Abstract
Lung cancer (LC) is a number one killer of cancer-related death among men and women worldwide. Major advances have been made in the diagnosis, staging and use of surgery for LC, but systemic chemotherapy and radiotherapy alone or in combination with some targeted agents remains the core treatment of advanced LC. Unfortunately, in spite of improved diagnosis, surgical methods and new treatments, mortality is still extremely high among LC patients. To understand the precise functioning of signaling pathways associated with resistance to current treatments in LC, as well as to identify novel treatment regimens, a holistic approach to analyze signaling networks should be applied. Here, we describe systems biology-based approaches to generate biomarkers and novel therapeutic targets in LC, as well as how this may contribute to personalized treatment for this malignancy.
Keywords: lung cancer, systems biology, therapeutic strategies, personalized cancer medicine
Facts
Lung cancer is divided into small and non-small cell lung carcinomas (SCLC and NSCLC, respectively).
Combined radiotherapy and chemotherapy is the main treatment regimen of lung cancer.
Targeted therapies have revolutionized the treatment of adenocarcinomas driven by mutated EGFR or EML4-ALK.
Systems biology of lung cancer has potential to reveal prognostic and treatment-predictive factors and novel signaling pathways for innovative therapeutic approaches.
Open Questions
To identify new targets of both NSCLC and SCLC to increase their sensitivity to conventional treatment.
To further understand targeted therapy resistance mechanisms and how to circumvent this.
To sub-classify different lung cancer histologies, reveal their Achilles heels and in a systems biology manner identify novel therapeutic pathways.
Lung cancer (LC) has, for decades, remained the most common cause of cancer-related death. In total, 85% of all diagnosed LC are non-small cell lung cancer (NSCLC); the remaining 15% belong to small cell lung carcinomas (SCLC). NSCLC is divided into three subtypes: squamous cell carcinoma (about 40%), adenocarcinoma (about 30%), and large cell lung carcinoma (about 15%). The histological distinction between SCLC and NSCLC, as well as different histology subtypes of NSCL, is related to their diverse biological behavior, which has impact on prognosis and treatment responses.1, 2, 3 Major advances have been made in diagnosis, staging and use of surgery for LC and targeted therapies and have revolutionized the treatment for a subset of NSCLC cases (adenocarcinomas driven by mutated EGFR or echinoderm microtubule-associated protein like 4 (EML4)-Alk fusion proteins, respectively). However, systemic chemotherapy and radiotherapy (CT and RT, respectively) remain important treatment regimens. It is still hard to predict which patients will benefit from CT/RT; as the mechanism(s) of resistance remain elusive and the development of resistance towards the few existing targeted agents is multifactorial. However, the identification of biomarkers would enable a personalized cancer therapy approach to be taken in which the individual signaling cascades of each tumor would dictate the choice of therapeutic strategy.
One of the main mechanisms of RT/CT is the induction of cell death, and therefore differences in treatment sensitivity may depend on the susceptibility of LC cells to undergo cell death.4 Although we and others have identified certain molecular factors that may explain this difference, the complete image remains unclear. Thus, further research is required to define the abnormal regulation of gene and/or protein expression that may influence tumor therapy-induced cell death. To understand the precise functioning of signaling pathways associated with resistance to treatments, a holistic analyzing approach on signaling networks should be applied.
Systems biology analysis aims to understand the properties of a given system, which, in relation to cancer, could be tumor cell lines, xenografts, genetically engineered mice or primary cells from patients pre- or post-treatment. It includes analysis of the relationships among various components of the system and integrating experiments in sequential repetitive cycles with mathematical modeling, simulation and theory.5 Modeling is not the final goal but is a tool to increase understanding of the system, develop proof-of-concept experiments and enable predictions that can be experimentally validated.
The key steps in systems biology involves: (I) holistic measurement and quantification of alterations at the DNA, RNA, miRNA, protein or signaling level; (II) integration of the information obtained from measurements to obtain a global image of the system in question; (III) assessment of its dynamic changes, which in relation to tumors means its progression towards a more malignant phenotype, as well as its responses to CT, RT and targeted agent therapies; and (IV) modeling the system using the integrated data. Here, we focus on what a systems biology approach has generated for biomarkers and novel therapeutic targets in LC.
Methods to Support Systems Biology Analysis
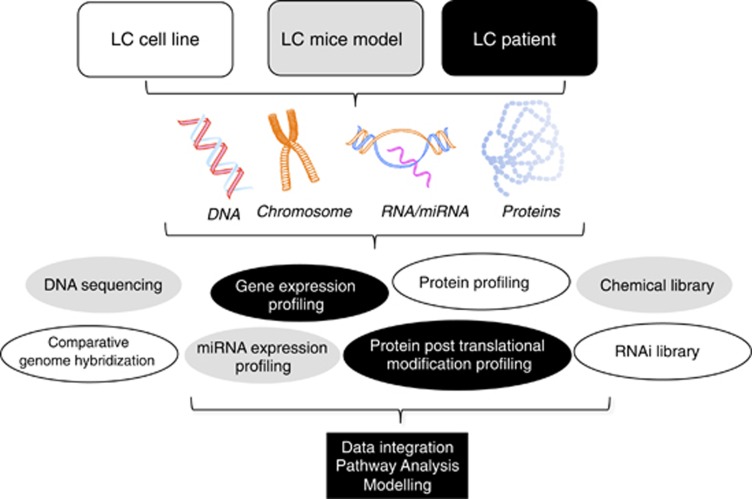
Several systems biology methods are currently used to reveal biomarkers and novel targets, as well as to pinpoint novel agents with antitumor and/or CT/RT capacity of LC tumors. These include: (I) DNA mutation and DNA copy number rearrangements; (II) gene, mRNA and miRNA expression; (III) proteomic profiling of total and post-translationally modified proteins; (IV) siRNA and chemical library screening; and (V) modeling and validation experiments using targeted strategies both in vitro and in vivo (Figure 1).
Figure 1.
Methods applied in systems biology of lung cancer. LC cell lines, animal models or patient-derived samples may be subjected to analysis of DNA, RNA or protein using the described methods followed by data integration, pathway analysis and modeling. For details, see text
The different techniques applicable for DNA sequencing, their limit of detection and shortcomings have recently been described.6 DNA microarrays are used to measure changes in gene expression levels, detect single-nucleotide polymorphisms (SNPs) and genotype or re-sequence mutant genomes. SNP detection can be used to measure predisposition to disease, identify drug candidates, evaluate germline mutations in individuals or somatic mutations within a tumor, assess loss of heterozygosity and perform genetic linkage analysis. Microarrays are also becoming widely used for profiling miRNAs, a class of regulatory RNAs that control mRNA translation and thereby regulate various biological processes.7
Systems biological analysis of protein expression and/or post-translational modifications (PTMs) in patient-derived tumor and normal tissue or body fluids (blood, serum, plasma and pleural effusions) offers great potential but is still associated with many hurdles. Two different proteomic approaches can be undertaken: global proteomic methods, in which protein expression, PTM, or their corresponding peptides are investigated using mass spectrometry (MS)-based analysis, or antibody-based analysis methods. The latter involved the ‘pathway walking', in which a signaling cascade is mapped in body fluids using enzyme-linked immunosorbent assay, multiplex bead array assay or reverse-phase protein array of tumor samples or tissue microarray for immunohistochemical staining and the assessment of protein expression in tumors versus normal tissues.8, 9
For global proteomics, two main analysis principles may be used, that is, a top–down approach, in which total proteins are investigated, or the bottom–up approach, in which proteins are enzymatically digested into peptides and used in the subsequent MS analysis.10
PTMs of proteins (phosphorylation, glycosylation, methylation, acetylation and sulfatation) alter protein function and thereby signaling capacity. Proteomic methods have been developed that allow a global image of PTMs in a particular system or during a specific treatment with phosphoproteomic profiling being one of them.11 RNA interference (RNAi) is a cellular process that controls gene expression at the post-translational level but can also be carried out using a global RNAi approach (by either a library of siRNA or a collection of vector-based shRNAs carried by plasmid or viruses). Global RNAi allows for the analysis of a particular biological event in a holistic manner but can also be used for validating ‘omics' data. RNAi has one disadvantage that primarily consists of off-target effects caused by unspecific gene silencing of the RNAi applied. Moreover, not all identified targets have clinical applicability, which is a shortcoming shared with targets identified by genomic or proteomic approaches. Thus, RNAi-identified targets need further animal and/or clinical validation to ascertain their clinical value as a therapeutic target or biomarkers of response.
