Abstract
Oxidative stress serves as an important regulator of both apoptosis and metabolic reprogramming in tumor cells. Chaetocin, a histone methyltransferase inhibitor, is known to induce ROS generation. As elevating basal ROS level sensitizes glioma cells to apoptosis, the ability of Chaetocin in regulating apoptotic and metabolic adaptive responses in glioma was investigated. Chaetocin induced glioma cell apoptosis in a ROS-dependent manner. Increased intracellular ROS induced (i) Yes-associated protein 1 (YAP1) expression independent of the canonical Hippo pathway as well as (ii) ATM and JNK activation. Increased interaction of YAP1 with p73 and p300 induced apoptosis in an ATM-dependent manner. Chaetocin induced JNK modulated several metabolic parameters like glucose uptake, lactate production, ATP generation, and activity of glycolytic enzymes hexokinase and pyruvate kinase. However, JNK had no effect on ATM or YAP1 expression. Coherent with the in vitro findings, Chaetocin reduced tumor burden in heterotypic xenograft glioma mouse model. Chaetocin-treated tumors exhibited heightened ROS, pATM, YAP1 and pJNK levels. Our study highlights the coordinated control of glioma cell proliferation and metabolism by ROS through (i) ATM-YAP1-driven apoptotic pathway and (ii) JNK-regulated metabolic adaptation. The elucidation of these newfound connections and the roles played by ROS to simultaneously shift metabolic program and induce apoptosis could provide insights toward the development of new anti-glioma strategies.
Keywords: Glioblastoma, ROS, ATM, YAP, JNK, metabolism
Chemotherapeutic agents can induce apoptosis in cancer cells by elevating oxidative stress.1 We have previously reported induction of apoptosis in glioma cells through elevation in ROS generation and perturbation of redox homeostasis.2, 3 Oxidative stress also regulates histone methylation to induce epigenetic dysregulation.4 Trimethylation of histone 3 lysine 9 (H3K9me3) is a marker of repressed transcription, and H3K9me3 positivity is found in glioblastomas.5 Inhibition of SUV39H1, an H3K9me3-specific methyltransferase, by Chaetocin prevents leukemia cell growth in a ROS-dependent manner.6 As ROS-inducing agents effectively kill cancer cells through elevation of intracellular ROS,1 we investigated whether Chaetocin could also affect survival and growth of glioma cells through oxidative stress induction.
The Yes-associated protein 1 (YAP1), which is the downstream target of the hippo pathway, is overexpressed in several solid tumors including glioma.7 DNA damage induces YAP1 phosphorylation,8 and ROS-mediated JNK activation-induced DNA damage causes mitochondrial dysfunction-related apoptosis.9 ATM which is well known for its role in the cellular response to DNA breaks also regulates metabolic diseases through JNK.10 Besides, JNK regulates YAP1 function in apoptosis.11 We have reported that ATM-induced metabolic modeler TIGAR affects oxidative stress and survival responses in glioma cells,12 and that inhibition of JNK rescues glioma cells from Oncrasin-induced apoptosis.13 Besides, IFNβ and temozolomide combination enhances p73/YAP-mediated apoptosis in glioblastoma.14 As ROS regulates several effectors associated with survival responses, we investigated whether Chaetocin-mediated oxidative stress links JNK, ATM and YAP1 to affect glioma cell viability.
The altered cellular energy metabolism in tumor cells characterized by aerobic glycolysis or the ‘Warburg effect' is regarded as a hallmark of cancer.15 Hexokinase 2 (HK2), an isoform of the enzyme HK that catalyzes the first step of the glycolytic pathway, is elevated in GBM and loss of HK2 redirects GBM to normal oxidative glucose metabolism.16 Importantly, HK2 limits the elevation of ROS levels.17 Besides HK2, the glycolytic enzyme pyruvate kinase (PK), which catalyzes conversion of phosphoenolpyruvate to pyruvate and serves as a key regulatory node in glycolysis, is altered in GBM.18 PK-mediated feedback activation of the pentose phosphate pathway prevents ROS accumulation.19 PK activity is low in gliomas, and activators of PKM2 (PKM2 – the catalytically inactive isoform of PK) regulate sensitivity of cells to oxidative stress induced death.20
As oxidative stress regulates both apoptosis and metabolic reprogramming in tumor cells and as these events are intertwined, the use of compounds that subserve the dual purpose of enhancing tumor killing through increased cellular ROS generation while concomitantly affecting the activity of key metabolic enzymes would be greatly advantageous. As targeting aberrant metabolic program is regarded as a potential antiglioma strategy,21 the role of Chaetocin in regulating metabolic and survival adaptive responses in glioma cells through altered redox homeostasis was investigated.
