Abstract
Species and subspecies of the Oncomelania hupensis species complex are recognized as intermediate hosts of Schistosoma japonicum. Of these species and subspecies, O. quadrasi is distributed throughout the Philippines. This study used 12S ribosomal RNA sequences to explore the genetic structure of O. quadrasi populations in the Philippines. Three subspecies, O. h. hupensis, O. h. formosana, and O. h. chiui of this group were also examined. The phylogenetic tree and haplotypes network showed that O. quadrasi separated from the subspecies. Ten O. quadrasi haplotypes (Oq1–Oq10) clustered in relation to their geographic origin. Genetic differentiation (FST) and estimated gene flow (Nm) among populations showed significant differences, ranging from 0.556–1.000 to 0.00–0.74, respectively. Genetic differences among groups (FCT = 0.466), populations within a group (FSC = 0.727), and populations (FST = 0.854) were observed. These results indicate that the O. quadrasi populations in the Philippines have a substructure associated with their geographic origin.
Introduction
Snails of the Oncomelania hupensisis group are recognized as the intermediate hosts of Schistosoma japonicum, a major causative agent of schistosomiasis in countries in Asia. There are many species and subspecies in the O. hupensis complex that act as the intermediate hosts of S. japonicum, i.e., O. hupensis hupeneis, O. h. tangi and O. h. robertsoni in China, O. h. nosophora in Japan, O. h. lindoensis in the Indonesian islands, O. h. formosana, O. h. chiui in Taiwan, and O. quadrasi in the Philippines.1,2 During the 1970s, S. japonicum was still endemic to many areas in the Philippines. Currently 560,000 persons are estimated to be infected and more than 6.7 million are at risk of infection in 28 of the 79 provinces in the country, particularly among rice farmers and fishermen.1,2 Moreover, not only are humans infected, but there are many animals that act as final reservoir hosts of S. japonicum in the Philippines, including dogs, cats, pigs, rats, goats, cattle, and water buffalos.3,4
The genetic diversity of O. hupensis complex, especially O. h. hupensis, O. h. tangi, and O. h. robertsoni from mainland China, has been intensively investigated.5,6 Evidence of close co-evolutionary relationships between S. japonicum and its intermediate snail host O. hupensis has been demonstrated. This finding has led to an increased interest in the phylogenetics and population genetic structure of the snail.5,7 Because there are co-evolutionary relationships and close genetic interactions between the snails and their parasites, a snail population may reflect population genetic parameters of the parasite and vice versa.8,9 Genetically diverse snail populations appear to be more susceptible to infection with S. japonicum than homogeneous populations.10
In spite of a considerable amount of research on the genetic diversity within populations of O. quadrasi, only a limited assessment has been made on the significance of this diversity in a population structure or phylogenetic context. For instance, there are some reports of genetic diversity of O. quadrasi based on several genetic markers, including allozymes, nuclear DNA, and mitochondrial DNA sequences.11–13 These markers could be potentially used to demonstrate genetic diversity within O. quadrasi and its differentiation from other species and subspecies within the O. hupensis complex. However, knowledge of the genetic structure of O. quadrasi populations in the Philippines is virtually unknown.
The taxonomic status of variously designated subspecies of O. hupensis is controversial because some authors favor subspecific characterization while others consider certain previously designated subspecies, including O. quadrasi, to be full species based on morphologic (e.g., shell sculpture, operculum) and molecular characteristics and geographic distributions.14,15 Recent studies strongly support the specific status of O. quadrasi.13,14 For a better understanding of the population genetic structure of O. quadrasi in the Philippines, this study investigated 12S ribosomal RNA sequence variation from six localities in three provinces on three islands. Three subspecies, O. h. hupensis from China, and O. h. formosana and O. h. chiui from Taiwan, were also included in the analysis to determine the levels of interspecific genetic variation and phylogenetic relationships.
Materials and Methods
Sample collection.
Snails were collected from the field from different countries: O. quadrasi from the Philippines, O. h. hupensis from China, O. h. chiui, and O. h. formosana from Taiwan (Table 1). Collections from the Philippines were from three provinces: Cagayan, Oriental Mindoro, and Leyte (Table 1), which are located on the islands of Luzon, Mindoro, and Leyte, respectively. Snails were washed in sterile 0.3% NaCl and cracked to break their shells before being placed in individual vials containing 80% ethanol to transport to the laboratory for genetic investigation. Each snail was washed several times in sterile 0.3% NaCl, and the head–foot muscle dissected under a microscope and dried using a vacuum dryer. The tissues were then used for DNA extraction.
Table 1.
