Abstract
Histoplasmosis usually occurs in specific endemic areas. Sporadic cases have also been reported in mainland China. Here, we described an indigenous case of disseminated histoplasmosis. Phylogenetic analysis revealed that the Histoplasma capsulatum isolated in our case belongs to the Australian clade. Combined with previous studies, it revealed high genetic diversity among Chinese H. capsulatum isolates.
Introduction
Histoplasmosis caused by the soil-based dimorphic fungus Histoplasma capsulatum is a common endemic mycosis in the midwestern United States and Central America.1 However, sporadic cases of autochthonous histoplasmosis have been encountered in China, which was traditionally considered non-endemic for H. capsulatum.2–4 Recently, a multilocus sequence typing (MLST) approach and a sequence database were available for H. capsulatum, which helped us to better understand the molecular epidemiology of this disease.5 Here, we reported a case of disseminated histoplasmosis in an human immunodeficiency virus (HIV)-negative patient who had never been outside China. The isolate was phylogenetically characterized.
In April of 2012, a 28-year-old male construction worker presented with right-side abdominal pain of 6 months in duration. He also complained of intermittent fevers (up to 39°C), fatigue, and weight loss of 10 kg. The patient had been empirically treated with antibiotics at the local hospital with no apparent improvement. Other medical history was unremarkable. The patient was from Hunan Province, but he reported having lived in Fujian Province for 4 years before he started experiencing the aforementioned symptoms. He was a native-born Chinese individual and had never been outside China. He admitted to having used injectable drugs on several occasions in the past.
The patient presented with reduced general condition and a temperature of 41°C. Physical examination revealed cervical lymphadenopathy and hepatosplenomegaly. A complete blood cell count showed pancytopenia, a white blood cell count of 2.14 × 109/L (80.4% neutrophils, 9.4% lymphocytes, and 7.3% monocytes), hemoglobin level of 70 g/L, and platelet count of 57 × 109/L. Renal function and liver enzymes were within normal limits. Tests for HIV-1 and HIV-2 antibodies were negative. Abdominal computed tomography confirmed the hepatosplenomegaly. Chest radiograph showed bilateral reticulonodular infiltrates.
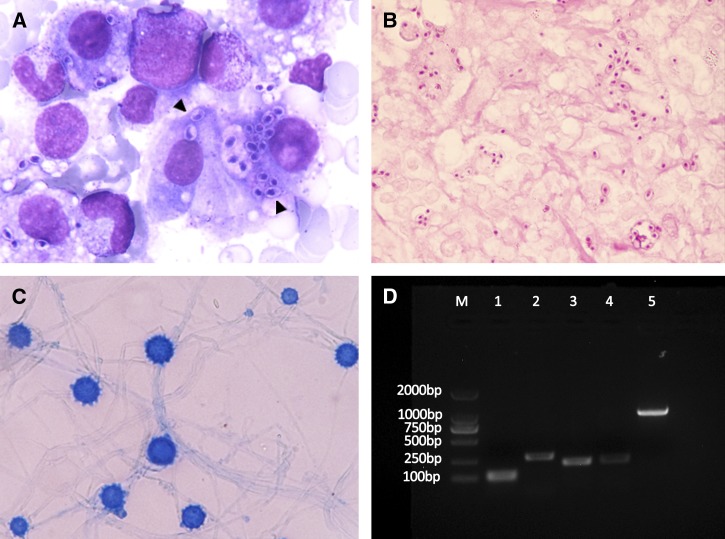
Bone marrow biopsy was performed because of pancytopenia. Wright-stained bone marrow aspirate revealed the presence of numerous intracellular oval or spherical encapsulated organisms, which is highly suggestive of H. capsulatum (Figure 1A, arrowheads). A biopsy of the cervical lymph node showed many small oval microorganisms with dotlike inclusions that tested positive by periodic acid-Schiff stain (PAS) (Figure 1B). The lymph node was cultured on Sabourauds dextrose agar at 30°C. Two weeks later, cottony white colonies with no red pigment were observed. Microscopic examination showed hyaline hyphae that produced tuberculated macroconidia, confirming the presence of H. capsulatum (Figure 1C). When the colonies were cultured on brain heart infusion agar at 37°C, the fungi converted to the yeast phase from the mycelial phase.
Figure 1.
(A) Bone marrow aspirate stained with Wright's stain shows numerous encapsulated oval or spherical intracellular and extracellular organisms (×1,000). (B) PAS staining of the cervical lymph node revealed ovoid-shaped microorganisms with dotlike inclusions (×1,000). (C) Phenol cotton blue stain of the isolated mold showing thick-walled and tuberculate macroconidia (×1,000). (D) Identification of H. capsulatum is confirmed by PCR assay based on the specific nucleotide sequence. Lanes 1 and 2, PCR products amplified with M-antigen probes Msp1F-Msp1R (111 bp) and Msp2F-Msp2R (279 bp)8; lanes 3 and 4, PCR products amplified with sequence-characterized amplified region markers 1281–1283220 (220 bp) and 1281–1283230 (230 bp)9; lane 5, ITS amplified with primers V9G and LS266.6 M = molecular size marker.