Results
Chaetocin inhibits glioma cell proliferation
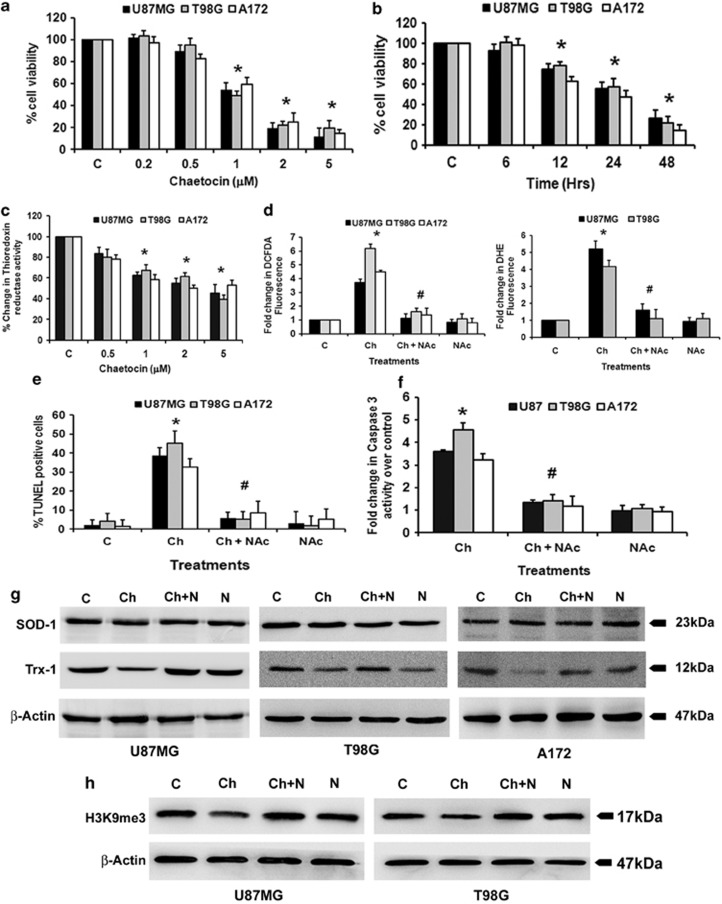
Chaetocin, an inhibitor of lysine-specific histone methyltransferase SUV39H1, induces apoptosis in leukemia cell lines in vitro and inhibits leukemia growth in vivo.6 As gliomas are refractory to current treatment strategies, we investigated whether Chaetocin could affect glioma cell proliferation. Treatment with Chaetocin induced glioma cell death in a dose- (Figure 1a) and time- (Figure 1b) dependent manner. An ∼40–50% decrease in cell viability was observed in A172, T98G and U87MG glioma cells upon treatment with a 1 μM concentration of Chaetocin for 24 h (Figure 1a). Treatment with 1 μM Chaetocin for 24 h was used to dissect its mechanisms of action in subsequent experiments.
Figure 1.