Localities of Oncomelania quadrasi and O. hupensis ssp., the Phillippines
| Sample code | Locality | Province* | Country | Subspecies | No. |
|---|---|---|---|---|---|
| AN | Aling Nene | Oriental Mindoro | Philippines | O. quadrasi | 10 |
| MB | Malabo | Oriental Mindoro | Philippines | O. quadrasi | 10 |
| DG | Dagami | Leyte | Philippines | O. quadrasi | 10 |
| MG | Magrafil | Cagayan | Philippines | O. quadrasi | 10 |
| SM | Sta. Maria | Cagayan | Philippines | O. quadrasi | 10 |
| TP | Tapel | Cagayan | Philippines | O. quadrasi | 9 |
| AH | Guichi | Anhui | China | O. h. hupensis | 5 |
| SC | Tianquan | Sichuan | China | O. h. hupensis | 5 |
| YN | Weishan | Yunnan | China | O. h. hupensis | 5 |
| KS | Yue-Mei | Kaohsiung | Taiwan | O. h. formosana | 5 |
| TA | Shimen | Taipei | Taiwan | O. h. chiui | 5 |
Oriental Mindoro, Leyte, and Cagayan are located on the islands of Mindoro, Leyte, and Luzon, respectively.
DNA extraction, PCR amplification, and DNA sequencing.
Total genomic DNA of each individual snail was extracted by using the E.Z.N.A® Mollusc DNA Kit (Omega Bio-Tek, Norcross, GA) following the manufacturer's instructions. The 12S ribosomal RNA gene was amplified by using primers and polymerase chain reaction (PCR) conditions as reported by Okamoto and others.12 All PCR products were gel-purified using GENECLEAN®II Kit (Q-BIO Gene, Carlsbad, CA). The purified PCR products were cycle-sequenced by using ABI BigDye v3.1 chemistry and run on an ABI Prism 377 automated sequencer (Applied Biosystems, Foster City, CA).
Data analyses.
Sequences were assembled and edited by eye using the software program ABI sequence scanner v1.0. Multiple sequence alignment was performed using a BioEdit version 5.0.6. Genetic differentiation (FST), gene flow estimation (Nm), and other parameters of DNA sequence polymorphism, including number of segregation site (S), haplotype diversity (Hd), nucleotide diversity (π), genetic diversity (θs), and the mismatch distribution under an expected population growth-decline model,16 were computed and generated by using DnaSP v5 software.17 Genetic structure, fixation indices, and evolutionary neutrality were calculated by using Arlequin 3.5.18 