After the diagnosis of disseminated histoplasmosis, intravenous amphotericin B (0.72 mg/kg daily) was administered. The patient's general condition improved after 1 month of treatment. H. capsulatum was not observed in bone marrow aspirate obtained after treatment. The patient was discharged and treated with oral itraconazole (200 mg two times per day) for 6 months. He was followed up for 1 year thereafter, and no evidence of relapse was noted.
The internal transcribed spacer (ITS) domains of the ribosomal ribonucleic acid (rRNA) gene of the isolate were amplified using V9G and LS266 primers and sequenced with ITS5 and ITS4.6 The sequence was deposited in the GenBank database under the accession number KF443065, which was found to have 99% identity with the Ajellomyces capsulatum isolate UAMH 7141. The isolate was identified as H. capsulatum var. capsulatum, because it lacked the characteristic ITS1 sequence of H. capsulatum var. duboisii.7 The identification was confirmed by using four sets of specific primers (Figure 1D).8,9
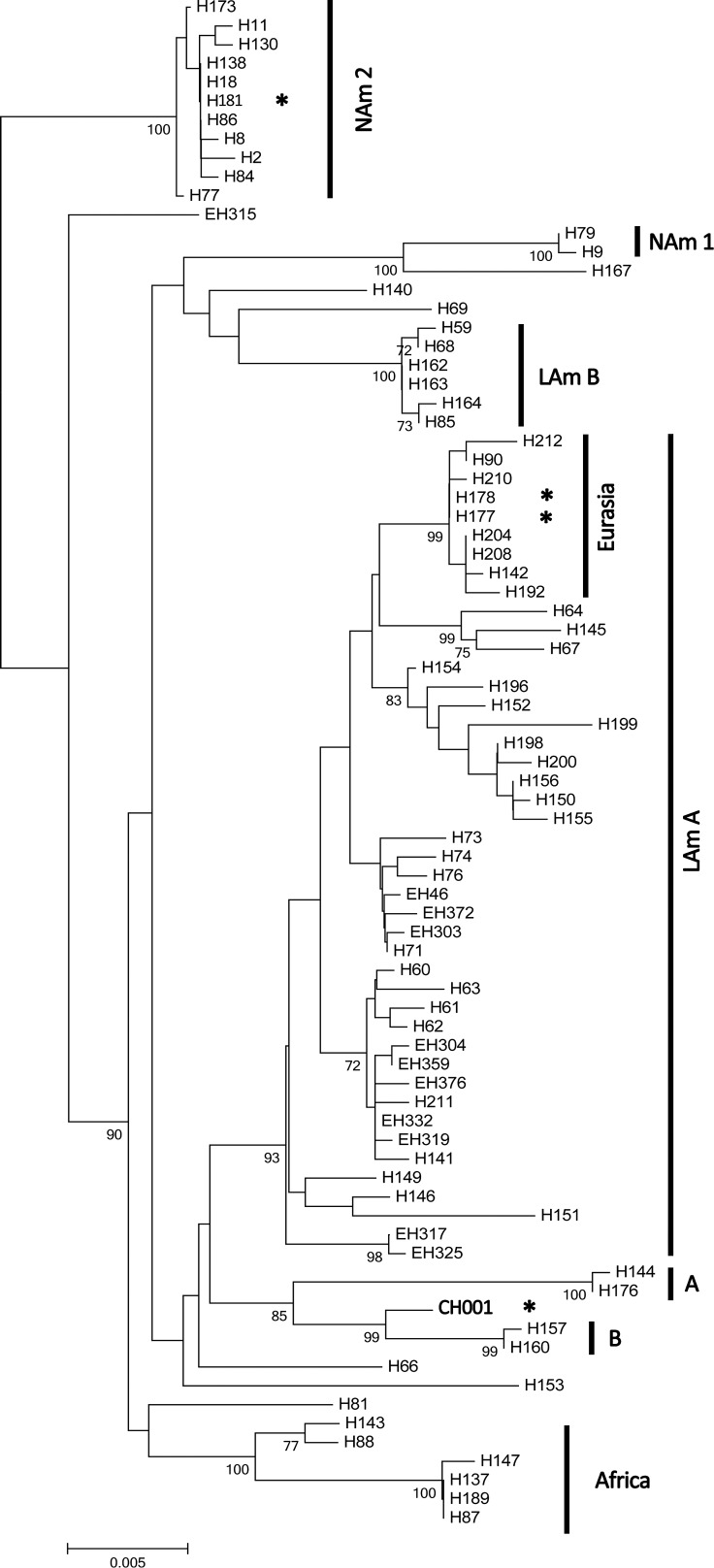
Four unlinked loci previously used for phylogenetic studies of Histoplasma species complex were applied to analyze the isolate according to the method of Kasuga and others.5 These four loci were ADP-ribosylation factor (arf), H-antigen precursor (H-anti), Δ9 fatty acid desaturase (ole), and α-tubulin (tub1). The polymerase chain reaction (PCR) conditions used had been reported elsewhere.10 MLST sequences from H. capsulatum strains were downloaded from the TreeBase database, and the DNA was aligned manually. The phylogenic tree was constructed with MEGA 5 using the neighbor-joining method. Branching pattern was statistically evaluated by bootstrap analysis of 1,000 replicates. The genotype of the isolate (CH001) from our patient belonged to the Australian clade, with bootstrap support of 99% (Figure 2). The maximum likelihood algorithm provided similar results. Four separate gene trees are also presented (Supplemental Figures 1–4). The rRNA gene sequence of our patient showed 100% identity to the isolate IFM47750 (GenBank) obtained from a Chinese male patient 25 years ago.7 However, no detailed information of that patient or the MLST sequence of IFM47750 was available.