Chemical library screening using small molecules is yet another technique if combined with other appropriate ‘omics' approaches can govern a systems biology approach in understanding tumor biology and drug applicability.
To be able to convert genetic or proteomic networks into an analytical tool, the data obtained should be translated into a mathematical language to describe the possible dynamic behaviors in specific conditions.12, 13, 14 The general strategy of building models of cell fate includes constructing a generic graph that describes the different processes under normal conditions. The resulting network recapitulates the current knowledge and should be verified by crossing it with genetic results, which might be obtained by using various mutants and/or drug treatments.12, 13, 14 Altogether, these results lead to the formulation of predictions on network topology, mutant phenotypes and alterations in pathways.
Systems Biology Approach to Identify Genetic Abnormalities in Lung Cancer
Systems biology analyses of both NSCLC and SCLC have recently been applied to reveal prognostic and treatment predictive factors as well as to explore and decipher novel signaling pathways that might be used to develop innovative therapeutic approaches (Figure 1). The methods described above have all been applied in different cell systems in vitro, in animal models and in LC patient samples (tumor tissue and plasma/serum/pleural effusions). Prognosis assessment has become a major concern in the area of personalized medicine, and gene expression studies in various LC cohorts have overwhelmed the scientific community. Some of the generated profiles have indeed shown the ability to predict the prognosis of LC patients in an accurate way.15, 16
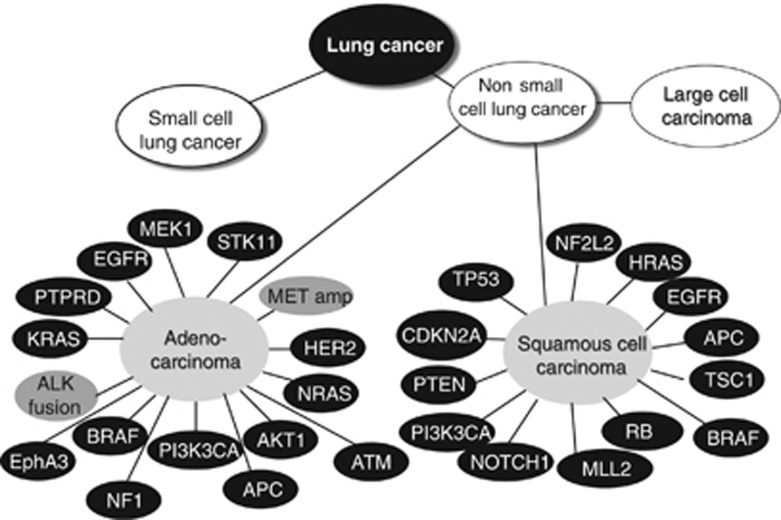
The two most commonly exploited genetic aberrations of NSCLC adenocarcinoma are the epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) gene rearrangements17 (Figure 2). EGFR mutations account for about 5–10% of all mutations found in adenocarcinomas, with a deletion in exon 19 and a point mutation in exon 21 being the two most common; these render the patients' tumors hypersensitive to EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib (Iressa1, AstraZeneca, Wilmington, DE, USA) and erlotinib (Tarceva1, F. Hoffmann-La Roche, Basel, Switzerland), the two currently approved drugs against locally advanced or metastatic NSCLC.17 ALK rearrangements are found in about 5–7% of adenocarcinomas in which a fusion between the EML4 gene and ALK render a constitutively active kinase18 (Figure 2). An ALK-driven tumor is currently treated with TKI crizotinib (Xalkori1; Pfizer, Groton, CT, USA) causing G1-S phase arrest and/or apoptotic signaling resulting in reduced tumor growth.19 It is clear that we just are at the beginning of the targeted therapy era for LC, where inhibitors towards other driving genetic alterations, such as ROS1 and RET translocations, MET amplification or HER2, BRAF, PIK3CA and CTNNB1 mutations, are in trials or in development and in which KRAS-driven tumors can be treated with synthetic lethality approaches using a mix of MEK, PI3KCA and AKT1 inhibitors20 (Figure 2). To identify further combinations of targeted approaches and understand the mechanisms of resistance, a systems biology approach has already proven to be of value.
Figure 2.
Genetical alterations in adenocarcinoma and squamous cell carcinoma of the lung. LC is primarily divided into SCLC and NSCLC, respectively. NSCLC is further subdivided into adenocarcinoma, squamous cell carcinoma and large cell lung carcinoma. Based on published data from references,2,21,27,91 some of the genetical alterations (mutations, deletions or amplifications) described in adenocarcinoma and squamous cell carcinoma are shown
A major focus in NSCLC research is on the role of EGFR as a driver of LC signaling and how different EGFR mutations and other signaling aberrations render LC tumors sensitive to EGFR inhibitors.17 In other NSCLC cases which are not driven by EGFR mutations, both EGFR- and IGF1R-mediated pathways have been demonstrated to have altered signaling, as illustrated by the DNA sequencing studies of NSCLC clinical specimens, where multiple kinase pathways were found to be changed.2, 17, 21
It is almost inevitable for any LC patient to develop resistance to TKIs in part caused by second mutation in the EGFR receptor, for example, T790M mutation.22, 23 Whereas the T790M mutation is found in 50% of EGFR-TKI-resistant NSCLC cases, another 20% are likely to be driven by MET gene amplification.22, 23 A systems biology approach to search for putative EGFR-TKI resistance mechanisms by comparing biopsies from NSCLC patients before and after the development of EGFR-TKI resistance revealed that EGFR gene amplification and PIK3CA mutations resulting in hyperactivation of the PI3K signaling pathway also may drive EGFR-TKI resistance in vivo.22 Moreover, in pre-clinical LC models of EGFR-TKI resistance, epithelial-to-mesenchymal (EMT) transition was observed in clones that did not show MET amplification, a signaling aberration also associated with EGFR-TKI resistance in vivo.22 EGFR TKI resistance was also in about 14% of the cases shown to involve a conversion from a lung adenocarcinoma into SCLC with a neuroendocrine marker expression and initial CT sensitivity.22 Thus, if the selection pressure of EGFR-TKI was taken off, the T790M and PIK3CA mutations as well as the SCLC phenotype disappeared, and patients recovered their response to EGFR-TKI treatment. Hence, these molecular dissection data support the idea to not only personalize but also to ‘tumoralize' patient treatment, that is, follow tumor evolution during the course of treatment and adjust the therapy according to the genomic and phenotypic make-up of the tumor. It was also suggested that the inhibition of NF-κB signaling might help to improve EGFR-TKI therapy outcome.24 Using RNAi, it was demonstrated that inhibition of either Fas or several components of the NF-κB pathway enhanced the apoptotic effect of EGFR-TKI. Delineating whether such an approach might be relevant to NSCLC in vivo during EGFR-TKI treatment, the authors showed that increased expression of the NF-κB inhibitor IκB was predictive for the improved EGFR-TKI response and survival of NSCLC patients with a EGFR-mutant tumor. These results again illustrate the usefulness of a systems biology approach to reveal novel targeted therapy combinations.