Minimum spanning haplotype network was constructed by using the Network 4.6.1.0 program based on median-joining algorithm.19 A phylogenetic tree was constructed based on an unweighted pair group method with arithmetic mean analysis by using Phylip program version 3.6.20 The relative support for clades was determined by using 1,000 bootstrap replicates.
Results
Interspecific analyses of O. hupensis ssp. and O. quadrasi.
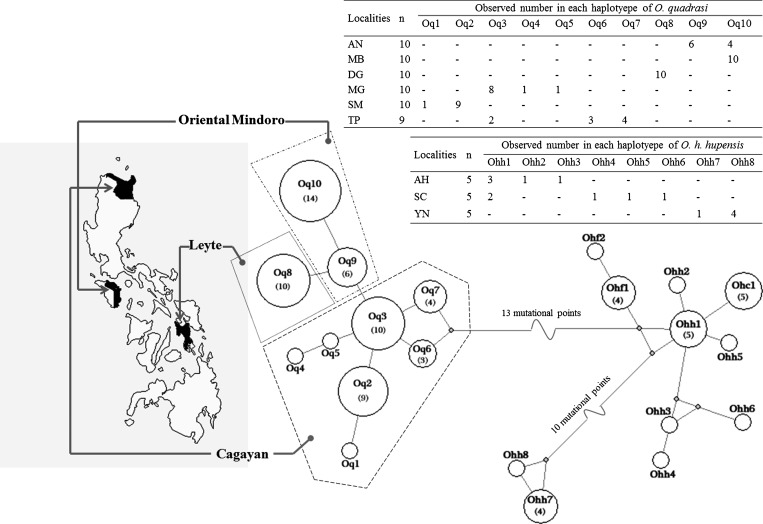
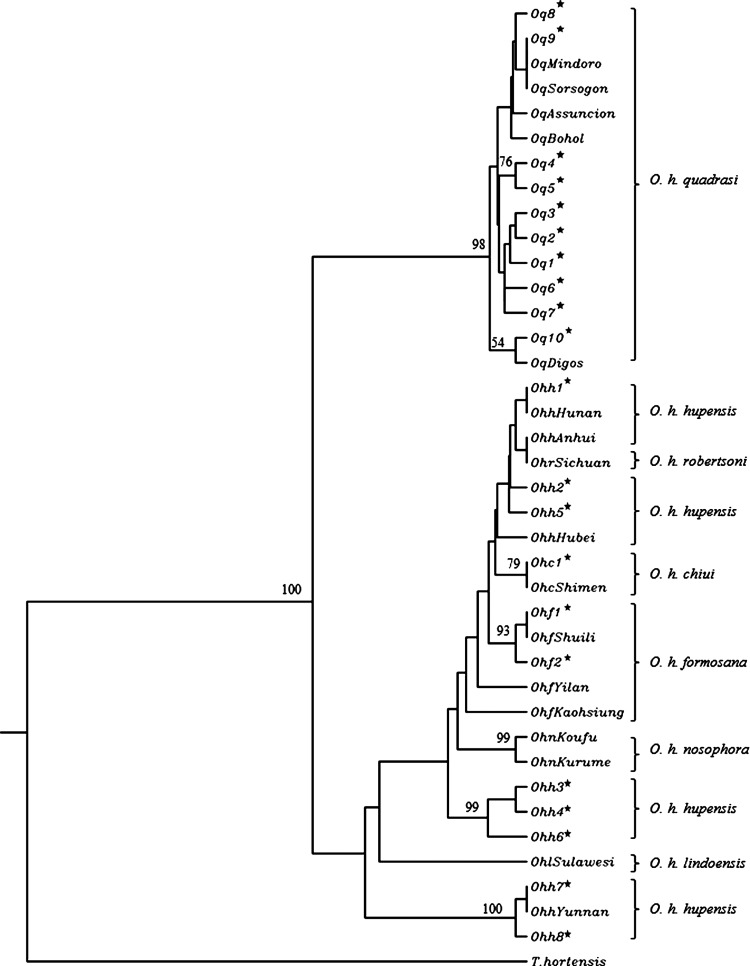
The 357 bp partial sequence of the mitochondrial 12S ribosomal RNA gene was compared within and between populations of O. quadrasi, O. h. hupensis, O. h. formosana, and O. h. chiui. In total, 40 variable positions consisting of 34 transitions, 2 transversions, 1 insertion/deletion, and 3 multiple mutational changes were observed (Table 2). Of these positions, 1–16 (0.28–4.48%) variable positions were observed within each taxon, which were classified into 21 haplotypes: 10 haplotypes of O. quadrasi (Oq1–Oq10), 8 haplotypes of O. h. hupensis (Ohh1–Ohh8), 2 haplotypes of O. h. formosana (Ohf1–Ohf2), and 1 haplotype of O. h. chiui (Ohc1) (Table 2 and Figure 1). The nucleotides sequences of all haplotypes detected in this study were submitted to GenBank under the following accession numbers: KC763451–60 are haplotypes of O. quadrasi, KC763461–68 are O. h. hupensis, KC763470–71 are O. h. formosana, and KC763469 is O. h. chiui. A minimum spanning haplotype network showed that the network of 10 haplotypes of O. quadrasi was clearly separated from the other haplotypes at 13 mutational points (Figure 1). The haplotypes of subspecies O. h. hupensis, O. h. formosana, and O. h. chiui were grouped in the same network except the two haplotypes of O. h. hupensis from Yunnan, which were distinctly separated from the others at 10 mutational points (Figure 1 ). No common haplotype was shared among/between populations of the four subspecies. The phylogenetic tree also showed that O. quadrasi formed a distinct cluster from the other subspecies, whereas O. h. hupensis, O. h. chiui, O. h. formosana, O. h. nosophara, and O. h. lindoensis were clustered as a monophyletic group (Figure 2 ).
Table 2.
