Abstract
The involvement of Axl kinase in non-small cell lung cancer's (NSCLC) acquired resistance to tyrosine kinase inhibitors (TKIs) gefitinib or erlotinib has been identified recently, but the mechanism by which Axl contributes to TKI resistance is largely unknown. MicroRNAs (miRNAs) repress gene expression and their critical role in tumorigenesis has been implicated. To investigate the role of miRNAs in the Axl-mediated acquired gefitinib resistance, we examined the Axl-mediated miRNA changes in gefitinib-resistant lung cancers. A panel of Axl kinase-altered miRNAs was identified. In this study, we validate and report that miR-374a and miR-548b modulated by Axl have essential roles in cell cycle arrest, gefitinib-induced apoptosis, epithelial-to-mesenchymal transition, migration and tumorigenesis of gefitinib-resistant lung cancer cells in vitro and in vivo by targeting Wnt5a and CCNB1 genes, respectively. Of clinical significance, high expression of Axl and miR-374a and low expression of miR-548b are associated with poor disease-free survival postoperatively. These findings indicate that the modulation of specific miRNAs may provide a therapeutic target to treat or reverse gefitinib resistance in NSCLC with high expression of Axl in the future.
Keywords: Axl, microRNA, non-small cell lung cancer, gefitinib, epithelial-to-mesenchymal transition
Compared with chemotherapy, epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, have been implicated to improve clinical outcome and quality of life for advanced non-small cell lung cancer (NSCLC) patients whose tumors harbor an EGFR-activating mutation.1, 2 Although they responded to TKIs at initiation, the majority of these patients will develop acquired resistance after ∼12 months of disease control.3 The molecular mechanism of acquired resistance to EGFR-TKI has been deeply investigated and a few candidates such as T790M (a secondary EGFR mutation) and c-MET amplification have been found.4, 5, 6 Recently, Axl – a tyrosine kinase – has been reported to be a novel mechanism that causes acquired resistance in NSCLC.7 However, the mechanism by which Axl contributes to TKI resistance in NSCLC is not clear.
Originally identified as a transforming gene in the cancer cells of chronic myeloid leukemia patients, Axl is a transmembrane tyrosine kinase receptor, activated by either its ligand-growth arrest specific 6 or extracellular domain-mediated dimerization or cross talk with human EGFR2.8, 9 Axl deregulation has been found in a number of cancers including metastatic breast cancer, prostate cancer, colon cancer and NSCLC.10 It is known to induce cancer cell survival, growth, proliferation and cell–cell adhesion.11, 12, 13 Recently, it has been found to be associated with chemoresistance or resistance to TKIs in lung cancer cells.7 But little is known about the Axl regulation network in TKI-resistant NSCLC.
MicroRNAs (miRNAs) negatively regulate gene expression by binding to 3′UTR of mRNAs, either inhibiting mRNA translation or promoting its degradation.14 Altered miRNA expression has been implicated in a variety of cancers including NSCLC.15 The important role of miRNAs in cancer pathogenesis and drug sensitivity has been observed. A number of miRNAs, including miR-30b-c and miR-221/222, are altered by EGFR or MET in gefitinib-resistant NSCLC, resulting in the regulation of TKI resistance and NSCLC tumorigenesis.16 Whether or not Axl can modulate miRNA expression in NSCLC is not clear. Here, we identified a panel of Axl kinase-altered miRNAs and validated that miR-374a and miR-548b were altered by Axl in TKI-resistant cancer cells. By targeting Wnt5a and CCNB1, these two miRNAs regulate cell cycle, gefitinib-induced apoptosis, epithelial-to-mesenchymal transition (EMT), migration and tumorigenesis of TKI-resistant lung cancer cells. Our findings suggest that miR-374a and miR-548b may provide novel therapeutic targets to treat or reverse TKI resistance in NSCLC with high expression of Axl in the future.
Results
miRNAs modulated by Axl kinase in gefitinib-resistant lung cancer cells
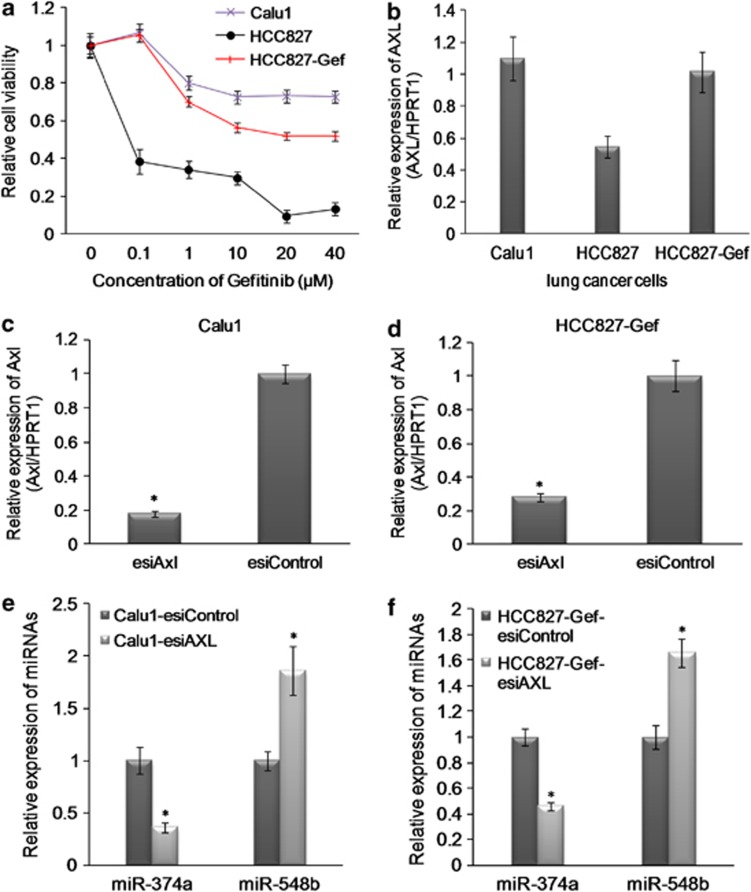
To identify Axl-modulated miRNAs in NSCLC, we first generated a gefitinib-resistant cell line HCC827-Gef (Figure 1a)17 and examined the Axl expression both in HCC827-Gef and in Calu1 (EGFR wild type and resistant to TKI) cells. Axl was significantly upregulated both in HCC827-Gef and in Calu 1 cells (Figure 1b). Next, we silenced Axl in both HCC827-Gef and Calu1 cells using endoribonuclease-prepared siRNA (esiRNA) targeting Axl. The expression of Axl was significantly silenced by real-time quantitative reverse transcription PCR (RT-qPCR) and western blotting analysis (Figures 1c and d, Supplementary Figures S1A and S1B). The microarray analysis was performed at 48 h on pooled Calu1-esiAxl- and HCC827-Gef-esiAxl- or esiControl-treated cells. A number of deregulated miRNAs were identified (Supplementary Table S2). We validated a group of miRNAs using RT-qPCR and identified that miR-374a and miR-548b were mostly deregulated in gefitinib-resistant NSCLC cells, Calu1 and HCC827-Gef, with silencing of Axl. MiR-374a was significantly downregulated and miR-548b was upregulated both in Calu1 and in HCC827-Gef cells (Figures 1e and f). To further demonstrate whether signaling through Axl is directly related to miR-374a and miR-548b expression, we overexpressed18 the Axl expression using the pWZL-Neo-AXL plasmid (Addgene) in HCC827 cells and examined the expression level of miR-374a and miR-548b. We also found that overexpression of Axl significantly increased the expression level of miR-374a and decreased the expression of miR-548b (Supplementary Figures S1C and D).
