this observation did not correlate with the parasite detection outcome. A plausible explanation for this observation may be the fact that blood samples are treated with a running buffer in the RDT case, whereas for FP, blood is directly spotted with no pre-treatment. Chemicals like sodium azide, present in the running buffer, are likely to help the preservation of DNA integrity and enhance PCR efficiency.
We tested the applicability of the developed RDT-PCR method in two venues: (1) using it as a quality control tool to evaluate RDT diagnostic performance and (2) evaluating minicircle kDNA sequence diversity in relation to species identification and geolocation. Regarding the latter, in our pilot study, the high genetic diversity of kDNA and the reduced number of samples analyzed did not allow the depiction of a clear genotype/location association. Nevertheless, we show that the proposed system can be used to evaluate the regional prevalence of VL subspecies, which is important information for clinical and public health use for monitoring Leishmania parasite fluctuation and transmission routes in time and space in a scaled-up manner.
The estimated predictive RDT diagnostic capacity assessed by the RDT-PCR method is in agreement with more conservative published results that sensitivity ranges from 67% to 100% in various studies.13–20 More research is needed to clearly establish the correlation between PCR positivity and positive RDT, because we did not use microscopy for parasitological detection. However, the presence of PCR-positive samples among negative RDTs strongly suggests that some infections are not being diagnosed by the RDTs studied.
An important characteristic of our study is that RDTs were performed in a non-controlled, real-life routine setting at governmental health institutions and account for all variables inherent with it (e.g., RDT reader, technical issues, infrastructures, etc.). Thus, the proposed Leishmania PCR-RDT monitoring and evaluation system increases the effectiveness of the RDT, whereas most previous studies relate with RDT affectivity and were performed under controlled circumstances.
An advantage of using PCR for a quality control purpose is that other analytic material (DNA) is used instead of an additional comparable immunologic reactions approach. Previously RDTs results were compared with other immunological/serological tests, which have similar indirect analytic background and might bias the final results. We specifically refer to the cases of false positives, which are most probably not able to be discriminated when using immune detection tests for evaluation.
kDNA sequencing showed divergent homology within the L. donovani complex. Some of the analyzed sequences shared preferential homology with not previously reported Leishmania subspecies in Nepal, like L. donovani infantum/chagasi. Despite the fact that RDTs were capable of diagnosing these divergent parasites, it is known that different RDTs sensitivities and specificities were obtained when used in different geographic locations.21 The present article paves the way to perform future investigations in this field, which previously was not possible to do in a feasible manner.
Elimination of VL from Nepal and its neighboring countries mainly depends on strict adherence to the regional strategic plan that strongly recommends the use of RDT for diagnosis of VL. Here, we propose a method that generates added value to the use and deployment of RDTs through its reuse and consequent molecular analysis of harvested DNA. This method has the potential for other future application beyond the proof of concepts herein presented. We are now starting its usage for molecular surveillance of parasite resistance to antileishmanial drugs and/or monitoring of insidious parasite reservoirs in different interventions, such as VL elimination programs. Reuse of RDT is safe, cost-effective, and field-adapted, it does not require highly invasive procedures, and it is easy to use for molecular surveillance methods. Up-scaled surveillance programs—based on the asset of the already large coverage of the RDT itself—have the potential to provide pivotal information needed for VL elimination programs.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Dr. Jose Pedro Gil for the fruitful discussions and considerations regarding this work. We also acknowledge all patients, medical staff, and local authorities of Nepal for their assistance.