Variable positions of 12S ribosomal RNA sequences of Oncomelania quadrasi (Oq), O. h. hupensis (Ohh), O. h. chiui (Ohc) and O. h. formosana (Ohf) haplotypes, the Philippines*
| Haplotype | Nucleotide sequence | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | |||||||
| 3 | 4 | 5 | 6 | 6 | 6 | 1 | 2 | 2 | 3 | 4 | 4 | 5 | 5 | 6 | 6 | 0 | 0 | 0 | 2 | 3 | 3 | 3 | 5 | 5 | 5 | 6 | 6 | 6 | 6 | 6 | 6 | 8 | 9 | 2 | 2 | 4 | 4 | 5 | 5 | |
| 3 | 4 | 8 | 1 | 3 | 4 | 8 | 2 | 3 | 6 | 0 | 2 | 3 | 4 | 2 | 3 | 0 | 1 | 6 | 5 | 0 | 2 | 7 | 4 | 7 | 8 | 0 | 1 | 2 | 4 | 5 | 8 | 9 | 9 | 0 | 2 | 6 | 8 | 0 | 6 | |
| Oq1 | A | A | A | T | T | C | T | G | T | A | A | T | T | C | T | T | A | T | G | G | T | C | C | T | G | G | G | G | G | G | A | G | C | G | T | G | A | T | T | G |
| Oq2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq3 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq4 | . | . | . | . | . | . | . | . | C | . | . | . | C | . | . | C | . | . | . | . | . | . | T | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq5 | . | . | . | . | . | . | . | . | C | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq6 | . | . | . | . | . | . | . | . | . | . | . | . | C | T | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq7 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | A | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq8 | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | A | . | . | . | . | . | . |
| Oq9 | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Oq10 | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | A | A | . | . | . | − | . | . | . | . | . | . | . | . | . | . |
| Ohh1 | . | G | . | C | . | T | . | A | . | . | G | . | C | T | . | C | . | . | A | A | . | T | . | . | A | A | A | A | . | − | . | . | T | . | A | . | . | . | C | . |
| Ohh2 | . | G | . | C | . | T | . | A | . | . | G | . | C | T | . | C | G | . | A | A | . | T | . | . | A | A | A | A | . | − | . | . | T | . | A | . | . | . | C | . |
| Ohh3 | . | G | . | C | . | T | . | A | . | G | G | C | C | T | . | C | . | . | A | A | . | T | . | C | A | A | A | A | . | − | . | . | T | . | A | . | G | . | C | . |
| Ohh4 | . | G | . | C | . | T | . | A | . | G | G | C | C | T | . | C | . | C | A | A | . | T | . | C | A | A | A | A | . | − | . | . | T | . | A | . | G | . | C | . |
| Ohh5 | . | T | . | C | . | T | . | A | . | . | G | . | C | T | . | C | . | . | A | A | . | T | . | . | A | A | A | A | . | − | . | . | T | . | A | . | . | . | C | . |
| Ohh6 | . | G | . | C | C | T | . | A | . | G | G | C | C | T | . | C | . | . | A | A | . | T | . | A | A | A | A | A | . | − | . | . | T | . | A | . | G | C | C | . |
| Ohh7 | . | G | G | . | . | T | . | A | . | . | T | G | . | T | C | C | . | . | A | A | C | T | . | . | A | A | A | A | . | − | . | A | T | . | . | A | . | . | . | A |
| Ohh8 | . | G | G | . | . | T | . | A | . | . | T | A | . | T | C | C | . | . | A | A | C | T | . | . | A | A | A | A | . | − | . | A | T | . | . | A | . | . | . | A |
| Ohc1 | . | G | . | C | . | T | . | A | . | . | G | . | C | T | . | C | . | . | A | A | . | . | . | A | A | A | A | A | − | . | . | T | . | A | . | . | . | C | . | |
| Ohf1 | . | G | . | C | . | T | C | A | . | . | . | . | C | T | . | C | . | . | A | A | . | T | . | . | A | A | A | A | . | − | . | . | T | . | A | . | . | . | C | . |
| Ohf2 | . | G | . | C | . | T | C | A | . | . | . | . | C | T | . | C | . | . | A | A | . | T | . | . | A | A | A | A | . | − | G | . | T | . | A | . | . | . | C | . |
. = identity.
Figure 1.
Minimum spanning haplotype network of the three subspecies of Oncomelania hupensis generated in based on 12S ribosomal RNA sequence. A network of O. quadrasi corresponds to their geographic localities in three provinces of the Philippines. The area of the circles represents the proportion of sample number (indicate in parentheses) found in each haplotype. Oq = O. quadrasi; Ohh = O. h. hupensis; Ohf = O. h. formosana; and Ohc = O. h. chiui.
Figure 2.
Phylogenetic tree generated from 12S ribosomal RNA sequences of Oncomelania snails. The star represents the haplotype samples examined in this study; other haplotype samples were obtained from Okamoto and others.12 Tricula hortensis was obtained from GenBank under accession no. NC_013833 and used as an outgroup.
Intraspecific analyses of O. quadrasi.