Figure 1.
miRNAs modulated by Axl kinase in gefitinib-resistant lung cancer cells. (a) The sensitivity of HCC827, HCC827-Gef and Calu1 cells to gefitinib was examined by MTS analysis. (b) The relative expression of Axl in HCC827, HCC827-Gef and Calu1 cells by RT-qPCR analysis. The expression level was normalized by HPRT1. (c and d) The knockdown effects of Axl by esiRNA by RT-qPCR analysis. The expression level was normalized by HPRT1. (e and f) The relative expression of miR-374a and miR-548b in gefitinib-resistant lung cancer cells with silencing of Axl by RT-qPCR analysis. The expression level was normalized by U6 snRNA. Data were shown as the mean±S.D. of three independent experiments. *P<0.05
Clinical significance of Axl-modulated miRNAs in NSCLC
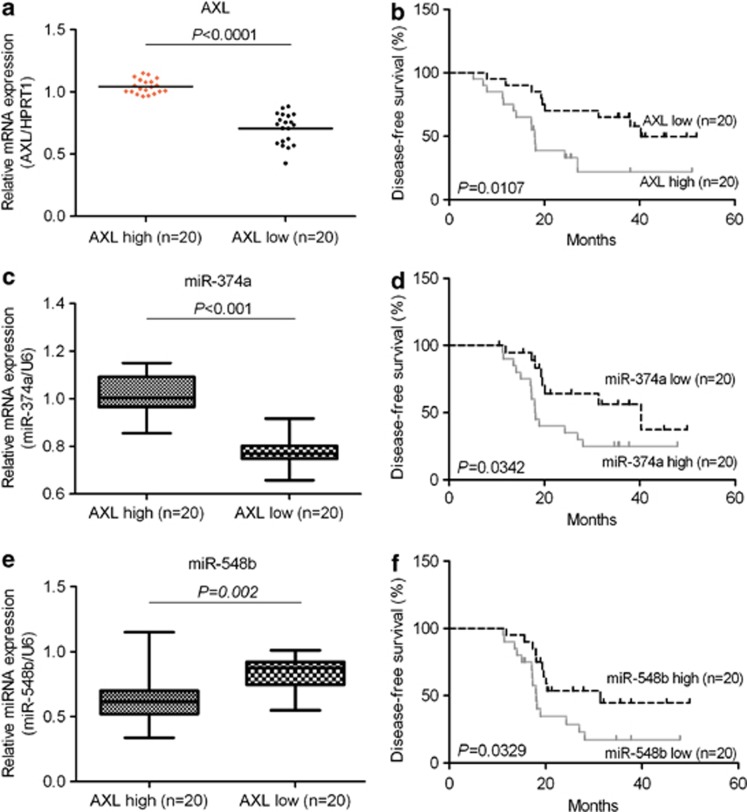
To investigate the clinical significance of Axl and Axl-modulated miRNAs in NSCLC, the expression of Axl mRNA in 40 samples of NSCLCs was compared by RT-qPCR. The clinical characteristics of the NSCLC patients have been summarized in the Supplementary Table S3. Interestingly, we found that NSCLC patients with low Axl had longer disease-free survival (DFS) compared with patients with high Axl (P=0.0107, Student's t-test, Figures 2a and b). Next, we detected miR-374a in these 40 tumor samples. Consistent with results of cell lines, expression of miR-374a was significantly higher in high-Axl tumors. Moreover, low expression of miR-374a was significantly correlated with longer DFS of patients (P=0.0342; Figures 2c and d). MiR-548b was lower in high-Axl tumors (Figure 2e). Also, we found that high expression of miR-548b was significantly correlated with longer DFS of patients (P=0.0342; Figure 2f). In NSCLC, shorter DFS means that patients easily developed resistance and underwent recurrence or metastasis.19 According to these results, Axl, along with its significantly modulated miRNAs miR-374a and miR-548b, may have a critical role in the development of therapeutic resistance and metastasis of NSCLC. Thus, they may serve as prognostic biomarkers or therapeutic targets for treating NSCLC.
Figure 2.