A link between apoptotic signaling and response to TKI in EGFR-mutant-dependent lung adenocarcinomas has also been shown in which low expression level of the Bcl-2 family protein Bim in the primary tumor was linked to shorter progression-free survival (PFS) in NSCLC patients treated with EGFR-TKI.25 Moreover, a common intronic deletion polymorphism in the Bim gene was discovered, resulting in a different splicing pattern of Bim into an isoform that lacked the pro-apoptotic Bcl-2-homology domain 3 (BH3).26 Expression of this Bim isoform conferred TKI resistance in vitro and patients with EGFR-driven NSCLC harboring the polymorphism experienced significantly inferior responses to TKIs than individuals without the polymorphism. Thus, for a subgroup of EGFR-driven NSCLC, BH3 mimetics may be used to overcome Bim-polymorphism-associated TKI resistance.
Global DNA Sequencing Approach to Identify Genetic Alterations in LC Influencing Therapy Response
A systems biology approach by performing global DNA sequencing efforts in large scientific consortia has been carried out on different LC histologies to decipher the underlying genetic aberrations, which may drive LC cell survival and impact therapy response. In the first study, about 200 lung adenocarcinomas were analyzed for mutations in almost 600 cancer-associated genes and >1000 different somatic mutations were identified some of which were associated with smoking status, clinical features and DNA repair defects of the LC patients.2 In addition to EGFR, multiple ephrin receptor genes (notably EPHA3), NTRK genes and vascular endothelial growth factor receptor KDR were identified to have high mutation frequencies as did several tumor-suppressor genes, for example, APC, NF1, RB1 and ataxia-telangiectasia mutated (ATM). With respect to squamous cell lung cancer, the TCGA project examined treatment of naive tumors from 178 patients with stage I–IV disease and revealed a complex genotype with a mean of 360 exonic mutations, 165 genomic rearrangements and 323 segments of copy number alterations per tumor.21 Recently, SCLC was also assessed on a global scale and a large number of cases were screened for mutations.27 Strikingly, SCLC was found to have a much higher mutation frequency than NSCLC as well as other solid tumors examined, with approximately seven mutations identified in every million base pairs of DNA. Among them were mutations in p53, RB1 and in multiple genes regulating histone modifications (CREBP, EP300 and MLL), which may also offer a novel therapeutic approach for these tumors. A systems biology approach with combined genomic and pharmacological vulnerability screen of SCLC cell lines in vitro was used with the aim of revealing novel targeted therapeutic approaches.28 A chemical library screen of almost 300 compounds with different scaffolds and thereby different targets were performed across the 44 SCLC cell lines. Results showed that a clear vulnerability towards Aurora kinase inhibitors existed in SCLC driven by Myc amplification and that such inhibition resulted in inactivation of PI3K signaling, G2/M arrest and induction of apoptosis. Although SCLC driven by Myc only accounts for a fraction of cases, this study clearly illustrates the capacity of a systems biology approach in the identification of novel therapeutic avenues.
Combined Technologies in Molecular Profiling of LC
Advances in both global and targeted proteomic technologies have opened up molecular profiling at the protein level, not only to interrogate tumor genotypes and phenotypes but also to tailor treatments that may improve clinical outcomes. By applying a combined proteomic and genomic profiling approach where gene and phosphoprotein ‘signatures' associated with the invasive and metastatic propensities of LC were studied, a metastatic signature was revealed.29 By using unsupervised hierarchical clustering of the RNA expression data obtained from a transgenic model of Kras-induced adenocarcinoma, which becomes invasive and metastatic to lymph nodes and distal organs when combined with a deletion of the pluripotent serine/threonine kinase Lkb1, three distinct groups were identified: primary lung tumors with activated KrasG12D, primary tumors with activated KrasG12D and Lkb1 loss, and metastases from lung tumors with activated-rasG12D and Lkb1 loss. A 750 gene signature was found to be associated with Lkb1 loss or metastasis. The loss of Lkb1 resulted in a significantly altered transcriptional profile of the primary tumors, which were further altered when the tumors became metastatic. The genetic changes included alterations in multiple signaling pathways that previously have been linked to metastasis, for example, focal adhesion kinase (FAK), EMT, TGF-β and β-catenin, as well as in genes associated with embryonic stem cell signaling (the transcription factors TCF3 and OCT4). By integrating the gene profiles data of metastasis or Lkb1 deletion tumors with publicly available gene expression sets from two NSCLC patient cohorts, a gene profile of associated metastasis within the animal model was shown to match gene expression patterns in NSCLC clinical specimens from patients with lower overall survival. The metastasis-associated gene signature showed significant overlap with profiles observed in clinical specimens containing both Kras and Lkb1 alterations.30 Examination of the tyrosine phosphoproteome of the Lkb1 deleted LC cells by MS analysis showed a critical role for Src activation, as the majority of differentially phosphorylated proteins upon Lkb1 deletion had this kinase as an upstream regulating kinase.31 A combined inhibition of PI3K–mTOR, MEK and Src family kinases caused a synergistic in vivo response in LKb1-defective LC tumors. By taking the metastatic-associated gene expression signature generated by Ji et al.,30 in a bioinformatics approach, we discovered a role for miRNA-214 in regulating metastasis of NSCLC and identified four genes (pregnancy-associated plasma protein A, alpha-protein kinase 2, cyclin-dependent kinase 6 and tumor necrosis-factor alpha-induced protein 3) to be involved in this process.32 Analyses of the corresponding proteins in a clinical NSCLC samples revealed a high-to-moderate expression in the tumor specimens. However, we failed to demonstrate an association to metastasis, which may indicate that these proteins are altered early during tumor development, before metastasis. Still, this study illustrates the potential use of systems biology data from the public domain to further interrogate global ‘omics' data.
In an attempt to make genetic profiles predictive for response to TKIs, a global systems biology-based approach was undertaken on NSCLC cell lines, which represented different histologies and whose genomic landscape (amplifications/deletions, mutations and gene expression patterns) matched clinical LC specimens.33 Apart from confirming a role for mutated EGFR as the strongest predictor for erlotinib treatment response and Kras mutation to corroborate erlotinib responsiveness, a number of new treatment strategies were generated and validated. Using hierarchical clustering of genomic data from sensitive and resistant NSCLC cell lines, a response signature was generated, as exemplified by the MEK inhibitor U0126, which was linked to chromosomal gains of ART and RAB13. By applying the same data analysis on a broader kinase inhibitor, dasatinib, the authors showed that copy number gain of ephrin receptor kinases (EPHA3, EPHA5 and EPHA8), Src kinases (Src, FRK, YES, LCK and BLK) and ABL2 loci all predicted dasatinib treatment response, some of which were also validated in vitro and in vivo. Interestingly, Src and EPHA3 has both been shown to be deregulated in about 10% of NSCLC cases, suggesting that a therapeutic approach involving dasatinib might have a clinical impact. Corroborating a role of Eph kinase signaling in NSCLC, EphA2 deregulation was found in a sub-fraction of NSCLC.34 Expression of EphA2 was positively correlated with activated but not mutated EGFR and had increased level in clinical specimens from patients harboring Kras mutations. In vitro analyses of NSCLC cells revealed that EphA2 regulated both proliferation and invasion, and in clinical specimens, high tumor expression of EphA2 was found to be an independent prognostic factor for time to recurrence and also for time to metastasis.34 In searching for putative resistance pathways for RT, we, by genomic profiling, identified Ephrin B3 to impede RT-induced cell death and to alter cell cycle regulation.35 A pronounced effect on NSCLC cell proliferation was observed when Ephrin B3 siRNA was applied, which encouraged our search for the signaling pathways involved.36 Using phosphoproteomic analysis (TiO2-based fractionation followed by nano-LC MS) 150 proteins showed Ephrin B3-dependent phosphorylation, including EphA2, Akt, HSP90AA1 and CK2. This shows that the Ephrin/Eph signaling axis is involved in NSCLC proliferation and therapy response and also illustrates the power of global proteomic approaches for identifying critical pathways.