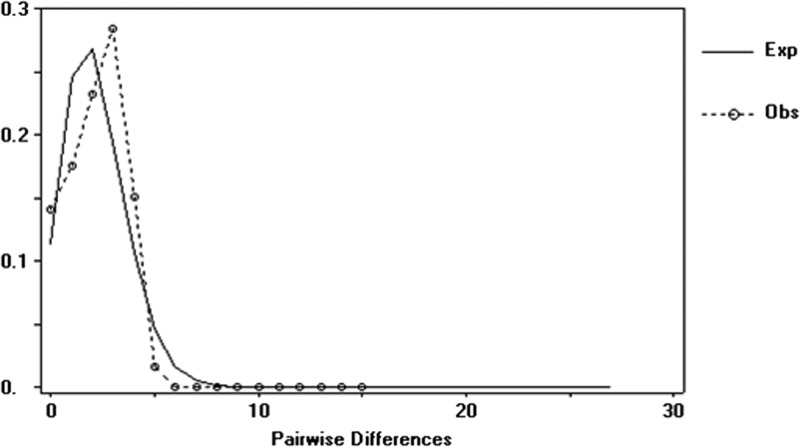
Of the 10 haplotypes of O. quadrasi, 1–2 unique haplotypes existed in each population. Two haplotypes were shared between two populations: Oq3 and Oq10 shared between the populations MG with TP, and AN with MB, respectively (Table 2 and Figure 1). The haplotype, nucleotide, and genetic diversity were 0.859 ± 0.019, 0.218 ± 0.012, and 0.215 ± 0.087, respectively. In the neutrality test, the observed values of both Tajima's D (0.034, P = 0.721) or Fu's Fs (−1.279, P = 0.100) suggest that natural selection may have occurred (Table 3). Significant genetic differentiation (FST) and estimated gene flow (Nm) between populations ranged from 0.556 to 1.000 and from 0.00 to 0.74, respectively (Table 4). Substantial genetic subdivision and low gene flow existed among the six populations (FST = 0.854; P < 0.001 and Nm = 0.06, P < 0.001), respectively (Table 4). The network of O. quadrasi haplotypes shows a star-like pattern and is significantly separated into three groups according to three provinces/islands by FCT = 0.466 (P < 0.05). A significant FSC = 0.727 (P < 0.001) was also observed among populations within groups (Table 5). The mismatch distribution of 12S ribosomal RNA sequences was unimodal and almost fitted the expected distribution (Figure 3 ).
Table 3.
Summary statistics of molecular variation and neutrality analysis in six populations of Oncomelania quadrasi, the Philippines*
| Population | n | S | H | Uh | Hd | π ± SD | θs ± SD | Tajima's D | Fu's Fs |
|---|---|---|---|---|---|---|---|---|---|
| AN | 10 | 2 | 2 | 1 | 0.533 ± 0.095 | 0.107 ± 0.019 | 0.107 ± 0.054 | 1.641 (0.970) | 2.338 (0.854) |
| MB | 10 | 0 | 1 | 0 | 0.000 ± 0.000 | NA | NA | NA | NA |
| DG | 10 | 0 | 1 | 1 | 0.000 ± 0.000 | NA | NA | NA | NA |
| MG | 10 | 2 | 3 | 2 | 0.378 ± 0.181 | 0.056 ± 0.028 | 0.071 ± 0.054 | −0.691 (0.218) | −0.594 (0.105) |
| SM | 10 | 1 | 2 | 2 | 0.200 ± 0.154 | 0.020 ± 0.015 | 0.035 ± 0.035 | −1.112 (0.179) | −0.339 (0.164) |
| TP | 9 | 2 | 3 | 2 | 0.722 ± 0.097 | 0.106 ± 0.016 | 0.074 ± 0.056 | 1.494 (0.958) | 0.453 (0.527) |
| All populations | 59 | 10 | 10 | 8 | 0.859 ± 0.019 | 0.218 ± 0.012 | 0.215 ± 0.087 | 0.034 (0.721) | −1.279 (0.100) |
n = number of samples; S = number of segregating sites; H = number of haplotypes; Uh = unique haplotype; Hd = haplotype diversity; π = nucleotide diversity; θs = genetic diversity estimated from segregating sites; NA = not analyzed. P values are indicated in parentheses.
Table 4.
Genetic differentiation (FST) (lower triangle) and estimated gene flow (Nm) (upper triangle) among Oncomelania quadrasi populations, the Philippines*
| Population | AN | MB | DG | MG | SM | TP |
|---|---|---|---|---|---|---|
| AN | − | 0.20† | 0.11 | 0.16 | 0.07 | 0.17 |
| MB | 0.556† | − | 0.00 | 0.02 | 0.01 | 0.04 |
| DG | 0.704 | 1.000 | − | 0.03 | 0.01 | 0.06 |
| MG | 0.614 | 0.916 | 0.879 | − | 0.09 | 0.74 |
| SM | 0.838 | 0.980 | 0.976 | 0.843 | − | 0.13 |
| TP | 0.588 | 0.868 | 0.820 | 0.258 | 0.789 | − |
Among all populations, FST = 0.854 and Nm = 0.06 (significant at P < 0.001). Comparison values were significant at P < 0.001.
P < 0.01.
Table 5.