Clinical significance of Axl and Axl-modulated miRNAs in NSCLC. (a, c, e) The relative expression of Axl and altered miR-374a and miR-548b in a group of NSCLC patients' samples was examined by RT-qPCR analysis. The expression level of Axl was normalized by HPRT1. The expression level of miRNAs was normalized by U6 snRNA. (b, d, f) The Kaplan–Meier method was used to analyze survival in patients with lung cancer. The average value of Axl, miR-374a and miR-548b in all samples was chosen as the cutoff point
miR-374a and miR-548b regulate gefitinib sensitivity in vitro
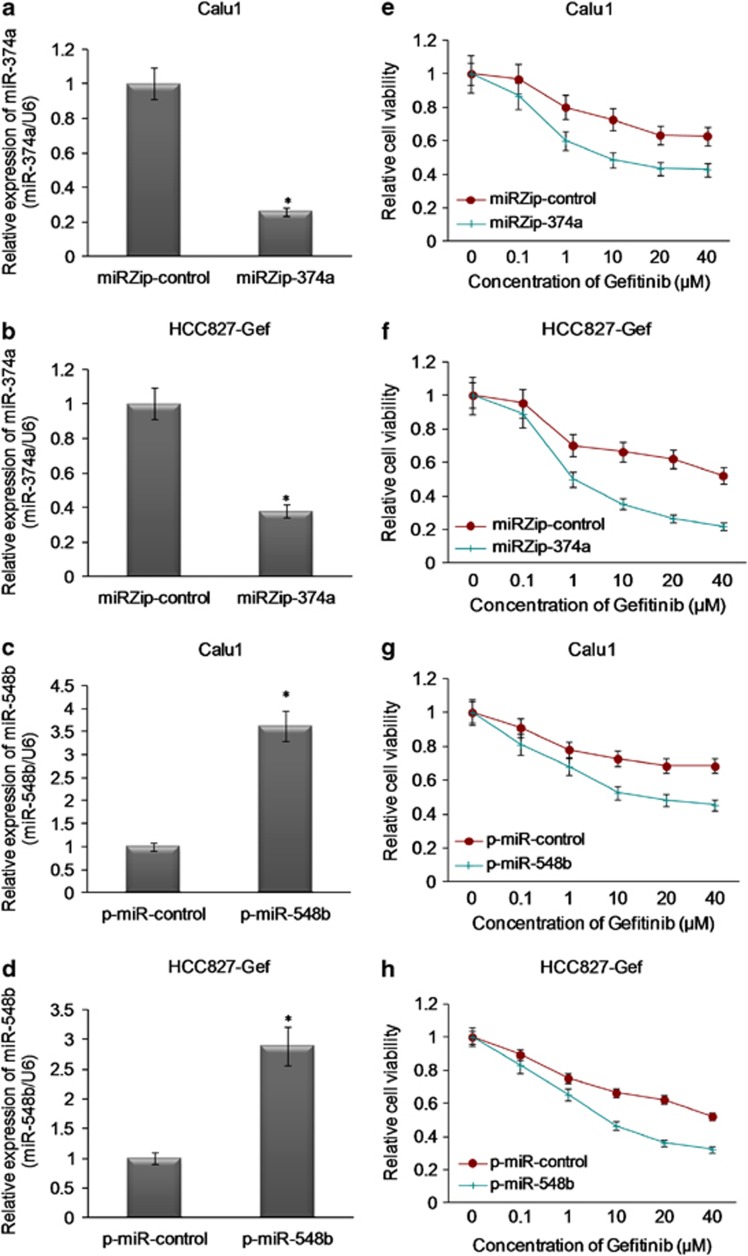
To investigate the role of these miRNAs in gefitinib resistance of NSCLC, the expression of miR-374a was knocked down using miRZip-374a vector or the expression of miR-548b was overexpressed using p-miR-548b vector in both Calu1 and HCC827-Gef cells. As Figures 3a and b showed, miR-374a was significantly repressed by miRZip-374a transfection in both Calu1 and HCC827-Gef NSCLC cells. Also, miR-548b was significantly upregulated by p-miR-548b transfection in these two cell lines (Figures 3c and d). When miR-374a was silenced, we observed that the sensitivity of Calu1 and HCC827 cells to gefitinib was significantly increased by transfection of miRZip-374a (Figures 3e and f). Of note, the sensitivity of Calu1 and HCC827-Gef cells to gefitinib was also restored, whereas miR-548b was overexpressed by p-miR-548b transfection (Figures 3g and h). These findings partly suggest that the expression of miR-374a and miR-548b can regulate gefitinib sensitivity in NSCLC cells in vitro.
Figure 3.