Chaetocin induces glioma cell apoptosis in a reactive oxygen species (ROS)-dependent manner. (a) Chaetocin reduces viability of glioma cells in a dose-dependent manner. Viability of glioma cells treated with different concentrations of Chaetocin was determined by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfopheny)-2H-tetrazolium) assay. The graph represents viable glioma cells upon treatment with 0.2–5 μM of Chaetocin for 24 h, expressed as the percentage of control. (b) Chaetocin reduces glioma cell viability in a time-dependent manner. Glioma cells treated with 1 μM of Chaetocin for various intervals of time were assessed for viability using MTS assay. The graph represents the viable cells, percentage of control, observed when glioma cells were treated with Chaetocin. (c) Chaetocin reduces thioredoxin reductase activity in a dose-dependent manner. Thioredoxin reductase enzyme activity was assessed in glioma cells treated with increasing concentrations of Chaetocin (0.5–5 μM) for 12 h. Graph represents the enzyme activity as percent change over control. (d) ROS generation in glioma cells treated with Chaetocin in the presence or absence of N-acetyl-cysteine (NAc) as determined by DCFDA and DHE fluorescence assay. The change in fluorescence intensity is plotted as fold change over control. The increase in DCFDA and DHE fluorescence induced upon Chaetocin treatment is significantly abrogated in the presence of NAc. (e) Chaetocin-induced increase in terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive cells is abrogated in the presence of NAc. Graph depicts the percent of TUNEL-positive cells treated with Chaetocin, NAc or both. (f) NAc abrogates the Chaetocin-induced increase in caspase-3 activity. Graph shows caspase-3 enzyme activity in glioma cells treated with Chaetocin in the presence or absence of NAc, expressed as fold change over control. (g) Western blot images showing the effect of Chaetocin on superoxide dismutase 1 (SOD-1) and thioredoxin-1 (Trx-1) expression and (h) H3K9me3 levels in glioma cells in the presence and absence of NAc. Blots were reprobed for β-actin to establish equivalent loading. Blots are representative images of three independent experiments showing similar results. Values in (a–f) represent the means±S.E.M. of three independent experiments. *Significant change from control. #Significant change from Chaetocin-treated group (P<0.05)
Chaetocin inhibits thioredoxin reductase activity
As Chaetocin is a competitive substrate and inhibitor of thioredoxin reductase,22 thioredoxin reductase activity was determined in Chaetocin-treated cells. Chaetocin treatment reduced thioredoxin reductase activity in a dose-dependent manner with ∼40% decrease in activity observed at 1 μM concentration (Figure 1c).
Chaetocin-induced glioma cell death is ROS mediated
Chaetocin-mediated increased ROS production induces leukemia cell death.6 As we have reported that elevated ROS level induces glioma cell apoptosis,2 we determined ROS levels in Chaetocin-treated cells with reduced thioredoxin reductase activity. A significant 4–6-fold increase in ROS production was observed in A172, T98G and U87MG glioma cells upon Chaetocin treatment indicated by increased DCFDA and DHE fluorescence intensity (Figure 1d). The increased DCFDA and DHE fluorescence intensity observed in Chaetocin-treated cells was abrogated to control levels in the presence of ROS inhibitor, NAc (Figure 1d).
As increased ROS production is critical in inducing glioma cell apoptosis,2 we next determined whether Chaetocin-induced death is ROS dependent. The ∼30–40% increase in TUNEL-positive cells observed upon treatment with 1 μM Chaetocin was significantly reduced when cells were co-treated with Chaetocin and NAc (Figure 1e). This ability of ROS inhibitor to abolish Chaetocin-induced cytotoxicity suggested that Chaetocin-induced cell death is ROS mediated.
Chaetocin-induced glioma cell apoptosis is caspase mediated
Treatment with Chaetocin resulted in ∼3–5-fold increase in caspase-3 activity (Figure 1f). This ability of Chaetocin to induce caspase activation was abrogated in the presence of ROS inhibitor NAc (Figure 1f). The ability of pan-caspase inhibitor and caspase-3 inhibitor to rescue glioma cells from Chaetocin induced apoptosis (Supplementary Figure 1b), confirmed the role of caspase-3 in Chaetocin-induced cell death.
Chaetocin decreases the expression of TRX-1 in glioma cells
We have previously reported that siRNA mediated knockdown of SOD-1 and TRX-1, which are crucial in maintaining cellular redox homeostasis, increases sensitivity of glioma cells to Kaempferol-induced apoptosis through elevation of ROS levels in glioma cells.2 We therefore determined the expression of SOD-1 and TRX-1 in Chaetocin-treated cells with heightened ROS levels. While Chaetocin treatment had no effect on SOD-1 expression, a decrease in TRX-1 level was observed in an NAc-dependent manner (Figure 1g). Thus, Chaetocin increases ROS accumulation via disabling the antioxidant defense mechanism of glioma cells.