Analysis of molecular variance of six populations of Oncomelania quadrasi classified into three groups defined by island, i.e., groups of Luzon, Mindoro, and Leyte islands, the Philippines*
| Source of variation | df | Sum of squares | Variance components | % Variation | Fixation indices |
|---|---|---|---|---|---|
| Among groups | 2 | 40.057 | 0.75 | 46.60 | FCT = 0.466† |
| Among populations within group | 3 | 19.012 | 0.62 | 38.83 | FSC = 0.727‡ |
| Within population | 53 | 12.422 | 0.23 | 14.57 |
Level of significance is based on 1,000 permutations. df = degrees of freedom.
P < 0.05.
P < 0.001.
Figure 3.
Mismatch distribution of the 59 partial 12S ribosomal RNA sequences (357 basepairs) of six Oncomelania quadrasi populations from the Philippines. The mismatch distribution is the distribution of the number of pairwise differences among sequences. The expected distribution under a model of expansion is given as a continuous line, and the observed distribution is given as a dashed line. Exp = expected; Obs = observed.
Discussion
We found that substructuring occurred between the six populations of O. quadrasi examined according to their geographic origin in the Philippines, similar to that described for the subspecies O. h. hupensis and O. h. robertsoni from mainland China.5,6 The genetic differences found for O. quadrasi populations from different localities in the Philippines islands may be caused by lack or low level of gene flow, especially between the three provinces located on separated islands. Furthermore, significant genetic differentiation was detected between populations within each province, e.g., three populations in Cagayan and two populations in Oriental Mindoro provinces, suggesting that the level of gene flow between these populations is currently low.
The mismatch distribution of the 12S ribosomal RNA haplotypes of O. quadrasi is markedly unimodal, which is observed when the genealogy of the sample resembles a star network, which in turn is observed during demographic expansions.16 A high level of haplotype and nucleotide diversity of O. quadrasi populations was observed in this study, in agreement with the results of previous studies of O. hupensis ssp. in mainland China.5,6 Because of the short generation times and large effective population sizes, invertebrates, including snails, usually show a high level of genetic diversity.21 Moreover, in the case of O. quadrasi, the observed high genetic variability is also potentially related to the low mobility of the species, leading to population subdivision and potential increase global genetic diversity. Unique haplotypes for each population were found, except for MB, for which only one haplotype was observed, which was shared with an adjacent AN population from Oriental Mindoro province. The results in this study strongly indicate that the O. quadrasi populations in the Philippines have a substructure based on their geographical origin. However, a larger sample size from a wider range of geographic areas is needed to confirm that all unique haplotypes are specific to each particular geographic area.
Genetic differences between some S. japonicum isolates in the Philippines have also been noted; the isolate from Mindoro is moderately genetically differentiated from the Leyte and Luzon populations.22 We have now determined that the genetic structure of O. quadrasi populations from Leyte, Oriental Mindoro, and Cagayan also show significant genetic differences from each other, which was in concordance with findings of a previous report.13 This finding may be caused by reciprocal evolution between the parasite and its snail hosts, as indicated in several host-parasite systems, e.g., O. viverrini and its Bithynia snail hosts,23,24 and S. mansoni and its Biomphalaria snail hosts.9 Moreover, the three provinces, i.e., Cagayan, Oriental Mindoro, and Leyte, are located on separated islands so that gene flow is likely to be highly restricted, eventually resulting in high genetic differentiation between the different geographic areas. This conclusion is suggested by the significant FCT differences. Such a situation has also been observed between populations of O. h. hupensis in mainland China, where the population genetic structure is related to geographic area and is correlated with differing shell characteristics.6
Another objective of our study was to establish whether O. quadrasi was genetically distinct from three subspecies of O. hupensis examined. A significant difference between O. quadrasi and the three subspecies was found by phylogenetic analysis of 12S ribosomal RNA. Our phylogenetic tree shows that O. quadrasi forms a distinct clade when compared with the O. hupensis ssp. Moreover, the genetic relationships of O. h. hupensis from Yunnan province are distinct from the other subspecies, which is in concordance with findings of previous studies.6,12 Although previous morphologic and biogeographic studies of O. hupensis ssp. across mainland China indicate that O. h. robertsoni and O. h. hupensis have the same shell growth allometry, the shell length is on average different.25 Oncomelania h. robertsoni has a patchy distribution in Sichuan and Yunnan provinces above the Three Gorges, whereas O. h. hupensis occurs in the Yangtze River drainage below the Three Gorges.26,27 Both snails are probably different at a specific level. In addition, S. japonicum from Yunnan also showed significant genetic differences from the other geographic isolates.28 However, subspecies validity and the assignment of O. hupensis in mainland China and O. quadrasi in the Philippines remain controversial in these areas and need to be further clarified by intensively analyzing snail morphology and using more powerful molecular markers.