Although EGFR ablation therapy and targeted approaches against other oncogenic pathways have or will generate novel therapies with great potential, a large fraction of LC patients with disseminated disease will still rely on conventional CT/RT. With the aim of revealing candidate signaling networks that may predict outcome to certain CT/RT regimens, gene and miRNA expression profiling, DNA sequencing of LC genomes and different proteomic approaches have all been applied, hence allowing a tailored approach with old CT drugs to patients who will benefit the most.15, 16 Along the same line, the necessity of predicting the relapse status as well as metastatic propensity of LC patients was stated, thereby directing CT for those with a high risk of relapse in which a personalized approach of CT application should be taken.15, 16 In two studies, gene expression analyses was made on NCI 60 tumor cell lines and/or their corresponding xenografts in mice in relation to responsiveness of about 100 000 different compounds, including natural compounds from different sources, targeted agents and all currently applied CTs.37, 38 These studies generated large gene expression data sets that can be used in data mining and has enabled target prediction of compounds with an unknown mechanism of action.
Prognostic and Treatment Predictive Gene Signatures
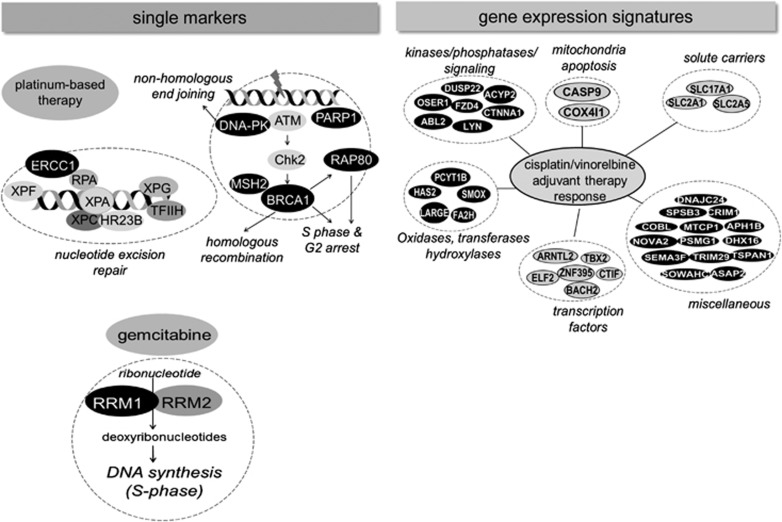
Analysis of treatment response of primary human tumor-derived xenografts (from either NSCLC or SCLC patient samples) has become a way of understanding tumor alterations during treatment, especially in tumors where biopsy is an obstacle.39, 40, 41 These patient-derived xenografts are good complements to the genetically modified mouse models of NSCLC and SCLC, which have been developed and applied by Jacks' group,42, 43 Bern's group44, 45 and Barbacid's group,46, 47 and together these tumor mice model approaches have significantly contributed to our understanding of LC tumorigenesis and will also continue to reveal novel therapeutic strategies. Both prognostic and treatment-predictive gene signatures have been generated when mRNA isolated from early stage resected NSCLC were analyzed in relation to outcome or cisplatin/vinorelbine adjuvant therapy response.48 Thus a prognostic algorithm was identified by analyzing genomic profiles from about 330 stage I–III adenocarcinomas. The signature was independently validated on a series of genomic profiles from NSCLC patients enrolled in a randomized controlled trial of cisplatin/vinorelbine versus observation alone. The functional sorting of the genes within the prognostic signature was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This analysis showed that genes involved in the control of cellular organization, metabolic processes, cell cycle and apoptosis were over-represented, as were genes linked to MAPK signaling. Using various bioinformatic tools and statistics, a 37-gene signature predictive of adjuvant CT response to cisplatin/vinorelbine was generated, and it was found that NSCLC patients with this gene expression profile had a 25% improvement in the 5-year-survival, compared with observation alone, whereas non-expressing NSCLC patients had no benefit of treatment. The genes included among other kinases/phosphatases, enzymes involved in metabolism and different transcription factors (Figure 3). This work is promising, although it waits to be seen if it has predictive capacity in other NSCLC materials and further refinements are required in order to implement it into clinical practice.
Figure 3.
Chemotherapy predictive markers of NSCLCs. Chemotherapy predictive markers are divided into single markers (left) and gene expression signatures (right). Platinum or gemcitabine markers (black) are shown based on data from references. 49,50,51,54 The gene expression signature shown is adopted from Van Laar48
It was recently proposed that several DNA-repair-associated proteins, such as ERCC1, BRCA1, RAP80, RRM1, PARP1, MSH2 or DNA-PK, could be used to customize NSCLC therapy and substantially improve patient outcomes49 (Figure 3). One of them is RRM1, an enzyme which have a role in both DNA synthesis/repair and in gemcitabine metabolism. In order to assess whether RRM1 mRNA expression could be a biomarker of gemcitabine-combined treatment regimes, expression levels were analyzed in relation to cisplatin/gemcitabine response in NSCLC patients. It was demonstrated that those patients whose tumors had low RRM1 mRNA expression levels had significantly longer median survival than those with high level, implicating a biomarker role.50 In the same study, a strong correlation between RRM1 and ERCC1 mRNA expression levels were found suggesting that combined RRM1 and ERCC1 levels might be used as biomarkers for cisplatin/gemcitabine treatment response.
Differences in ERCC1 expression were examined in a meta-analysis study in relation to objective response and median survival of NSCLC patients with advanced disease and undergoing platinum-based CT.51 The obtained data showed that the response to platinum-based CT was significantly higher in patients with ERCC1 low/negative expression. However, the ERCC1 predictive value for cisplatin treatment outcome in NSCLC is still not fully resolved as IHC with different antibodies against ERCC1 as well as mRNA analysis generated inconsistent results.49, 52
Working further in understanding the impact of ERCC1 on NSCLC signaling, Soria and his research team carried out global molecular analysis of genomic instability, global gene and miRNA expression as well as sequencing of certain key genes involved in lung carcinogenesis in ERCC1-positive and -negative tumors.53 ERCC1-negative tumors showed a higher rate of genomic abnormalities compared with ERCC1-positive tumors. Gene expression profiling of the same samples revealed the overexpression of genes linked to DNA-damage response, whereas miRNA expression analysis of the same tumor cohorts identified the downregulation of miR-375 in ERCC1-positive tumors, suggesting that ERCC1 status is not linked to a specific mutation pattern.