Chaetocin-mediated inhibition of histone H3 methylation is ROS dependent
In addition to being directly regulated by Chaetocin, SUV39H1 is also modulated by chaetocin-induced ROS.6 As Chaetocin elevated ROS generation in glioma cells, its effect on H3K9me3 levels in these cells was determined. The decrease in H3K9me3 levels observed upon Chaetocin treatment was reverted in the presence of ROS inhibitor NAc (Figure 1h). This suggested that Chaetocin inhibits H3K9me3 in a ROS-dependent manner.
Chaetocin induced ROS regulates YAP1 expression
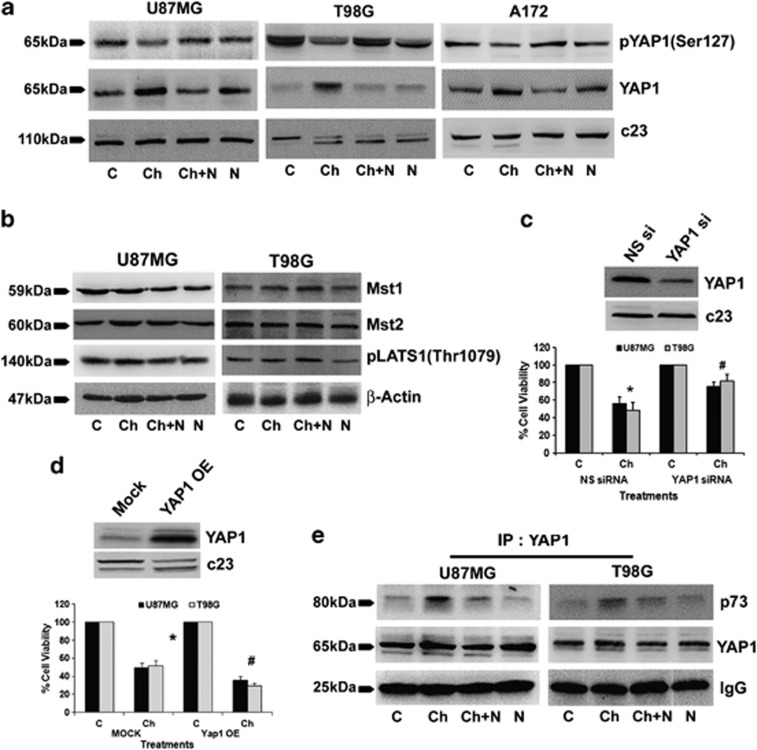
As YAP1 overexpression rescues proliferative response in cells exposed to oxidative damage,23 we investigated whether elevated ROS levels affected YAP1 expression in Chaetocin-treated cells. An increase in YAP1 level and decrease in YAP1 phosphorylation was observed upon Chaetocin treatment (Figure 2a). This ability of Chaetocin to induce YAP1 was abrogated in the presence of ROS inhibitor NAc. Treatment with NAc alone had no significant effect on YAP1 expression (Figure 2a).
Figure 2.
Chaetocin-induced reactive oxygen species (ROS)-dependent increase in YAP1 regulates glioma cell death (a) N-acetyl-cysteine (NAc) reverses the effects of Chaetocin on phospho- and total YAP1 levels. Western blots showing phosphorylated YAP1 and total YAP1 levels in nuclear extracts of glioma cells treated with Chaetocin in the presence and absence of NAc. Blots were reprobed for c23 to establish equivalent loading. (b) Chaetocin-induced YAP1 is independent of classical hippo pathway. Western blots indicating Mst1, Mst2 and LATS1 phosphorylation (Thr1079) levels in glioma cells treated with Chaetocin, NAc or both. β-Actin was used as loading control. (c) Small interfering RNA (siRNA)-mediated knockdown of YAP1 rescues glioma cells from Chaetocin-induced cell death. Graph represents the viability of glioma cells, transfected with either YAP1 siRNA or scrambled (NS) siRNA, and treated with Chaetocin. Values are expressed as the percentage of control. Inset shows the knockdown efficiency of YAP1 siRNA. (d) YAP1-overexpressing cells show increased responsiveness towards Chaetocin treatment. The graph represents the viable glioma cells, percentage of control, transfected with either YAP1 overexpression construct (YAP1 OE) or empty vector (mock) and treated with Chaetocin, as determined by MTS assay. Inset confirming the overexpression of YAP1 in glioma cells. Values (c and d) are means±S.E.M. of three independent experiments. *Significant change from non-silencing (NS) siRNA or mock control group. #Significant change from NS siRNA or mock transfected cells treated with Chaetocin (P<0.05). (e) NAc abrogates Chaetocin-induced increase in p73–YAP1 interaction. Nuclear extracts (200 μg) from glioma cells treated with Chaetocin or NAc or both were subjected to immunoprecipitation with YAP1. The representative blot of the pulled down fractions shows increased p73–YAP1 interaction upon Chaetocin treatment. Similar amounts of YAP1 were precipitated under different conditions. Efficiency of precipitation was established by IgG levels in each conditions. Blots shown in (a, b and e) are representative images of three independent experiments showing similar results
Chaetocin-mediated YAP1 induction is independent of Hippo signaling
As activation of YAP1, the main downstream effecter of the Hippo signaling pathway, results from the inactivation of LATS (large tumor suppressor) and Mst1/2 genes of the Hippo pathway,24 we evaluated the status of these molecules in Chaetocin-treated samples. No change in the expression of Mst1 and Mst2 and the phosphorylation of LATS1 was observed upon Chaetocin treatment (Figure 2b). This suggested that Chaetocin induced YAP1 expression involves mechanisms independent of the Hippo pathway.