References
- 1. Desjeux P, 2004. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis 27: 305–318 [DOI] [PubMed] [Google Scholar]
- 2. Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S, Peeling RW, Alvar J, Boelaert M, 2007. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 5: 873–882 [DOI] [PubMed] [Google Scholar]
- 3. Zijlstra EE, Ali MS, el-Hassan AM, el-Toum IA, Satti M, Ghalib HW, Kager PA, 1992. Kala-azar: a comparative study of parasitological methods and the direct agglutination test in diagnosis. Trans R Soc Trop Med Hyg 86: 505–507 [DOI] [PubMed] [Google Scholar]
- 4. Gatti S, Gramegna M, Klersy C, Madama S, Bruno A, Maserati R, Bernuzzi AM, Cevini C, Scaglia M, 2004. Diagnosis of visceral leishmaniasis: the sensitivities and specificities of traditional methods and a nested PCR assay. Ann Trop Med Parasitol 98: 667–676 [DOI] [PubMed] [Google Scholar]
- 5. Antinori S, Calattini S, Longhi E, Bestetti G, Piolini R, Magni C, Orlando G, Gramiccia M, Acquaviva V, Foschi A, Corvasce S, Colomba C, Titone L, Parravicini C, Cascio A, Corbellino M, 2007. Clinical use of polymerase chain reaction performed on peripheral blood and bone marrow samples for the diagnosis and monitoring of visceral leishmaniasis in HIV-infected and HIV-uninfected patients: a single-center, 8-year experience in Italy and review of the literature. Clin Infect Dis 44: 1602–1610 [DOI] [PubMed] [Google Scholar]
- 6. Pandey K, Pant S, Kanbara H, Shuaibu MN, Mallik AK, Pandey BD, Kaneko O, Yanagi T, 2008. Molecular detection of Leishmania parasites from whole bodies of sandflies collected in Nepal. Parasitol Res 103: 293–297 [DOI] [PubMed] [Google Scholar]
- 7. Aransay AM, Scoulica E, Tselentis Y, 2000. Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol 66: 1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Srivastava P, Mehrotra S, Tiwary P, Chakravarty J, Sundar S, 2011. Diagnosis of Indian visceral leishmaniasis by nucleic acid detection using PCR. PLoS One 6: e19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chargui N, Haouas N, Jaouadi K, Gorcii M, Pratlong F, Dedet JP, Mezhoud H, Babba H, 2012. Usefulness of a PCR-based method in the detection and species identification of Leishmania from clinical samples. Pathol Biol (Paris) 60: e75–e79 [DOI] [PubMed] [Google Scholar]
- 10. Salam MA, Khan MG, Bhaskar KR, Afrad MH, Huda MM, Mondal D, 2012. Peripheral blood buffy coat smear: a promising tool for diagnosis of visceral leishmaniasis. J Clin Microbiol 50: 837–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kent RJ, Norris DE, 2005. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg 73: 336–342 [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brandonisio O, Fumarola L, Maggi P, Cavaliere R, Spinelli R, Pastore G, 2002. Evaluation of a rapid immunochromatographic test for serodiagnosis of visceral leishmaniasis. Eur J Clin Microbiol Infect Dis 21: 461–464 [DOI] [PubMed] [Google Scholar]
- 14. Sundar S, Pai K, Sahu M, Kumar V, Murray HW, 2002. Immunochromatographic strip-test detection of anti-K39 antibody in Indian visceral leishmaniasis. Ann Trop Med Parasitol 96: 19–23 [DOI] [PubMed] [Google Scholar]
- 15. Carvalho SF, Lemos EM, Corey R, Dietze R, 2003. Performance of recombinant K39 antigen in the diagnosis of Brazilian visceral leishmaniasis. Am J Trop Med Hyg 68: 321–324 [PubMed] [Google Scholar]
- 16. Bern C, Jha SN, Joshi AB, Thakur GD, Bista MB, 2000. Use of the recombinant K39 dipstick test and the direct agglutination test in a setting endemic for visceral leishmaniasis in Nepal. Am J Trop Med Hyg 63: 153–157 [DOI] [PubMed] [Google Scholar]
- 17. Sarker CB, Momen A, Jamal MF, Siddiqui NI, Siddiqui FM, Chowdhury KS, Rahman S, Talukder SI, 2003. Immunochromatographic (rK39) strip test in the diagnosis of visceral leishmaniasis in Bangladesh. Mymensingh Med J 12: 93–97 [PubMed] [Google Scholar]
- 18. Zijlstra EE, Nur Y, Desjeux P, Khalil EA, El-Hassan AM, Groen J, 2001. Diagnosing visceral leishmaniasis with the recombinant K39 strip test: experience from the Sudan. Trop Med Int Health 6: 108–113 [DOI] [PubMed] [Google Scholar]
- 19. Boelaert M, Rijal S, Regmi S, Singh R, Karki B, Jacquet D, Chappuis F, Campino L, Desjeux P, Le Ray D, Koirala S, Van der Stuyft P, 2004. A comparative study of the effectiveness of diagnostic tests for visceral leishmaniasis. Am J Trop Med Hyg 70: 72–77 [PubMed] [Google Scholar]
- 20. Sundar S, Reed SG, Singh VP, Kumar PC, Murray HW, 1998. Rapid accurate field diagnosis of Indian visceral leishmaniasis. Lancet 351: 563–565 [DOI] [PubMed] [Google Scholar]
- 21. WHO , 2011. Visceral Leishmaniasis Rapid Diagnostic Test Performance. Geneva: World Health Organization [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.