miR-374a and miR-548b regulate gefitinib sensitivity in vitro. (a and b) The knockdown effect of miR-374a with transfection of miRZip-374a was detected by RT-qPCR analysis. (c and d) The overexpression of miR-548b with transfection of p-miR-548b was detected by RT-qPCR analysis. (e–h) The sensitivity of HCC827-Gef and Calu1 cells with knock down of miR-374a or overexpression of miR-548b to gefitinib was examined by MTS analysis *P<0.05
miR-374a targets Wnt5a and miR-548b targets CCNB1
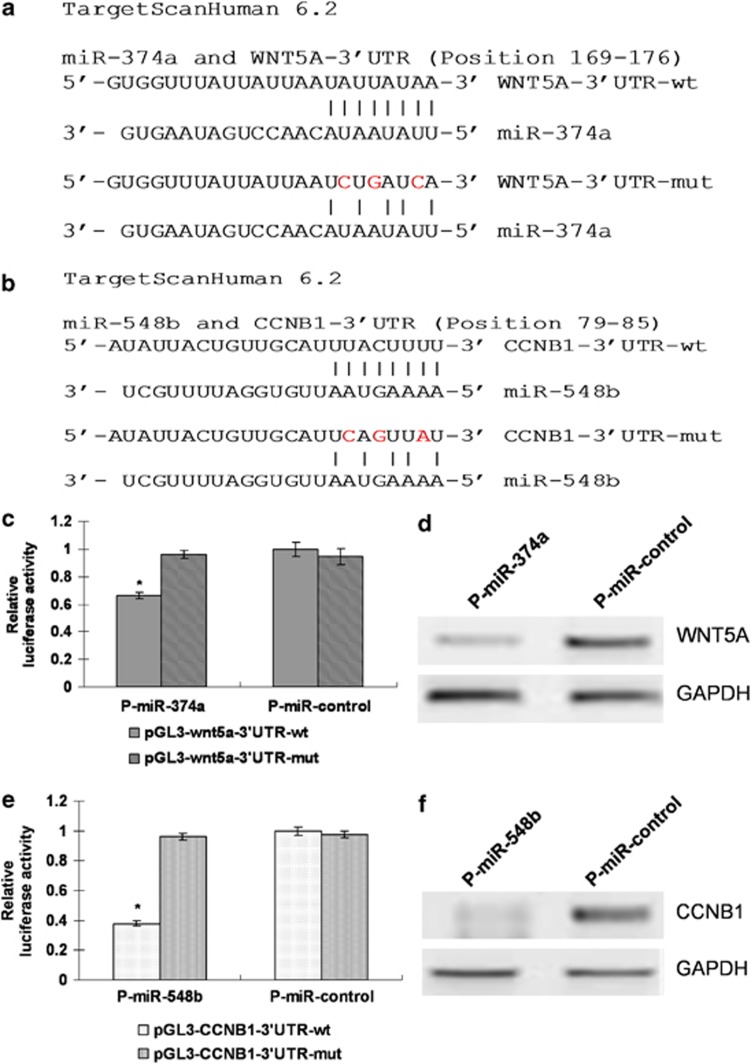
We searched for mRNA targets of miR-374a and miR-548b using computational tools including TargetScan and RNAhybrid.20 We screened a group of predicted potential targets and identified that Wnt5a and CCNB1 contained conserved binding sites specific for these two miRNAs (Figures 4a and b). To investigate whether these miRNAs directly interact with the two computational searched target genes, we co-transfected pGL3 vectors with p-miR-374a and p-miR-548b or controls in HEK293 cells. The significant decreasing luciferase activity was indicated by interaction between pGL3-Wnt5a-3′ UTR and miR-374a, and pGL3-CCNB1-3′ UTR and miR-548b co-transfection (Figures 4c and e). We next investigated whether these two miRNAs repressed the protein expression of Wnt5a and CCNB1 in lung cancer cells. The western blot study showed that overexpression of miR-374a by p-miR-374a in HCC827 cells significantly decreased the protein expression of Wnt5a. Meanwhile, overexpression of miR-548b in HCC827-Gef cells also inhibits the protein expression of CCNB1 (Figures 4d and f), indicating that, on the transcriptional level, miR-374a can repress Wnt5a and miR-548b represses CCNB1 in lung cancer cells. Next, we further investigated the effects of Wnt5a and CCNB1 on the gefitinib resistance/sensitivity phenotype of lung cancer cells. We found that overexpression of Wnt5a in HCC827-Gef cells partially restored the cell sensitivity to gefitinib (Supplementary Figures S2A and B); the re-expression of CCNB1 levels in p-miR-548b-transfected HCC827-Gef cells also recapitulated the cell sensitivity to gefitinib (Supplementary Figures S2C and D).
Figure 4.
miR-374a targets Wnt5a and miR-548b targets CCNB1. (a and b) The predicted binding sites of mRNA targets with miR-374a and miR-548b using computational tools including TargetScan and RNAhybrid were shown. (c and e) Effects of co-transfection of p-miR-374a with wild-type (wt) and mutant (mut) wnt5a 3'UTR pGL3 constructs, or p-miR-548b with wild-type (wt) and mutant (mut) CCNB1 3'UTR pGL3 constructs on luciferase reporter assays. Data were normalized by the ratio of Firefly and Renilla luciferase activities measured at 48 h posttransfection. The bar graph shows the mean±S.D. in three independent transfection experiments. *P<0.05. (d and f) The expression of WNT5a and CCNB1 in P-miR-control- and P-miR-374a or P-miR-548b-transfected NSCLC cells by western blotting analysis
Silencing of miR-374a inhibits migration and invasion in gefitinib-resistant NSCLC
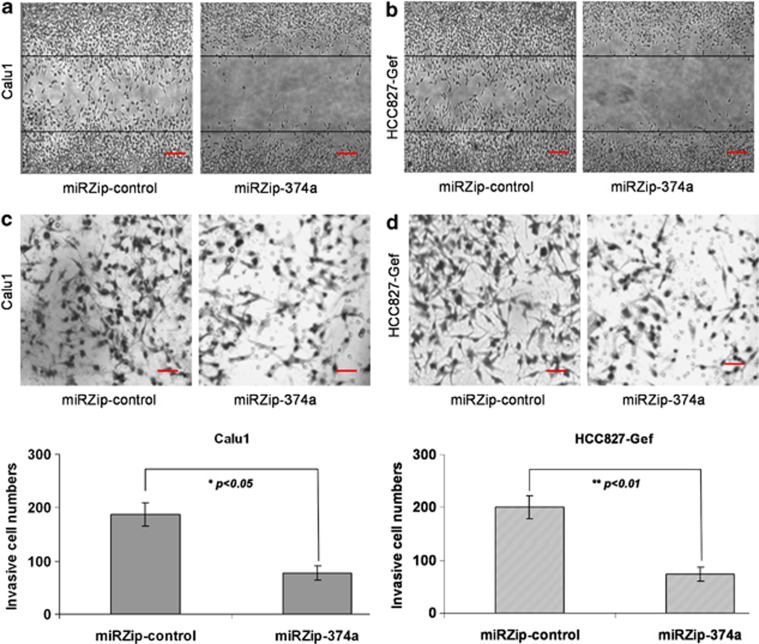
MiR-374a has been reported to be associated with EMT in breast cancer, thus it has an important role in invasion and migration of breast cancer cells.21, 22 To understand the specific functional role of miR-374a in gefitinib-resistant NSCLC cells, we performed a loss-of-function assay. MiR-374a was downregulated by miRZip-374a in HCC827-Gef and Calu1 cell lines. Next, we performed the wound healing and in vitro Matrigel invasion assays. Both HCC827-Gef and Calu1 cells migrated much more slowly when miR-374a was downregulated after 24 h in culture (Figures 5a and b). Invasion is a direct measure of metastatic potential and requires cell migration and proteolytic properties that allow cells to pass through a solid extracellular matrix, such as Matrigel. Consistent with the results of the migration assay, the percentage of invasive cells was also significantly decreased when miR-374a was downregulated in both HCC827-Gef and Calu1 cells (Figures 5c and d). This evidence demonstrates that the silencing of miR-374a inhibits migration and invasion in gefitinib-resistant NSCLC cells.