YAP1 has a functional role in Chaetocin-induced cell death
To investigate the role of YAP1 in Chaetocin-induced glioma cell death, the viability of cells transfected with either scrambled siRNA or YAP1-specific siRNA, was determined upon Chaetocin treatment. siRNA-mediated YAP1 knockdown was able to rescue Chaetocin-induced glioma cell death to a significant extent (Figure 2c), confirming the involvement of YAP1 in Chaetocin-induced apoptosis. As YAP overexpression increases p73-mediated apoptosis,25 we further confirmed the role of Chaetocin-induced YAP1 in regulating the viability of glioma cells by overexpressing YAP1. Chaetocin-induced death was significantly greater in cells transfected with YAP1 overexpression construct as compared with mock-transfected Chaetocin-treated cells (Figure 2d).
Chaetocin-mediated increased interaction of YAP1 with p73 involves ROS
YAP phosphorylation leads to its interaction with 14-3-3, which promotes its loss from the nucleus where it functions as a coactivator of p73.25 Importantly, p73–YAP interaction leads to apoptosis in glioma treated with a combination of IFNβ and temozolomide.14 We therefore performed co-immunoprecipitation to determine whether elevated YAP1 levels in Chaetocin-treated cells is accompanied by its altered interaction with p73. An increased interaction between p73 and YAP1 was observed in Chaetocin-treated U87MG and T98G cells (Figure 2e). This increased association between YAP1 and p73 was abrogated in the presence of NAc, suggesting the involvement of ROS in the interaction.
Chaetocin induces ATM phosphorylation in a ROS-dependent manner
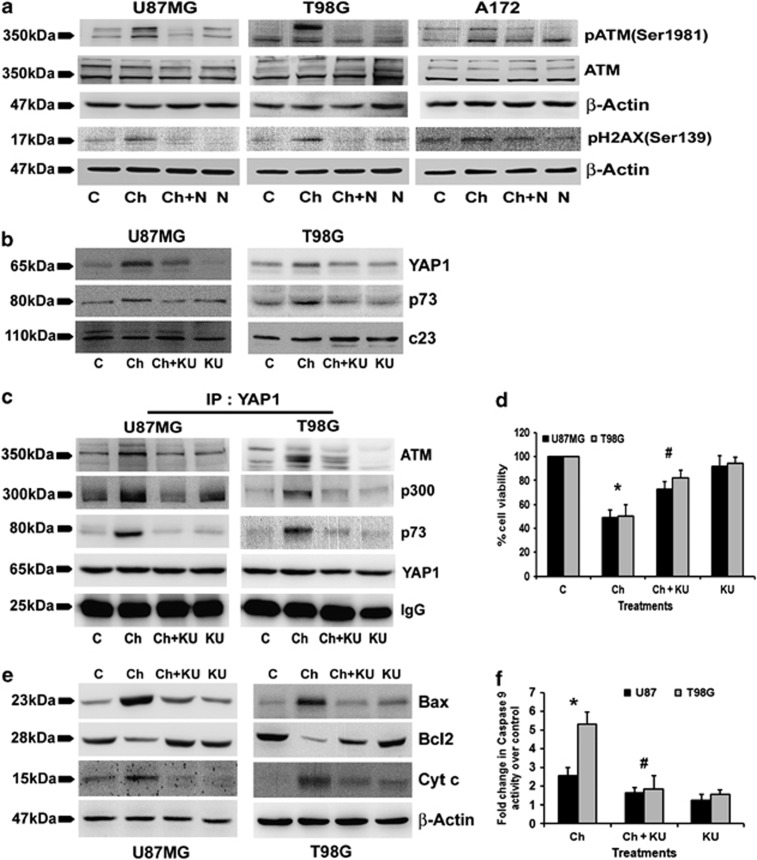
Emodin-induced ROS leads to ATM-dependent apoptosis in lung cancer cells.26 We have shown that ATM-induced metabolic modeler TIGAR affects oxidative stress and prosurvival responses in glioma cells.12 Besides, ATM phosphorylates H2AX27 and ROS induction is partly mediated by increasing γH2AX.28 We therefore determined pATM and γH2AX levels in Chaetocin-treated cells with elevated ROS. Chaetocin induced an increase in pATM and γH2AX expression in a ROS-dependent manner (Figure 3a).