In vitro studies have shown that BRCA1-deficient tumor cells, in contrast to the BRCA1-proficient ones, are very sensitive to cisplatin. A prospective non-randomized phase II clinical trial was performed where therapy was customized according to the EGFR mutation status of the tumors in which mutant-carrying patients received erlotinib, whereas patients whose tumors expressed wtEGFR were allocated to cisplatin plus gemcitabine.54 Results showed that choosing CT based on BRCA1 expression levels improved survival for at least a subset of NSCLC patients. Moreover, low BRCA1 mRNA expression alleviated the inhibitory effect of the T790M mutation on EGFR-TKI erlotinib response,54 suggesting further analysis of T790M mutation baseline assessment and BRCA1 expression to predict outcome and alternative treatment approaches including customized CT. Cisplatin resistance of NSCLC has in some but not in all cases been found to be associated with hyperactivation of PARP.55 Importantly, for cisplatin-resistant cases governed by hyperactivation of PARP ablation of its activity resulted in a restored cisplatin-induced DNA damage response and apoptosis. The effect of two different PARP inhibitors on DNA repair functionality was also analyzed in a panel of cisplatin-treated NSCLC cell lines.56 Pharmacological inhibition of PARP induced a 1.7- to 2.3-fold increase in platinum adduct accumulation, indicating impaired platinum DNA-adduct repair capacity. Examination of PARP1 expression in whole tumor sections from the International Adjuvant Lung cancer Trial (IALT)-bio study showed that whereas PARP1 expression level did not predict patient survival or the effect of platinum-based postoperative CT, a combination of high expression of PARP1/MSH2/ERCC1 did. Hence, all of these studies call for PARP inhibitors in NSCLC to revert platinum resistance and suggest broader systems biology approach to understand DNA repair alterations in LC. Of note, trials of NSCLC in which combination of PARP inhibitors (NCT01082549 and NCT01086254) and standard CT are currently ongoing.49
Systemic Profiling of miRNAs in LC
Analysis of miRNA may generate both prognostic- and treatment-associated signatures and reveal putative signaling networks with CT/RT-sensitizing potential. Thus, by regulating gene expression at the post-transcriptional level, miRNAs profoundly impact on oncogenic pathways in NSCLC and SCLC.57 The first miRNA to be linked to LC prognosis was let-7, which target oncogenic Ras, and it was reported that let-7 had a reduced expression in LC tumor material and was associated with shorter survival after surgical resection.58 Following these findings, the global analysis of miRNA expression in various LC cell lines and in clinical specimen of NSCLC with different histology identified a number of miRNA to be associated with certain subclasses and linked to prognosis and early disease recurrence.59, 60 Analysis of miRNA in squamous lung carcinomas identified 15 miRNAs to be differentially expressed between normal lung and tumor tissue, including members of the miR-17-92 cluster, miR-155 and let-7, which previously have been linked to prognosis in adenocarcinoma, and miR-146b, which had the strongest prediction accuracy for stratifying prognostic groups.61 This signature of miRNAs was found to be superior to a previously described 50-gene prognostic signature of squamous lung carcinoma. Although the authors did not find an overlap between the mRNAs targeted by the prognostic miRNAs and the 50-gene expression signature, they observed a substantial overlap among the pathways involved, which included fibroblast growth factor (FGF) and interleukin-6 signaling. Analysis of 165 adenocarcinoma and 125 squamous cell carcinoma tissue samples from the Environment and Genetics in Lung Cancer Etiology study was performed in order to determine whether miRNA expression profiles could differentiate histological subtypes and predict survival for NSCLCs.62 This study clearly distinguished adenocarcinoma from squamous cell carcinoma, and the latter was characterized by downregulation of most miRNAs, including all members of the let-7 family. A miRNA signature predicting the post-operative recurrence of stage I NSCLC has also been described.63 These results showed that miRNA profiles from different patient cohorts were almost non-overlapping, similarly to when gene expression profiling was compared straight off between different studies. It could be explained by the different experimental platforms used (qRT-PCR versus different miRNA array systems), the batch effects that are inherent in the microarray experiment itself or the potential ethnic differences in the study populations.
Recently, miRNA expression profiles of lung tumors, normal lung tissues and plasma samples from cases with variable prognosis identified in a completed spiral-CT screening were examined.64 The authors identified an association between different miRNA expression with (i) tumors from normal lung tissues, (ii) tumor histology and growth rate, (iii) clinical outcome, and (iv) year of LC CT. By repetitive plasma sampling, miRNA signatures with strong predictive, diagnostic and prognostic potential were identified. Further profiling of circulating miRNAs demonstrated that approximately 400 miRNAs could be detected in human serum, and out of these, a group of six miRNAs (miR-30c-1* miR-616*, miR-146b-3p, miR-566, miR-550 and miR-939) had significantly higher expression in serum from NSCLC adenocarcinoma patients than in serum from healthy controls.65
MiRNA profiling showed that the miRNA-200 family was involved in the regulation of EMT and metastasis in tumor cell lines derived from mice that develop metastatic lung adenocarcinoma.66 In clinical material, this miRNA was also found to be overexpressed, suggesting its function as an oncogenic miRNA (oncomir) in LC. An increased expression of miR-31 in lung adenocarcinoma tissues from patients with lymph node metastases compared with those without such metastases was recently demonstrated.67 MiR-31 was validated as a marker for lymph node metastasis in an external validation cohort of 233 lung adenocarcinoma cases of the TCGA and was demonstrated to be a significant predictor of survival in a multivariate cox regression model, even when controlling for tumor stage. As mentioned above, miRNA-214 is expressed in at least a subfraction of NSCLC and negatively regulates invasion potential.32 Whether miRNA-214 expression holds the potential to predict metastasis in vivo in clinical NSCLC specimens waits to be determined.
In searching for miRNAs whose aberrant expression may influence LC therapy response, a number of important findings has been revealed. Mir-7 was found to negatively regulate the EGFR pathway, and miR-21 was found to be upregulated when EGFR signaling was activated, especially, in tumors with EGFR-activating mutations,68 and may hence influence EGFR ablative therapy outcome. Similarly, in a global profiling of miRNAs in a cohort of stage 1 NSCLC and adjacent uninvolved lung tissues, miR-486 was found to be the most downregulated miRNA in tumor specimens.69 In functional studies, miR-486 was found to directly target components of insulin growth factor (IGF) signaling, including insulin-like growth factor 1 (IGF1), IGF1 receptor (IGF1R), and phosphoinositide-3-kinase, regulatory subunit 1 (alpha) (PIK3R1 or p85a), and in this way suppress LC growth both in vitro and in vivo.
MiRNA expression has also been analyzed in relation to LC CT/RT response with the aim of identifying biomarkers of response or novel therapeutic approaches. Among a number of miRNAs expressed in a panel of NSCLC and SCLC cell lines with different RT sensitivity, miRNA-214 was characterized by an increased expression in RT-resistant NSCLC cells.70 MiRNA-214 expression impaired RT-induced apoptotic signaling and involved p38MAPK, as downregulation of this kinase reversed miRNA-214 overexpression-induced RT resistance. Comparative analysis of miRNA-21 in 60 NSCLC samples and adjacent histologically normal tissues using RT-qPCR revealed a correlation between its expression and lymph node metastasis, clinical stage and poor prognosis.71 Ablation of miRNA-21 inhibited proliferation and cell cycle progression of A549 cells and sensitized for RT by increasing RT-induced apoptotic signaling.
The Kroemer's research team analyzed miRNAs that were upregulated in A549 cells in response to cisplatin.72 Among them were miR-181a and miR-630, which regulated cisplatin-induced cell death via the involvement of multiple cisplatin-induced DNA damage response proteins, including ATM kinase and two of its substrates, H2AX and p53, as well as modulation of mitochondrial/post-mitochondrial steps of the intrinsic apoptosis pathway. In another study, it was shown that silencing of miR-155, a miRNA which is overexpressed in LC, resulted in the overexpression of Apaf-1 and greatly increased the sensitivity of A549 cells to cisplatin-induced DNA damage and apoptotic signaling.73 Moreover, it was reported that miR-451 inhibits growth and induces apoptosis in A549 cells, leading to increased cisplatin sensitivity with a concomitant downregulation of Akt.74 Thus, miR-181a, miR-630 and miR-155 were all identified as drivers of cisplatin resistance of NSCLC, whereas miR-451 could reverse such resistance, illustrating that multiple miRNAs likely govern the cisplatin responsiveness of NSCLC. Moreover, expression of miR-21was significantly increased in platinum-based CT-resistant patients and was associated with shorter disease-free survival. Importantly, 70% of the 32 patients analyzed had similar expression patterns of miR-21 in their plasma as in their tumors, illustrating that miR-21may indeed be a suitable plasma-associated biomarker for cisplatin responsiveness. In a clinical study, a two miRNA signature (miR-149 and miR-375) in NSCLC tumors was found to be predictive for first-line cisplatin and vinorelbine CT response and was associated with PFS.75 A signature of four miRNAs (miR-200c, miR-424, miR-29c and miR-124) could delineate patients with good and poor CT responses with an approximately three-fold difference in median survival time, suggesting that miRNA may also be used in the clinical setting to predict CT response.