Figure 5.
Silencing of miR-374a inhibits migration and invasion in gefitinib-resistant NSCLC. (a and b) The representative images show the migration ability of Calu1 and HCC827-Gef cells with transfection of miRZip-374a or miRZip-control. (c and d) The invasive cells were stained and counted under the microscope. Quantitative results for the transmembrane ability of each group of cells. All data are given as mean ±S.D. of three independent experiments. Significant differences are indicated as follows: one-sample t-test, *P<0.05
MiR-548b induces cell cycle arrest in gefitinib-resistant lung cancer cells
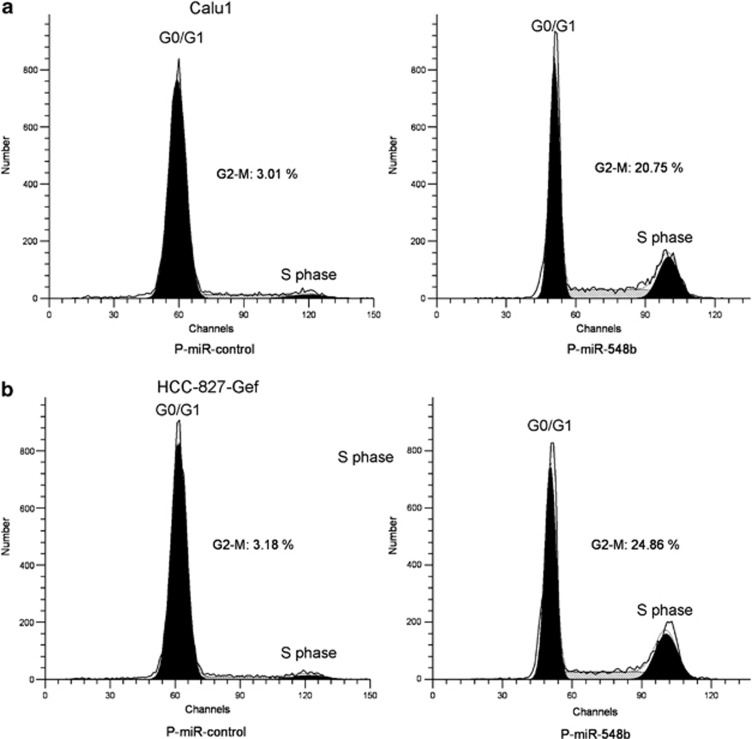
The cell cycle is regulated by a series of cytoplasmic proteins such as cyclins. Cyclin B1 is essential for the initiation of mitosis and is an important cell cycle regulator. We have showed that CCNB1 is a downstream target of miR-548b in gefitinib-resistant NSCLC cells. To investigate the role of miR-548b in cell cycle regulation of gefitinib-resistant NSCLC cells, we overexpressed miR-548b in both HCC827-Gef and Calu1 cells. We found that compared with control vector-transfected cells, levels of G2-M in HCC827-Gef cells increased from 3.28 to 24.86% and in Calu1 cells from 3.01 to 20.75% when miR-548b was upregulated by p-miR-548b vector transfection (Figures 6a and b). This evidence suggests that miR-548b can induce cell cycle arrest in gefitinib-resistant lung cancer cells.
Figure 6.
MiR-548b induces cell cycle arrest in gefitinib-resistant lung cancer cells. (a and b) The representative images show miR-548b overexpression on cell cycle progression. The G2/M phase arrest was observed in both Calu1 and HCC827-Gef cells tranfected with p-miR-548b compared with p-miR-control tranfection
MiR-374a induces EMT, colony formation and drug resistance in gefitinib-sensitive lung cancer cells
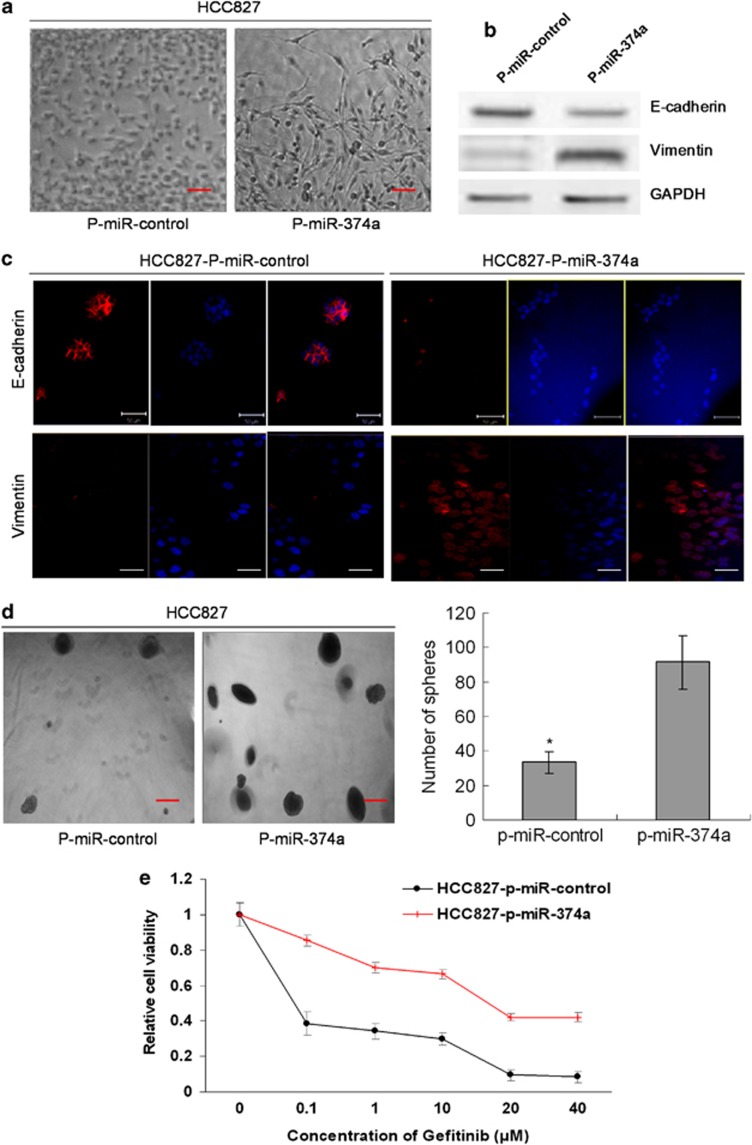
We found that downregulation of miR-374a can suppress migration and invasion in gefitinib-resistant NSCLC. Recently, the association of miR-374a with EMT has been reported in breast cancer cells.21, 22 Next, we assessed the possibility of miR-374a inducing EMT in NSCLC. As it was shown in Figure 7a, when miR-374a was overexpressed in HCC827, most cells were neither adherent to each other nor apically polarized. We postulated that HCC827 may have gone through EMT on the basis of this morphological change. The western blot was performed to evaluate the expression of EMT-related markers in HCC827 cells when miR-374a was overexpressed. As expected, the epithelial marker E-cadherin was downregulated, whereas the mesenchymal marker Vimentin was upregulated in HCC827 cells with overexpression of miR-374a, as compared with HCC827 tranfected with p-miR-control (Figure 7b). Consistent with the result of the western blot, we validated the expression of E-cadherin and Vimentin using immunofluorescence analysis and further confirmed that E-cadherin was downregulated, whereas Vimentin was upregulated when miR-374a was overexpressed in HCC827 cells (Figure 7c). Surprisingly, we also found that sphere formation ability increased approximately threefold when miR-374a was overexpressed in HCC827 cells (Figure 7d). Meanwhile, the expression of stem cell markers, Nanog and Oct4, was examined by immunofluorescence. The high expression of these stem cell markers in p-miR-374a-transfected cells compared with p-miR-control transfected cells was also observed (Supplementary Figure S3). Of note, when miR-374a was overexpressed, the sensitivity of HCC827 cells to gefitinib was significantly decreased compared with HCC827-p-miR-control cells (Figure 7e). These results indicate that miR-374a has an important role in EMT, stemness and resistance to gefitinib of sensitive NSCLC cells with an EGFR-activating mutation.