Figure 3.
ATM regulates YAP1- and Chaetocin-induced glioma cells apoptosis. (a) N-acetyl-cysteine (NAc) abrogates Chaetocin-induced increase in phospho-ATM and phospho-H2AX levels. Western blots demonstrating increased phosphorylation of ATM (Ser 1981) and H2AX (Ser139) in Chaetocin-treated glioma cells. Blots were reprobed for β-actin to establish equivalent loading. (b) ATM inhibitor (KU) abrogates Chaetocin-induced increased expression of YAP1 and p73. c23 was used to establish equal loading. (c) Chaetocin-mediated increased interaction of YAP1 with ATM, p300 and p73 was inhibited by KU. YAP1 was immunoprecipitated from the nuclear extracts (200 μg) of glioma cells treated with Chaetocin, KU or both. Efficiency of precipitation across the treatment conditions was established by immunoglobulin G (IgG). (d) KU abrogated Chaetocin-induced glioma cell death. Graph represents the viable population of glioma cells treated with Chaetocin or KU or both, expressed as the percentage of control. (e) ATM inhibition reverses Chaetocin-induced effects on proteins associated with apoptosis induction. Western blot demonstrating expression of proapoptotic proteins Bax, cytochrome c and antiapoptotic protein Bcl2 in glioma cells, treated with Chaetocin in the presence and absence of ATM inhibitor. Blots were reprobed for β-actin to establish equivalent loading. Blots shown in (a–c) and (e) are representative of three independent experiments showing similar results. (f) KU inhibits Chaetocin-induced increased caspase-9 activity in glioma cells. Graph represents the change in caspase-9 activity expressed as fold change over control in glioma cells treated with Chaetocin, KU or both. Graph (d and f) represent values means±S.E.M. pooled from experiments repeated at least three times. *Significant change from control and #significant change from Chaetocin-treated cells (P<0.05)
ATM regulates YAP1/p73 interaction and cell death in Chaetocin-treated cells
YAP1 induces apoptosis in response to DNA damage.8 As ATM that has pivotal roles in DNA damage-induced responses is necessary for YAP accumulation,29 we investigated the role of ATM in Chaetocin-mediated YAP1 induction. Chaetocin-induced ATM was involved in YAP1 regulation, as ATM inhibitor KU60019 decreased YAP1 and p73 levels in Chaetocin-treated glioma cells (Figure 3b). YAP1 regulates DNA damage-induced accumulation of p73 and potentiates the p300-mediated acetylation of p73.30 As ATM regulates both YAP1 and p73 levels, we determined whether it also affects their mutual interaction as well as their association with p300. An increased interaction of YAP1 with ATM, p73 and p300 was observed upon Chaetocin treatment (Figure 3c). Moreover, this increased interaction between YAP1 and its partners was abrogated in the presence of ATM inhibitor (Figure 3c). Further, the ability of ATM inhibitor KU60019 to significantly protect glioma cells from Chaetocin-induced cell death (Figure 3d) suggested the functional importance of ATM in Chaetocin-induced apoptosis.