The levels of miR-21, miR-29b, miR-34a/b/c, miR-155 and let-7a were determined by PCR in NSCLC specimens from over 600 of the IALT patients and examined in relation to survival as well as other clinicopathological prognostic factors.76 Apart from miR-21, none of these miRNAs were associated with survival, whereas low miR-21 had a deleterious prognostic effect.
Expression levels of Dicer 1 and Drosha, two miRNA biogenesis enzymes, were linked to NSCLC histology in NSCLC patients.77 Analysis of mRNA expression levels of Drosha in clinical specimens revealed that low expression was associated with an increased median survival (154.2 versus 39.8 months). However, in certain subgroups (adenocarcinoma, grade III tumors and low stage) the opposite was true: high Drosha expression was associated with decreased median survival. We found that Drosha and Dicer had higher expression in RT-resistant NSCLC cells; however, knockdown of these proteins did not influence RT response, suggesting that although miRNA biogenesis is clearly altered in NSCLC, it might not be a way for therapy sensitization as tumor cells have likely adapted to this altered signaling in multiple ways.78 In conclusion, profiling of miRNA in relation to CT and RT has highlighted certain miRNAs that might explain resistance pathways in LC, which might be important for the prediction of treatment outcome. However, therapies and markers of clinical utility based on miRNA remain to be established.
Proteomic Approach in Analysis of LC Response to Treatment
Recent advances in global and phosphoproteomic methods have been integrated in a systems biology manner to reveal putative signaling cascades driving LC and its treatment response. We have applied different MS-based methodologies with the aim of linking the expression of single proteins, protein groups or complete proteomes to CT/RT response of both NSCLC and SCLC.79, 80, 81 Using the SELDI technique, S100A6 was identified as a protein linked to RT response of lung adenocarcinoma.80 In NSCLC patient materials S100A6 protein was reported to be an independent prognostic factor in p53-negative stage I NSCLC.81 Recently, we demonstrated that S100A4 interacts with p53 within cell nuclei, likely causing its degradation, and thereby influencing both CT-induced apoptosis and cell cycle arrest; furthermore, this illustrates that proteomic profiling can reveal novel signaling cascades to already existing tumor-driving networks.82
A proteomic approach to study the plasma proteome of four different LC mouse models (TetO-EGFRL858R/CSSP-rtTA (Lung-EGFR), TetO-Kras4bG12d/CSSP-rtTA (Lung-Kras), urethane treated (Lung-Urethane) and an SCLC neuroendocrine model (AdCre-infected Trp53lox/lox; Rblox/lox) was taken and compared with those from breast, prostate, pancreatic and colon cancers.83 Plasma proteomes of the four LC models clustered together, as did the breast cancer model, suggesting that the plasma proteome of each tumor type is composed of a tumor-specific pattern of proteins. By filtering out the specific plasma proteome proteins, a 13 proteins signature was found in several of the lung adenocarcinomas. A 16 protein signature was found in the other epithelial-derived breast, prostate, pancreatic and colon tumors, of which WFDC2, a secreted protease inhibitor protein, was one. In the same study,83 using a panel of 21 lung adenocarcinoma cell lines, the total proteome, the plasma membrane or their secreted proteome were studied by MS; 25 out of the 39 proteins identified in the LC mouse model's plasma proteome were found in the lung adenocarcinoma cells and an overlapping set of another 26 proteins was found in the plasma membrane proteome. Two of the identified proteins, Stfpb and its transcriptional regulator Tift/Nkx2-1, a lineage-specific determinant of survival in LC, were further analyzed in the tumor specimens of the LC models and found to be expressed. Moreover, the authors also examined the plasma proteome alterations in response to erlotinib in the EGFR-driven LC mouse model, and two markers were identified (NPC2 and Adam10), demonstrating that plasma proteomes may be useful to monitor erlotinib treatment response.83 Finally, these findings were validated in plasma samples from LC patients, in which SFTB, WFCD2 and ANGPTL3 were found to have increased expression, whereas EGFR expression was decreased. Thus, this systems biology approach towards the plasma proteome represents an excellent example of diagnostic biomarker discovery of LC.
Another proteomic application was demonstrated where MALDI imaging was used to describe the spatial localization of erlotinib and gefitinib without chemical labeling in lung adenocarcinoma and large cell carcinomas.84 High resolution was achieved, allowing detection of drug concentrations down to attomolar levels and enabling drug monitoring in whole-lung tumor tissue. It will be interesting to see whether this technique can also be applied to monitor CT drugs in situ and reveal mechanisms of resistance.
Technological advances in the phosphoproteomic field have enabled global phosphoproteomic profiling of LC cell lines and tumor specimens in different contexts.31, 36, 85, 86, 87, 88, 89, 90 Among the kinases identified were c-MET, EGFR, DDR2 and various ephrin receptor kinases, including EphA2. With respect to the clinical samples analyzed, five major groups were distinguished on the basis of their phosphotyrosine protein pattern, that is, tumors driven by (I) one oncogenic kinase, for example, EGFR, (II) FAK in combination with Src and Abl, (III) DDR1 together with Abl and Src kinases, (IV) Src in combination with EGFR, and (V) Src or Abl kinases alone. Notably, in line with data from DNA sequencing,2 several Eph receptor kinases were among the kinases identified both in NSCLC cell lines and in patient-derived LC specimens. The phosphoproteome has also been analyzed in relation to EGFR mutations using either a targeted approach or global phosphotyrosine profiling followed by LC/MS/MS.85, 86, 87 Thus, the tyrosine phosphoproteome of NSCLC cell lines with different EGFR and Kras genomic status was analyzed on a global scale.86, 87 In mutant EGFR-expressing cells, other receptor tyrosine kinases, such as c-MET and IGF-1R, also showed a higher degree of phosphorylation than that observed in cells with wtEGFR, suggesting that multiple kinases are operative. By comparing the phosphoproteomic data from EGFR- and c-Met-dependent NSCLC cells following treatment with gefitinib or Su11274, respectively, a core network of about 50 proteins, which was affected by either of the oncogenic kinases in the different NSCLC cell lines, was revealed. Again, the Eph kinases were identified to be phosphorylated in response to EGFR and c-Met blockade, further emphasizing the role of this receptor family in LC treatment response. Recently, phosphoproteomic analysis of two clinical NSCLC cases was made.88 It was reported that these NSCLC tumors had different phosphoproteomes, likely reflecting their diverse pathways. Apart from signaling cascades directly linked to initial membrane signal (e.g., ErbB2, ITG5A, c-Met), other multi-functional hubs (e.g., AKT, Raf, ATR, PI3K) and transcriptional regulators (e.g., Mef2D, TP53, ELK3, ATF7) were identified, most of which were regulated in a tumor case-specific manner.
Another approach, activity-based probe profiling (ABPP), was taken to understand important pathways for dasatinib signaling in NSCLC.89 In addition to Src family kinases, cyclin G-associated kinases, DDR1 and several Eph receptor kinases, including EphA2, EphB2 and EphB4, from NSCLC cell extracts were found to bind to dasatinib. Phosphotyrosine proteomic profiling revealed that the majority of these kinases were active in NSCLC cells at the basal level. Repeating the same phosphoprofiling after dasatinib treatment showed a dramatic inhibition of the phosphorylation of some of these kinases, including Ephs. These data are in line with the observations concerning the link between Eph receptor overexpression and dasatinib responsiveness.3
Conclusions
It is becoming increasingly clear that the activation of multiple oncogenes or inactivation of tumor-suppressor genes results in a complex deregulation of cell signaling pathways in tumors in general and in LC in particular. Accumulating evidence suggests that a gene expression signature-based readout of pathway activation is more appropriate than relying on a single gene of each pathway, thus reflecting a complex interplay between the different components that require a more complex analytical approach in order to be visible. By using a systems biology approach, integrating data obtained from clinically relevant LC samples or mice models using gene expression, DNA sequencing, CGH analysis, miRNA profiling, protein or protein phosphorylation signaling network assessments and combining them with data generated from siRNA library screens, true progress in understanding LC biology has been achieved. This has indeed resulted in improved prognosis and treatment predictive strategies for NSCLC, whereas for SCLC the clinical utility of the obtained results is yet to be shown. Although collaborative efforts between different research institutions and consortium, as well as between different professionals within the biomedical research field, have enabled significant progress in understanding the complexity of LC, further joint strategies are warranted to improve attempts to individualize LC treatment and turn this serious disease into at least a chronic disorder and, eventually, if possible, find a cure.