Figure 7.
MiR-374a induces EMT, colony formation and drug resistance in gefitinib-sensitive lung cancer cells. (a) The representative images of significant morphological change from an epithelial cobblestone phenotype to an elongated fibroblastic phenotype, indicative of EMT. (b) The expression of epithelial marker E-cadherin and mesenchymal marker vimentin by western blot. (c) The stable transfected cells were stained for E-cadherin, vimentin and Hoechst 33342 and analyzed by confocal microscopy. The red signal represents staining for E-cadherin or vimentin. Nuclear DNA was detected by staining with Hoechst 33342. (d) Representative images of spheres. Stable overexpression of miR-374a promotes sphere formation of HCC827 cells. The results have been summarized in the bar graph. Error bars represent S.D. and asterisks denote statistical significance<0.05. (e) The sensitivity of HCC827 transfected with p-miR-374a or p-miR-control to gefitinib was examined by MTS analysis. The result showed that overexpression of miR-374a significantly decreased the sensitivity to gefitinib, P<0.05
MiR-374a and miR-548b regulate tumorigenesis of gefitinib-resistant lung cancer cells in vivo by modulating apoptosis
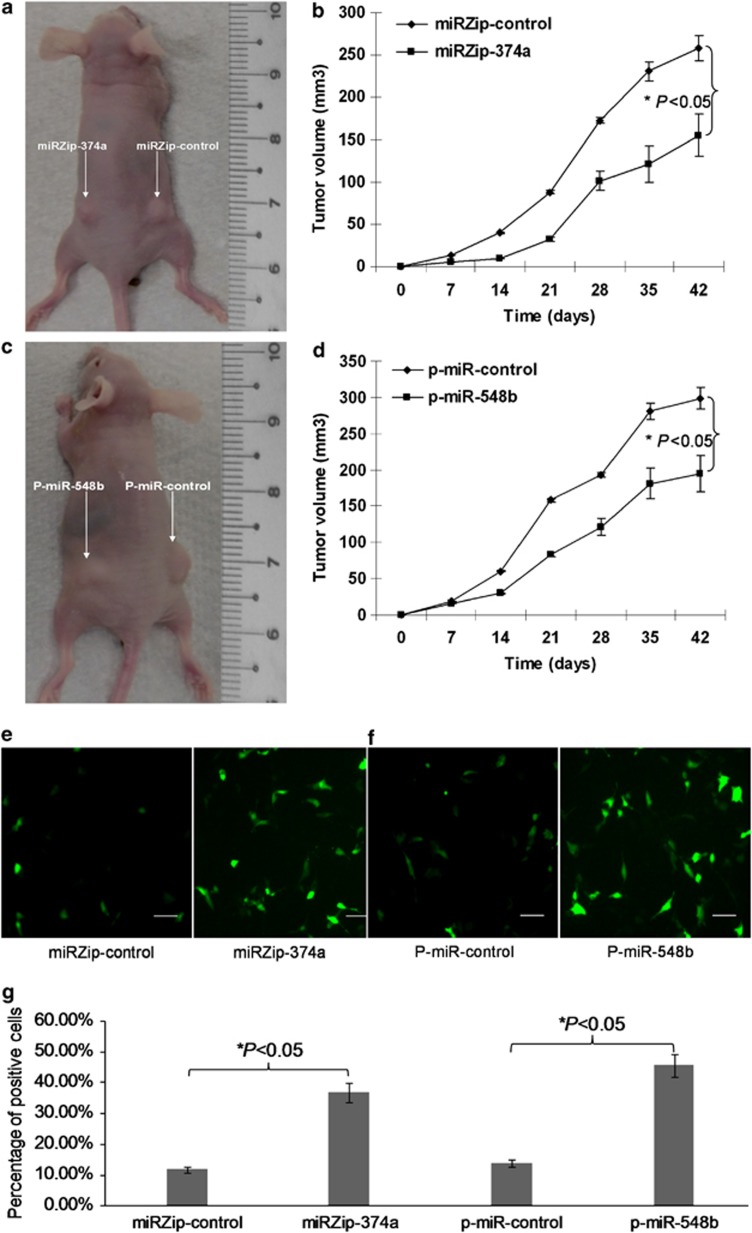
Next, we assessed whether miR-374a and miR-548b can modulate tumorigenesis of resistant NSCLC cells in vivo. We stably knocked down the expression of miR-374a and stably overexpressed the expression of miR-548b in gefitinib-resistant NSCLC HCC827-Gef cells. Stably transfected HCC827-Gef-miRZip-374a and HCC827-Gef-miRZip-control cells, or HCC827-Gef-p-miR-548b and HCC827-Gef-p-miR-control cells were implanted into the right and left flanks (2 × 106 cells per flank) of BALB/c athymic mice by subcutaneous injection. Tumor volumes were determined every week by measuring the length (a) and the width (b). The tumor volume (V) was calculated according to the formula V=ab2/2. At 6 weeks post-injection, the mean volumes of tumors generated from HCC827-Gef-miRZip-374a and HCC827-Gef-p-miR-548b cells were significantly smaller than those originating from the control cells (Figures 8a–d). The tissue sections from each group were collected and assayed by terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) analysis to detect apoptotic cells. The results showed that the apoptotic percentage was significantly increased in tumor tissues derived from HCC827-Gef-miRZip-374a and HCC827-Gef-p-miR-548b tumors compared with those from the control cells (Figures 8e–g). These results indicate that miR-374a and miR-548b regulate tumorigenesis of gefitinib-resistant lung cancer cells in vivo by modulating apoptosis.
Figure 8.