Figure 2.
Neighbor-joining tree from analysis of the combined data of the four loci. Branch lengths are proportional to Kimura's two-parameter distance. Numbers on the branches indicate bootstrap support of greater than 70%. The scale bar indicates the nucleotide substitutions per site. A = Netherlands clade; B = Australia clade; LAm A = Latin America group A clade; LAm B = Latin America group B clade; NAm 1 = North America 1 clade. *Isolates of H. capsulatum from China.
Conclusions
Since 1990, 300 cases of histoplasmosis had been reported in China, of which only 17 cases were possibly imported cases. Most of the patients were from regions through which the Yangtze River flows.4 These regions include the two provinces where our patient had resided. Our patient had never been abroad. He was a construction worker and usually worked in a dusty environment. Dusts containing H. capsulatum spores could be aerosolized during his work. He also had history of drug abuse, which could lead to host immunosuppression.11 These risk factors increased his chances of developing histoplasmosis.
It should be noticed that few reported histoplasmosis cases in China were acute pulmonary histoplasmosis or cave-associated cases. The lack of acute pulmonary cases can be explained by poor levels of awareness of this disease in China. One hospital in Manchester reported a case of acute histoplasmosis in a British spelunker who acquired the disease in a cave in Yunnan Province. Additionally, Yunnan Province accounted for more than 27% of the reported cases of histoplasmosis in China.12 There are many bat-inhabited caves in Yunnan Province. The environment there is also suitable for H. capsulatum growth. The knowledge of the geographic distribution of H. capsulatum is critical in making the diagnosis of histoplasmosis. However, no fungus has been isolated from the environment, and no outbreaks have been reported in China; therefore, the ecological niche of H. capsulatum in China remains unknown. Bat guano is thought to be a classic reservoir of H. capsulatum.13 However, it should be noticed that bat droppings, also called ye ming sha in Chinese, are a type of Chinese herbal medicine used to improve vision. People who have been exposed to this herbal medicine from H. capsulatum-endemic areas are potentially at risk of infection.
Phylogenetic analysis of the H. capsulatum MLST gene sequences showed that the CH001 strain belongs to the Australian clade, with bootstrap support of 99%, indicating a close phylogenetic relationship between them. The Australian clade contained one soil isolate and four clinical isolates from Australia.5 It also suggested a strong relationship between the clinical and environmental strains in the Australian clade. Although the isolate fell within the Australia clade, it should be noticed that the branch was typically deep for the individual loci, which implied that it was significantly diverged from the Australia clade members. Except for the CH001 strain in this study, there were three other strains of H. capsulatum isolated from China. However, no detailed clinical information of these strains was available. Two isolates, namely H177 and H178, were found in the Eurasian clade, and one isolate, H181, was found in the North America 2 (NAm 2) clade. According to the phylogenetic tree, based on ITS region sequence, the H177 and CH001 strains also belong to different clades.14 Among the four strains, H181 and CH001 were definitely isolated from patients who had never traveled outside China. Taken together, it suggests that a wide range of molecular types exists among Chinese H. capsulatum isolates. This genetic diversity could be explained by human activity or animal migration, vectoring H. capsulatum and consequently, causing its spread. However, additional studies are needed to draw a definite conclusion, because the number of the studied Chinese H. capsulatum strains is very small; also, no strains have been isolated from the environment in China.