Acknowledgments
The authors' work was supported by grants from the Stockholm Cancer Society, the Swedish Cancer Society, The Swedish Research Council, the Swedish Childhood Cancer Foundation, the Stockholm County Council (SLL), the Karolinska Institutet research funds and the EC FP-6 (Chemores) and FP-7 (Apo-Sys) programs.
Glossary
- ABL2
Abelson murine leukemia viral oncogene homolog 1
- ABPP
activity-based probe profiling
- APC
adenomatous polyposis coli
- ATM
ataxia telangiectasia mutated
- ATR
ATM- and RAD3-related
- Bcl-2
B-cell lymphoma 2
- Bim
B-cell lymphoma 2 interacting mediator of cell death
- BRCA1
breast cancer gene 1
- CGH
comparative genomic hybridization
- CREBP
cAMP-response element (CRE)-binding protein
- CT
chemotherapy
- DDR1
discoidin domain receptor tyrosine kinase 1
- DNA
deoxyribonucleic acid
- DNA-PK
DNA-dependent protein kinase
- EGFR
epidermal growth factor receptor
- ELK3
ETS domain-containing protein 3
- EML4-ALK
echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase
- EMT
epithelial to mesenchymal transition
- EPH
ephrin receptor
- ERCC1
excision repair cross-complementation group 1
- FAK
focal adhesion kinase
- FGF
fibroblast growth factor
- FRK
fyn-related kinase
- HER-2
human EGF (epidermal growth factor) receptor 2
- IGF1-R
insulin growth factor 1 receptor
- LC
lung cancer
- LCK
lymphocyte-specific protein-tyrosine kinase
- LKB1
liver kinase B1
- MALDI
matrix-assisted laser desorption/ionization
- MAPK
mitogen-activated protein kinase
- MEK
MAPK ERK kinase
- miRNA
microRNA
- mRNA
messenger RNA
- MS
mass spectrometry
- MSH2
MutS protein homolog 2
- mTOR
mammalian target of rapamycin
- NF1
neurofibromatosis type 1
- NFKβ
nuclear factor kappa B
- NSCLC
non-small cell lung carcinoma
- NTRK
neurotrophic tyrosine receptor kinase
- OCT4
octamer-binding transcription factor 4
- PAPP-A
pregnancy-associated plasma protein A
- PARP1
poly (ADP-ribose) polymerase 1
- PFS
progression-free survival
- PTMs
post translational modifications
- RAP80
receptor-associated protein 80
- RB1
retinoblastoma 1
- RET
rearranged during transfection
- RNAi
RNA interference
- ROS1
c-ros oncogene
- RRM1
ribonucleotide reductase, M1 subunit
- RT
radiotherapy
- SCLC
small cell lung carcinoma
- SELDI
surface enhanced laser desorption/ionization
- shRNA
short hairpin RNA
- SNP
single-nucleotide polymorphisms
- SRC
rous sarcoma oncogene cellular homolog
- TCF3
T-cell factor 3
- TCGA
the cancer genome atlas
- TGFβ
transforming growth factor beta
- TKI
tyrosine kinase inhibitor
- YES
Yamaguchi sarcoma virus oncogene
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Higgins MJ, Ettinger DS. Chemotherapy for lung cancer: the state of the art in 2009. Expert Rev Anticancer Ther. 2009;9:1365–1378. doi: 10.1586/era.09.115. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorsson K, Lewensohn R. Apoptotic signaling pathways in lung cancer. J Thorac Oncol. 2007;2:175–179. doi: 10.1097/JTO.0b013e318031cd78. [DOI] [PubMed] [Google Scholar]
- Arrell DK, Terzic A. Network systems biology for drug discovery. Clin Pharmacol Ther. 2010;88:120–125. doi: 10.1038/clpt.2010.91. [DOI] [PubMed] [Google Scholar]
- Ocak S, Sos ML, Thomas RK, Massion PP. High-throughput molecular analysis in lung cancer: insights into biology and potential clinical applications. Eur Respir J. 2009;34:489–506. doi: 10.1183/09031936.00042409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev Proteomics. 2011;8:61–79. doi: 10.1586/epr.10.99. [DOI] [PubMed] [Google Scholar]
- Pierobon M, Vanmeter AJ, Moroni N, Galdi F, Petricoin EF., 3rd Reverse-phase protein microarrays. Methods Mol Biol. 2012;823:215–235. doi: 10.1007/978-1-60327-216-2_14. [DOI] [PubMed] [Google Scholar]
- Indovina P, Marcelli E, Pentimalli F, Tanganelli P, Tarro G, Giordano A. Mass spectrometry-based proteomics: the road to lung cancer biomarker discovery. Mass Spectrom Rev. 2013;32:129–142. doi: 10.1002/mas.21355. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyev A, Fourquet S, Tournier L, Calzone L, Barillot E. Cell death and life in cancer: mathematical modeling of cell fate decisions. Adv Exp Med Biol. 2012;736:261–274. doi: 10.1007/978-1-4419-7210-1_15. [DOI] [PubMed] [Google Scholar]
- Cohen D, Kuperstein I, Barillot E, Zinovyev A, Calzone L. From a biological hypothesis to the construction of a mathematical model. Methods Mol Biol. 2013;1021:107–125. doi: 10.1007/978-1-62703-450-0_6. [DOI] [PubMed] [Google Scholar]
- Calzone L, Tournier L, Fourquet S, Thieffry D, Zhivotovsky B, Barillot E, et al. Mathematical modelling of cell-fate decision in response to death receptor engagement. PLoS Comput Biol. 2010;6:e1000702. doi: 10.1371/journal.pcbi.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosyan F, Daw H, Haddad A, Spiro T, Sood R.Gene expression profiling for early-stage NSCLC Am J Clin Oncol 2013. e-pub ahead of print 19 April 2013. [DOI] [PubMed]
- Rosell R, Taron M, Massuti B, Mederos N, Magri I, Santarpia M, et al. Predicting response to chemotherapy with early-stage lung cancer. Cancer J. 2011;17:49–56. doi: 10.1097/PPO.0b013e3182091fa3. [DOI] [PubMed] [Google Scholar]
- Alamgeer M, Ganju V, Watkins DN. Novel therapeutic targets in non-small cell lung cancer. Curr Opin Pharmacol. 2013;13:394–401. doi: 10.1016/j.coph.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12:1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- Network CGAR Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genetics. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos ML, Dietlein F, Peifer M, Schottle J, Balke-Want H, Muller C, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci USA. 2012;109:17034–17039. doi: 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Salim H, Arvanitis A, de Petris L, Kanter L, Haag P, Zovko A, et al. miRNA-214 is related to invasiveness of human non-small cell lung cancer and directly regulates alpha protein kinase 2 expression. Genes Chromosomes Cancer. 2013;52:895–911. doi: 10.1002/gcc.22085. [DOI] [PubMed] [Google Scholar]
- Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan JM, Sen B, Saigal B, Prudkin L, Behrens C, Solis L, et al. EphA2 in the early pathogenesis and progression of non-small cell lung cancer. Cancer Prev Res (Phila) 2009;2:1039–1049. doi: 10.1158/1940-6207.CAPR-09-0212. [DOI] [PubMed] [Google Scholar]
- Stahl S, Kaminskyy VO, Efazat G, Hyrslova Vaculova A, Rodriguez-Nieto S, Moshfegh A, et al. Inhibition of Ephrin B3-mediated survival signaling contributes to increased cell death response of non-small cell lung carcinoma cells after combined treatment with ionizing radiation and PKC 412. Cell Death Dis. 2013;4:e454. doi: 10.1038/cddis.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S, Branca RM, Efazat G, Ruzzene M, Zhivotovsky B, Lewensohn R, et al. Phosphoproteomic profiling of NSCLC cells reveals that ephrin B3 regulates pro-survival signaling through Akt1-mediated phosphorylation of the EphA2 receptor. J Proteome Res. 2011;10:2566–2578. doi: 10.1021/pr200037u. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Feng XD, Huang SG, Shou JY, Liao BR, Yingling JM, Ye X, et al. Analysis of pathway activity in primary tumors and NCI60 cell lines using gene expression profiling data. Genomics Proteomics Bioinformatics. 2007;5:15–24. doi: 10.1016/S1672-0229(07)60010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk J, Rolff J, Becker M, Leschber G, Fichtner I. Patient-derived xenografts of non-small-cell lung cancer: a pre-clinical model to evaluate adjuvant chemotherapy. Eur J Cardiothorac Surg. 2009;36:454–459. doi: 10.1016/j.ejcts.2009.03.054. [DOI] [PubMed] [Google Scholar]