Chaetocin-induced ATM regulates caspase-9 activation and mitochondrial ROS generation
ATM induces Bax-dependent cytotoxicity in response to ROS.26 Also, YAP1 induces Bax/Bcl2-dependent apoptosis in response to irradiation.31 We determined whether Chaetocin-induced ATM affects mitochondrial apoptotic regulators. Chaetocin elevated Bax and cytochrome c levels, and reduced Bcl2 expression in an ATM-dependent manner (Figure 3e). As caspase-9 is the initiator caspase associated with mitochondrial pathway of apoptosis, its activity in Chaetocin-treated cells was determined. The ability of ATM inhibitor to abrogate increased caspase-9 activity in Chaetocin-treated cells confirmed the involvement of ATM in Chaetocin-mediated apoptosis (Figure 3f). Chaetocin also elevated mitochondrial ROS generation in an ATM-dependent manner (Supplementary Figure 2).
YAP1 overexpression has no effect on ROS production or ATM activation
As YAP1 is involved in Chaetocin-induced glioma cell death, we investigated its role in the regulation of ROS and ATM – the two crucial regulators of Chaetocin-induced apoptosis. However, no change in either ROS production (Supplementary Figure 3a) or ATM activation (Supplementary Figure 3b) was observed in YAP1-overexpressing glioma cells upon Chaetocin treatment. This indicated that although YAP1 is induced downstream of ROS induced ATM in Chaetocin-treated cells; it has no role in their regulation.
ROS induced JNK activation regulates Chaetocin-mediated apoptosis
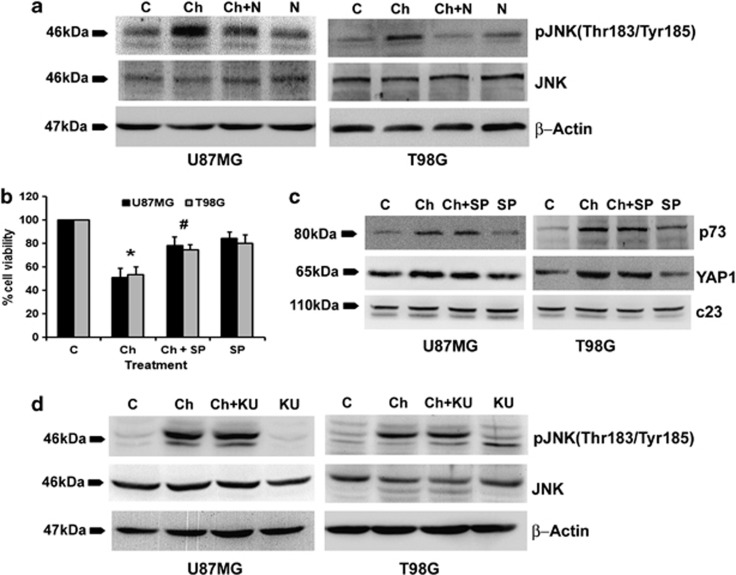
ATM regulates metabolic and cardiovascular diseases by inhibiting JNK.10 As JNK phosphorylates YAP1 to regulate its function in apoptosis,11 and as pATM levels are elevated in Chaetocin-treated cells, we determined the status of JNK activation in these cells. An increase in JNK phosphorylation was observed in Chaetocin-treated cells in a ROS-dependent manner (Figure 4a). As we have previously shown the involvement of JNK in glioma cell apoptosis,13 we questioned the role of JNK activation in Chaetocin-mediated glioma cell death. JNK inhibitor SP 600125 significantly reversed Chaetocin-induced cell death (Figure 4b), suggesting the involvement of JNK in Chaetocin-mediated apoptosis. As Chaetocin-induced cell death was both YAP1 and JNK dependent, and as ROS-mediated JNK activation induced apoptosis, we determined whether ROS regulates YAP1 through JNK. Inhibition of JNK phosphorylation had no significant effect on Chaetocin-induced YAP1 and p73 expression (Figure 4c). This indicated that Chaetocin-induced JNK affects glioma cell viability by other mechanisms independent of YAP1 induction. As Chaetocin induced both ATM and JNK activation and ATM is known to regulate JNK signaling,32 we questioned whether Chaetocin-mediated JNK activation is ATM dependent. ATM inhibitor KU60019 failed to revert Chaetocin-induced JNK phosphorylation (Figure 4d). This indicated that Chaetocin-mediated activation of JNK occurs independently of ATM activation.