ACKNOWLEDGMENTS
We thank the Deutsche Forschungsgemeinschaft (PE1611/1-3), the National Research Council of Thailand, and the International Excellence Fund of ASEAN-EU Year of Science, Technology and Innovation, 2012 for providing funding for cooperative workshops.
Footnotes
Financial support: This study was supported by the Japan Society for the Promotion of Science (grant no. L-11566) and TRF-CHE-MSU (grant no. MRG5480009) to Weerachai Saijuntha.
Authors' addresses: Weerachai Saijuntha, Walai Rukhavej Botanical Research Institute, Mahasarakham University, Maha Sarakham, Thailand, E-mail: weerachai.s@msu.ac.th. Blanca Jarilla and Takeshi Agatsuma, Division of Environmental Health Sciences, Kochi Medical School, Nankoku, Japan, E-mails: blancajarilla@yahoo.com and agatsuma@kochi-u.ac.jp. Alvin K. Leonardo and Lydia R. Leonardo, College of Public Health, University of the Philippines, Manila, Philippines, E-mail: leonardolydia7@gmail.com. Louie S. Sunico, Municipal Health Office, Municipality of Gonzaga, Cagayan, Philippines, E-mail: HRMO_gonzaga@yahoo.com. Ross H. Andrews and Paiboon Sithithaworn, Department of Parasitology, and Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand, E-mails: rhandrews@gmail.com and paib_sit@hotmail.com. Trevor N. Petney, Department of Ecology and Parasitology, Karlsruhe Institute of Technology, Karlsruhe, Germany, E-mail: petney@kit.edu. Masashi Kirinoki, Naoko Kato-Hayashi, and Yuichi Chigusa, Laboratory of Tropical Medicine and Parasitology, Dokkyo Medical University, Tochigi, Japan, E-mails: kirinoki@dokkyomed.ac.jp, nkato@dokkyo.med.ac.jp, and ychigusa@dokkyomed.ac.jp. Mihoko Kikuchi, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan, E-mail: mkikuchi@nagasaki-u.ac.jp.
References
- 1.Berger SA. Infectious Diseases of the Philippines. Gideon e-books; 2011. http://www.gideononline.com/ebooks/country/infectious-diseases-of-the-philippines/ Available at. [Google Scholar]
- 2.Berger SA. Schistosoma Japonicum: Global Status. Gideon e-books; 2011. http://www.gideononline.com/ebooks/disease/schistosoma-japonicum-global-status/ Available at. [Google Scholar]
- 3.Fernandez TJ, Jr, Tarafder MR, Balolong E, Jr, Joseph L, Willingham AL, 3rd, Belisle P, Webster JP, Olveda RM, McGarvey ST, Carabin H. Prevalence of Schistosoma japonicum infection among animals in fifty villages of Samar Province, the Philippines. Vector Borne Zoonotic Dis. 2007;7:147–155. doi: 10.1089/vbz.2006.0565. [DOI] [PubMed] [Google Scholar]
- 4.Tarafder MR, Balolong E, Jr, Carabin H, Belisle P, Tallo V, Joseph L, Alday P, Gonzales RO, Riley S, Olveda R, McGarvey ST. A cross-sectional study of the prevalence of intensity of infection with Schistosoma japonicum in 50 irrigated and rain-fed villages in Samar Province, the Philippines. BMC Public Health. 2006;6:61. doi: 10.1186/1471-2458-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauswald AK, Remais JV, Xiao N, Davis GM, Lu D, Bale MJ, Wilke T. Stirred, not shaken: genetic structure of the intermediate snail host Oncomelania hupensis robertsoni in an historically endemic schistosomiasis area. Parasit Vectors. 2011;4:206. doi: 10.1186/1756-3305-4-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao QP, Jiang MS, Littlewood DT, Nie P. Distinct genetic diversity of Oncomelania hupensis, intermediate host of Schistosoma japonicum in mainland China as revealed by ITS sequences. PLoS Negl Trop Dis. 2010;4:e611. doi: 10.1371/journal.pntd.0000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis GM, Wilke T, Zhang Y, Xu XJ, Qiu CP, Spolsky CM, Oiu DC, Li Y, Xia MY, Feng Z. Snail-Schistosoma, Paragonimus interaction in China: population ecology, genetic diversity, coevolution and emerging diseases. Malacologia. 1999;41:355–377. [Google Scholar]
- 8.Webster JP, Davies CM. Coevolution and compatibility in the snail-schistosome system. Parasitology. 2001;123((Suppl)):S41–S56. doi: 10.1017/s0031182001008071. [DOI] [PubMed] [Google Scholar]