- Krumbach R, Schuler J, Hofmann M, Giesemann T, Fiebig HH, Beckers T. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: activation of MET as one mechanism for drug resistance. Eur J Cancer. 2011;47:1231–1243. doi: 10.1016/j.ejca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Nemati F, Sastre-Garau X, Laurent C, Couturier J, Mariani P, Desjardins L, et al. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin Cancer Res. 2010;16:2352–2362. doi: 10.1158/1078-0432.CCR-09-3066. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- To MD, Quigley DA, Mao JH, Del Rosario R, Hsu J, Hodgson G, et al. Progressive genomic instability in the FVB/Kras(LA2) mouse model of lung cancer. Mol Cancer Res. 2011;9:1339–1345. doi: 10.1158/1541-7786.MCR-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–256. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laar K. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med Genomics. 2012;5:1–12. doi: 10.1186/1755-8794-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9:144–155. doi: 10.1038/nrclinonc.2012.3. [DOI] [PubMed] [Google Scholar]
- Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, Camps C, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang J, Wang R, Luo X, Chen H. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer. 2010;70:63–70. doi: 10.1016/j.lungcan.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368:1101–1110. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friboulet L, Barrios-Gonzales D, Commo F, Olaussen KA, Vagner S, Adam J, et al. Molecular characteristics of ERCC1-negative versus ERCC1-positive tumors in resected NSCLC. Clin Cancer Res. 2011;17:5562–5572. doi: 10.1158/1078-0432.CCR-11-0790. [DOI] [PubMed] [Google Scholar]
- Rosell R, Perez-Roca L, Sanchez JJ, Cobo M, Moran T, Chaib I, et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One. 2009;4:e5133. doi: 10.1371/journal.pone.0005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels J, Vitale I, Galluzzi L, Adam J, Olaussen KA, Kepp O, et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 2013;73:2271–2280. doi: 10.1158/0008-5472.CAN-12-3000. [DOI] [PubMed] [Google Scholar]
- Olaussen KA, Adam J, Vanhecke E, Vielh P, Pirker R, Friboulet L, et al. PARP1 impact on DNA repair of platinum adducts: preclinical and clinical read-outs. Lung Cancer. 2013;80:216–222. doi: 10.1016/j.lungcan.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genetics. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Pastorino U, Croce CM. MicroRNAs and lung cancer: from markers to targets. Cell Cycle. 2011;10:2045–2046. doi: 10.4161/cc.10.13.15712. [DOI] [PubMed] [Google Scholar]
- Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18:268–274. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S, Gately K, Crown J, O'Byrne K, O'Driscoll L. Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biol Ther. 2013;14:12. doi: 10.4161/cbt.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011;71:7670–7682. doi: 10.1158/0008-5472.CAN-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Ye Z, Cui R, Perry J, Dedousi-Huebner V, Huebner A, et al. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin Cancer Res. 2013;19:5423–5433. doi: 10.1158/1078-0432.CCR-13-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- Peng Y, Dai Y, Hitchcock C, Yang X, Kassis ES, Liu L, et al. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci USA. 2013;110:15043–15048. doi: 10.1073/pnas.1307107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim H, Akbar NS, Zong D, Vaculova AH, Lewensohn R, Moshfegh A, et al. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br J Cancer. 2012;107:1361–1373. doi: 10.1038/bjc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol. 2013;8:146. doi: 10.1186/1748-717X-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- Zang YS, Zhong YF, Fang Z, Li B, An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012;19:773–778. doi: 10.1038/cgt.2012.60. [DOI] [PubMed] [Google Scholar]
- Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549) J Exp Clin Cancer Res. 2011;30:20. doi: 10.1186/1756-9966-30-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans T, Ameye L, Willems L, Paesmans M, Mascaux C, Lafitte JJ, et al. Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer. 2013;82:340–345. doi: 10.1016/j.lungcan.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70:8288–8298. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Garcia CV, Agudo-Lopez A, Perez C, Lopez-Martin JA, Rodriguez-Peralto JL, de Castro J, et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: implications for outcomes and histologic classification. Carcinogenesis. 2013;34:1031–1038. doi: 10.1093/carcin/bgt022. [DOI] [PubMed] [Google Scholar]
- Surova O, Akbar NS, Zhivotovsky B. Knock-down of core proteins regulating microRNA biogenesis has no effect on sensitivity of lung cancer cells to ionizing radiation. PLoS One. 2012;7:e33134. doi: 10.1371/journal.pone.0033134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernemalm M, De Petris L, Branca RM, Forshed J, Kanter L, Soria JC, et al. Quantitative proteomics profiling of primary lung adenocarcinoma tumors reveals functional perturbations in tumor metabolism. J Proteome Res. 2013;12:3934–3943. doi: 10.1021/pr4002096. [DOI] [PubMed] [Google Scholar]
- Orre LM, Pernemalm M, Lengqvist J, Lewensohn R, Lehtio J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol Cell Proteomics. 2007;6:2122–2131. doi: 10.1074/mcp.M700202-MCP200. [DOI] [PubMed] [Google Scholar]
- De Petris L, Orre LM, Kanter L, Pernemalm M, Koyi H, Lewensohn R, et al. Tumor expression of S100A6 correlates with survival of patients with stage I non-small-cell lung cancer. Lung Cancer. 2009;63:410–417. doi: 10.1016/j.lungcan.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Orre LM, Panizza E, Kaminskyy VO, Vernet E, Graslund T, Zhivotovsky B, et al. S100A4 interacts with p53 in the nucleus and promotes p53 degradation. Oncogene. 2013;32:5531–5540. doi: 10.1038/onc.2013.213. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;20:289–299. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko-Varga G, Fehniger TE, Rezeli M, Dome B, Laurell T, Vegvari A. Drug localization in different lung cancer phenotypes by MALDI mass spectrometry imaging. J Proteomics. 2011;74:982–992. doi: 10.1016/j.jprot.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fang B, Liu RZ, Lin H, Kinose F, Bai Y, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, et al. Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci USA. 2008;105:14112–14117. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natil Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Rigas JR, Gerber SA. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J Proteomics. 2013;91C:286–296. doi: 10.1016/j.jprot.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci USA. 2013;110:12414–12419. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, He J. Molecular classification of non-small-cell lung cancer: diagnosis, individualized treatment, and prognosis. Front Med. 2013;7:157–171. doi: 10.1007/s11684-013-0272-4. [DOI] [PubMed] [Google Scholar]