MiR-374a and miR-548b regulate tumorigenesis of gefitinib resistance lung cancer cells in vivo by modulating apoptosis. (a and c) Representative image for tumor growth is shown. Nude mice were subcutaneously injected with 2 × 106 HCC827-Gef-miRZip-374a and HCC827-Gef-miRZip-control cells, or HCC827-Gef-p-miR-548b and HCC827-Gef-p-miR-control cells. (b and d) Determination of the tumor growth. Tumor volume was calculated every week after injection. Data are mean+S.D.s of three independent experiments. *P<005. (e and f) The representative images of TUNEL staining. The apoptotic percentage was significantly increased in tumor tissues derived from HCC827-Gef-miRZip-374a and HCC827-Gef-p-miR-548b tumors compared with those from the control cells. (g) The quantification of TUNEL-positive cells from different fields in the tissue section have been showed in the graph. *P<0.05
Discussion
In spite of recent improvements in NSCLC management, such as the use of targeted therapy including gefitinib and erlotinib, which have led to improved progression-free survival and quality of life in NSCLC patients with an activating EGFR mutation, progression or metastasis because of resistance to targeted therapy remains a major clinical problem.23 Apart from T790M and c-MET amplification, a recent study has reported that Axl kinase is upregulated in humans with acquired resistance to EGFR-TKI.7 Axl may drive cancer cell growth through activation of multiple pathways, such as MAPK, AKT and NF-κB, and Axl-mediated EGFR-TKI-acquired resistance may occur during EMT; however, Axl-mediated molecular alterations in TKI-resistant NSCLC are largely unknown. Therefore, a more complete understanding of how the downstream molecular mediators of Axl function will provide critical information to help design strategies to augment EGFR-TKI efficacy.
In our work, we mainly focus on Axl-altered miRNAs because one miRNA can target hundreds of mRNAs, thus controlling a wide range of biological functions, such as cellular proliferation, differentiation and apoptosis.24 We found that, in gefitinib-resistant cell lines, when we knocked down Axl by esiRNA several miRNAs were dysregulated. The significantly downregulated miRNA was miR-374a and the upregulated miRNA was miR-548b. Recent studies have demonstrated that miR-30b-c and miR-221-222, regulated by both EGFR and MET, had an important role in NSCLC tumorigenesis and gefitinib resistance.16 Together with our data, Axl may be correlated with NSCLC tumorigenesis and gefitinib resistance through regulating miR-374a and miR-548b. In addition, NSCLC patients with low Axl have longer DFS, indicating that it may serve as a prognostic biomarker for NSCLC.
MiR-374a, located on chromosome Xq13.2, has been previously shown to constitutively activate Wnt/β-catenin signaling in breast cancer cells.21, 22 Thus, by suppressing WIF1, PTEN or WNT5A, miR-374a may serve as an anti-metastasis therapeutic target in breast cancer. Consistent with this evidence, we found that miR-374a targeted Wnt5a in NSCLC. Moreover, miR-374a can regulate migration and invasion in gefitinib-resistant NSCLC cell lines and promote EMT, colony formation and drug resistance in gefitinib-sensitive NSCLC cells. In addition, downregulation of miR-374a resulted in tumor shrinkage of gefitinib-resistant NSCLC cells in xenograft mice. These results suggest that miR-374a may be a therapeutic target to reverse gefitinib resistance. Interestingly, we also found that miR-374a is consistently expressed with Axl and its low expression is associated with longer DFS, suggesting that it can also be a biomarker for NSCLC prognosis.