- 9.Webster JP, Shrivastava J, Johnson PJ, Blair L. Is host-schistosome coevolution going anywhere? BMC Evol Biol. 2007;7:91. doi: 10.1186/1471-2148-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi CH, Wilke T, Davis GM, Xia MY, Qiu CP. Population genetic, micro-phylogeography, ecology, and susceptibility to schistosome infection of Chinese Oncomelania hupensis hupensis (Gastropoda: Rissooidea: Pomatiopsidae) in the Miao river system. Malacologia. 2002;44:333–347. [Google Scholar]
- 11.Hope M, McManus DP. Genetic variation in geographically isolated populations and subspecies of Oncomelania hupensis determined by a PCR-based RFLP method. Acta Trop. 1994;57:75–82. doi: 10.1016/0001-706x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto M, Lo Chin Tson, Tiu WU, Qui Dong Chuan, Hadidjaja P, Upatham S, Sugiyama H, Taguchi T, Hirai H, Saitoh Y, Habe S, Kawanaka M, Hirata M, Agatsuma T. Phylogenetic relationships of snails of the genera Oncomelania and Tricula inferred from the mitochondrial 12S rRNA gene. Jpn J Trop Med Hyg. 2003;31:5–10. [Google Scholar]
- 13.Woodruff DS, Staub KC, Upatham ES, Viyanant V, Yuan HC. Genetic variation in Oncomelania hupensis: Schistosoma japonicum transmitting snails in China and the Philippines are distinct species. Malacologia. 1998;29:347–361. [Google Scholar]
- 14.Viyanant V, Upatham ES, Blas BL, Yuan HC. Analysis of allozymes by electrofocusing in schistosome snail hosts (Oncomelania hupensis) from China and the Philippines. Malacol Rev. 1987;20:91–96. [Google Scholar]
- 15.Woodruff DS, Carpenter MP, Upatham ES, Viyanant V. Molecular phylogeography of Oncomelania lindoensis (Gastropoda: Pomatiopsidae), the intermediate host of Schistosoma japonicum in Sulawesi. J Molluscan Stud. 1999;65:21–31. [Google Scholar]
- 16.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 19.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- 21.Thomas JA, Welch JJ, Lanfear R, Bromham L. A generation time effect on the rate of molecular evolution in invertebrates. Mol Biol Evol. 2010;27:1173–1180. doi: 10.1093/molbev/msq009. [DOI] [PubMed] [Google Scholar]
- 22.Woodruff DS, Merenlender AM, Upatham ES, Viyanant V. Genetic variation and differentiation of three Schistosoma species from the Philippines, Laos, and Peninsular Malaysia. Am J Trop Med Hyg. 1987;36:345–354. doi: 10.4269/ajtmh.1987.36.345. [DOI] [PubMed] [Google Scholar]
- 23.Kiatsopit N, Sithithaworn P, Saijuntha W, Petney TN, Andrews RH. Opisthorchis viverrini: Implications of the systematics of first intermediate hosts, Bithynia snail species in Thailand and Lao PDR. Infect Genet Evol. 2013;14:313–319. doi: 10.1016/j.meegid.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Saijuntha W, Sithithaworn P, Wongkham S, Laha T, Pipitgool V, Tesana S, Chilton NB, Petney TN, Andrews RH. Evidence of a species complex within the food-borne trematode Opisthorchis viverrini and possible co-evolution with their first intermediate hosts. Int J Parasitol. 2007;37:695–703. doi: 10.1016/j.ijpara.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis GM, Zhang Y, Guo YH, Spolsky CM. Population genetics and systematic status of Oncomelania hupensis (Gastropoda: Pomatiopsidae) throughout China. Malacologia. 1995;37:133–156. [Google Scholar]
- 26.Li SZ, Wang YX, Yang K, Liu Q, Wang Q, Zhang Y, Wu XH, Guo JG, Bergquist R, Zhou XN. Landscape genetics: the correlation of spatial and genetic distances of Oncomelania hupensis, the intermediate host snail of Schistosoma japonicum in mainland China. Geospat Health. 2009;3:221–231. doi: 10.4081/gh.2009.222. [DOI] [PubMed] [Google Scholar]
- 27.Zhou YB, Zhao GM, Jiang QW. Genetic variability of Schistosoma japonicum (Katsorada, 1904) intermediate hosts Oncomelania hupensis (Gredler, 1881) (Gastropoda: Rissooidea) Annal Zool. 2008;58:881–889. [Google Scholar]
- 28.Zhao QP, Jiang MS, Dong HF, Nie P. Diversifiation of Schistosoma japonicum in mainland China revealed by mitochondrial DNA. PLoS Negl Trop Dis. 2012;6:e1503. doi: 10.1371/journal.pntd.0001503. [DOI] [PMC free article] [PubMed] [Google Scholar]