This case of indigenous human histoplasmosis in mainland China emphasizes the need to determine likely reservoirs of H. capsulatum to improve the understanding of its epidemiology. As international travel becomes accessible, clinicians whose patients have resided in China should consider the diagnosis of this disease.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Stephen Craven (University of Cape Town) for providing valuable information of acute pulmonary histoplasmosis in China.
Footnotes
Financial support: This study was supported by National 973 Program Grant 2013CB531600, National Science and Technology Major Projects of China Grant 2013ZX10004612001, Shanghai Science and Technology Committee Grant 10dz2220100, and a key project from Chinese Military Logistics (AWS11L009).
Authors' addresses: Ying Wang, Jianhua Wu, Xinling Bi, and Jun Gu, Department of Dermatology, Changhai Hospital, Second Military Medical University, Shanghai, People's Republic of China, E-mails: wangying88_2@hotmail.com, 44434731@qq.com, bxlsmmu@gmail.com, and jungusmmu@gmail.com. Bo Pan, Wanqing Liao, and Weihua Pan, Department of Dermatology, Changzheng Hospital, Second Military Medical University, Shanghai, People's Republic of China, E-mails: simajiangtailangpb@hotmail.com, liaowanqing@sohu.com, and panweihua@medmail.com.cn.
References
- 1.Kauffman CA. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev. 2007;20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge L, Zhou C, Song Z, Zhang Y, Wang L, Zhong B, Hao F. Primary localized histoplasmosis with lesions restricted to the mouth in a Chinese HIV-negative patient. Int J Infect Dis. 2010;14((Suppl 3)):e325–e328. doi: 10.1016/j.ijid.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Cao C, Bulmer G, Li J, Liang L, Lin Y, Xu Y, Luo Q. Indigenous case of disseminated histoplasmosis from the Penicillium marneffei endemic area of China. Mycopathologia. 2010;170:47–50. doi: 10.1007/s11046-010-9295-4. [DOI] [PubMed] [Google Scholar]
- 4.Pan B, Chen M, Pan W, Liao W. Histoplasmosis: a new endemic fungal infection in China? Review and analysis of cases. Mycoses. 2013;56:212–221. doi: 10.1111/myc.12029. [DOI] [PubMed] [Google Scholar]
- 5.Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castaneda E, Da Silva Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancope-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol. 2003;12:3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 6.de Hoog GS, Vicente V, Caligiorne RB, Kantarcioglu S, Tintelnot K, Gerrits van den Ende AH, Haase G. Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J Clin Microbiol. 2003;41:4767–4778. doi: 10.1128/JCM.41.10.4767-4778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura M, Kasuga T, Watanabe K, Katsu M, Mikami Y, Nishimura K. Phylogenetic characterization of Histoplasma capsulatum strains based on ITS region sequences, including two new strains from Thai and Chinese patients in Japan. Nihon Ishinkin Gakkai Zasshi. 2002;43:11–19. doi: 10.3314/jjmm.43.11. [DOI] [PubMed] [Google Scholar]
- 8.Guedes HL, Guimaraes AJ, Muniz Mde M, Pizzini CV, Hamilton AJ, Peralta JM, Deepe GS, Jr, Zancope-Oliveira RM. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. J Clin Microbiol. 2003;41:535–539. doi: 10.1128/JCM.41.2.535-539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frias De Leon MG, Arenas Lopez G, Taylor ML, Acosta Altamirano G, Reyes-Montes Mdel R. Development of specific sequence-characterized amplified region markers for detecting Histoplasma capsulatum in clinical and environmental samples. J Clin Microbiol. 2012;50:673–679. doi: 10.1128/JCM.05271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasuga T, Taylor JW, White TJ. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J Clin Microbiol. 1999;37:653–663. doi: 10.1128/jcm.37.3.653-663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez LR, Mihu MR, Gacser A, Santambrogio L, Nosanchuk JD. Methamphetamine enhances histoplasmosis by immunosuppression of the host. J Infect Dis. 2009;200:131–141. doi: 10.1086/599328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan B, Deng S, Liao W, Pan W. Endemic mycoses: overlooked diseases in China. Clin Infect Dis. 2013;56:1516–1517. doi: 10.1093/cid/cit033. [DOI] [PubMed] [Google Scholar]
- 13.Lyon GM, Bravo AV, Espino A, Lindsley MD, Gutierrez RE, Rodriguez I, Corella A, Carrillo F, McNeil MM, Warnock DW, Hajjeh RA. Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998–1999. Am J Trop Med Hyg. 2004;70:438–442. [PubMed] [Google Scholar]
- 14.Murata Y, Sano A, Ueda Y, Inomata T, Takayama A, Poonwan N, Nanthawan M, Mikami Y, Miyaji M, Nishimura K, Kamei K. Molecular epidemiology of canine histoplasmosis in Japan. Med Mycol. 2007;45:233–247. doi: 10.1080/13693780601186069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.