Another significantly altered miRNA found in Axl-mediated gefitinib-resistant NSCLC was miR-548b. This miRNA has been reported to be dysregulated in malignant astrocytomas.25 Its functional role in NSCLC is largely unclear. In our work, we first noted that miR-548b was significantly upregulated when Axl was knocked down in gefitinib-resistant NSCLC. Next, we also found that miR-548b can target CCNB1 in NSCLC cells, thus controlling cell cycle progression. By regulating CCNB1, it may regulate resistance in gefitinib-resistant NSCLC cells. More importantly, stable overexpression of miR-548b inhibits tumorigenesis of gefitinib-resistant lung cancer cells in vivo. When we analyzed the expression of miR-548b and NSCLC patients' survival, we found that high miR-548b expression is associated with longer DFS, demonstrating that it could be a prognostic biomarker in NSCLC.
In conclusion, we observed Axl-altered miRNAs in gefitinib-resistant cells lines. We also found that two mostly dysregulated miRNAs, miR-374a and miR-584b, can regulate tumorigenesis and gefitinib resistance in NSCLC through Wnt5a and CCNB1, respectively, in vitro and in vivo. These two miRNAs may serve not only as therapeutic targets but also as prognostic biomarkers in NSCLC.
Materials and Methods
Cell lines and cell culture
Human NSCLC cell line HCC827 and Calu1 was bought from American Type Culture Collection and cultured in RPMI-1640 medium (Invitrogen, CA, USA) with 10% FBS and 100 U/ml penicillin/streptomycin (Sigma, St. Louis, MO, USA). HCC827-Gef-resistant cells were established as reported before.17
RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen). The concentration of isolated total RNA was measured by NanoDrop ND-1000 Spectrophotometer (Agilent, CA, USA). The RT-qPCR was performed using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) with hypoxanthine phosphoribosyltransferase 1 (HPRT1) as an internal control, as described previously26 and in Supplementary Materials and Methods.The primers are shown in Supplementary Table S1.
MTS assays
The cells were seeded into 96-well plates at a density of 5 × 103 per well (100 μl). For the MTS assay, the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI, USA) was used following the manufacturer's instruction. Briefly, at 2 h before each of the desired time points (24, 48 and 72 h), 20 μl of the MTS reagent was added into each well and cells were incubated at 37 °C for around 2 h. The absorbance was detected at 490 nm using a Wallac Victor 1420 Multilabel plate reader. All the experiments were repeated three times.
Luciferase reporter assay
For the reporter assay, HEK293 cells were plated onto 24-well plates and transfected with the constructs (pGL3-wnt5a-wt, CCNB1-wt or pGL3-wnt5a-mut, CCNB1-mut) and P-miR-374a, P-miR-548b or P-miR-control vectors using the FuGENE HD in vitro tranfection (Promega). A Renilla luciferase vector pRL-SV50 (Promega) was co-transfected to normalize the difference in the transfection efficiency. After 48 h, the cells were harvested and assayed using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Results were obtained from three independent experiments performed in duplicate.
Flow cytometry
The treated cells were fixed in ice-cold 70% ethanol and stained with the use of Coulter DNA-Prep Reagents kit (Beckman Coulter, Fullerton, CA, USA). Cellular DNA content of 5 × 105 cells from each sample was determined by PI staining using the BD FACSCanto II flow cytometer (BD Bioscience, Franklin Lakes, MA, USA). Cell cycle was analyzed with the use of FlowJo software using data obtained from two separate experiments.
Immunofluorescence analysis
The stable cells were seeded in the BD Falcon 8-well CultureSlide and incubated with primary antibodies, rabbit E-Cadherin (24E10) and Vimentin (Cell Signaling, MA, USA); and then incubated with Alexa Fluor 594 Goat Anti-Rabbit IgG (Invitrogen). The culture slides were counterstained with Hoechst 33342 and imaged with a confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany). Data were processed with Adobe Photoshop 7.0 software for analysis.
Xenograft models
The nude mice were 5 weeks old and 20 g in weight. They were bred in aseptic conditions and kept at a constant humidity and temperature (25–28 °C) according to standard guidelines under a protocol approved by Medical School of Nanjing University. All mice were injected subcutaneously with a 100 μl suspension (2 × 106) of stable transfected HCC827-Gef-miRZip-374a and HCC827-Gef-miRZip-control or HCC827-Gef-p-miR-548a and HCC827-Gef-p-miR-control cells. The size of the tumor was measured once a week with calipers, and the volume of tumor was determined using the simplified formula of a rotational ellipsoid (length × width2 × 0.5).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China; Grant Number: 81301882, 81172240.
Glossary
- NSCLC
non-small cell lung cancer
- TKI
tyrosine kinase inhibitor
- miRNA
microRNA
- EMT
epithelial-to-mesenchymal transition
- GAS-6
growth arrest specific gene-6
- DFS
disease-free survival
- TUNEL assay
terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling assay
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Janne PA, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C, et al. Axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. doi: 10.1128/mcb.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci USA. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafizi S, Dahlback B. Gas6 and protein S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 2006;273:5231–5244. doi: 10.1111/j.1742-4658.2006.05529.x. [DOI] [PubMed] [Google Scholar]
- Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC. Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol. 2005;204:36–44. doi: 10.1002/jcp.20265. [DOI] [PubMed] [Google Scholar]
- Song X, Wang H, Logsdon CD, Rashid A, Fleming JB, Abbruzzese JL, et al. Overexpression of receptor tyrosine kinase Axl promotes tumor cell invasion and survival in pancreatic ductal adenocarcinoma. Cancer. 2011;117:734–743. doi: 10.1002/cncr.25483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerdrum C, Tiron C, Hoiby T, Stefansson I, Haugen H, Sandal T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci USA. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Y, Wang Y, Xia H, Zhou S, SchmidBindert G, Zhou C. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev. 2012;13:255–260. doi: 10.7314/apjcp.2012.13.1.255. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Sasaki R, Komaki R, Macapinlac H, Erasmus J, Allen P, Forster K, et al. [18F] Fluorodeoxyglucose uptake by positron emission tomography predicts outcome of on-small-cell lung cancer. J Clin Oncol. 2005;23:1136–1143. doi: 10.1200/JCO.2005.06.129. [DOI] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34 (Suppl 2:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosa U, Vooder T, Kolde R, Fischer K, Valk K, Tonisson N, et al. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132:116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- Xia H, LLPJ Ooi, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.