Figure 4.
Chaetocin induces c-Jun N-terminal kinase (JNK) phosphorylation in a reactive oxygen species (ROS)-dependent manner independent of YAP1 and ATM. (a) N-acetyl-cysteine (NAc) abrogates Chaetocin-induced increase in JNK phosphorylation. Western blot demonstrating pJNK levels in glioma cells treated with Chaetocin in the presence and absence of NAc. (b) Chaetocin-induced glioma cell death is significantly abrogated by JNK inhibitor, SP. The graph represents the viable cells, percentage of control, observed when glioma cells were treated with Chaetocin, SP or both. Values represent means±S.E.M. from three independent experiments. *Significant change from control and #significant change from the Chaetocin-treated group (P<0.05). (c) Western blot demonstrating that Chaetocin-induced increase in p73 and YAP1 levels are independent of JNK activation. (d) Chaetocin-induced JNK phosphorylation is independent of ATM. Western blot analysis indicating comparable pJNK levels in glioma cells treated with Chaetocin both in the presence or absence of KU. Western blots shown in (a, c and d) are representative image of three independent experiments showing similar results. Blots were reprobed for β-actin or C23 to establish equivalent loading
Chaetocin affects glucose metabolism in glioma cells
HK2, which regulates aerobic glycolysis, is involved in glioma progression,16 and HK2 knockdown increases ROS accumulation.17 Interestingly, ATM activates the pentose phosphate pathway to promote antioxidative defense.33 As ROS levels were elevated in Chaetocin-treated cells, the status of genes associated with glucose metabolism was analysed in Chaetocin-treated cells using qRT-PCR-based metabolism gene array (Table 1). In U87MG cells, among the 84 genes tested, the transcript levels of 30 genes were differentially modulated by Chaetocin treatment. The mRNA levels of 16 genes (ALDOB, ENO3, FBP1, FBP2 G6PC, GCK GSK3B, GYS2, HK3, PCK1, PGK2, PGM3, PHKG1, PKLR, PRPS1L1 and PYGM) were elevated by more than 2 fold upon Chaetocin treatment. At the same time, transcript levels of 14 genes (AGL, ACLY, ALDOA, ALDOC, G6PD, HK2, IDH1, IDH2, PDHB, PDK2, PGM1, PRPS2, RPE and TKT) were downregulated upon Chaetocin treatment. Among these genes, we chose HK2, downregulated by 4.5 fold (Table 1) and PKLR, up-regulated by 20.9 fold (Table 1), for further validation as HK and PK constitute two important regulatory nodes in glycolysis.
Table 1. Quantitative real-time PCR analysis demonstrating the relative transcript levels of a panel of genes associated with glucose metabolism.
| Gene | Chaetocin |
|---|---|
| Fold upregulation | |
| ALDOB | 14.7026 |
| ENO3 | 16.8304 |
| FBP1 | 49.2824 |
| FBP2 | 243.5375 |
| G6PC | 527.49 |
| GCK | 4.9348 |
| GSK3B | 2.6262 |
| GYS2 | 43.3512 |
| HK3 | 10.6516 |
| PCK1 | 169.2486 |
| PGK2 | 6.489 |
| PGM3 | 2.0321 |
| PHKG1 | 4.7338 |
| PKLR | 20.9372 |
| PRPS1L1 | 22.8322 |
| PYGM | 107.1138 |
| Fold downregulation | |
| AGL | −1.9889 |
| ACLY | −2.6427 |
| ALDOA | −156.173 |
| ALDOC | −3.0674 |
| G6PD | −1.9889 |
| HK2 | −4.5065 |
| IDH1 | −2.1916 |
| IDH2 | −2.2222 |
| PDHB | −3.4153 |
| PDK2 | −3.2423 |
| PGM1 | −1.9279 |
| PRPS2 | −2.0097 |
| RPE | −2.1615 |
| TKT | −2.5438 |
Gene expression profiles of mRNA isolated from untreated and Chaetocin-treated U87MG glioma cells were analyzed by quantitative PCR for genes involved in glucose metabolism. Fold change in expression level of genes affected by Chaetocin treatment is shown. The table represents the average data from two independent experiments
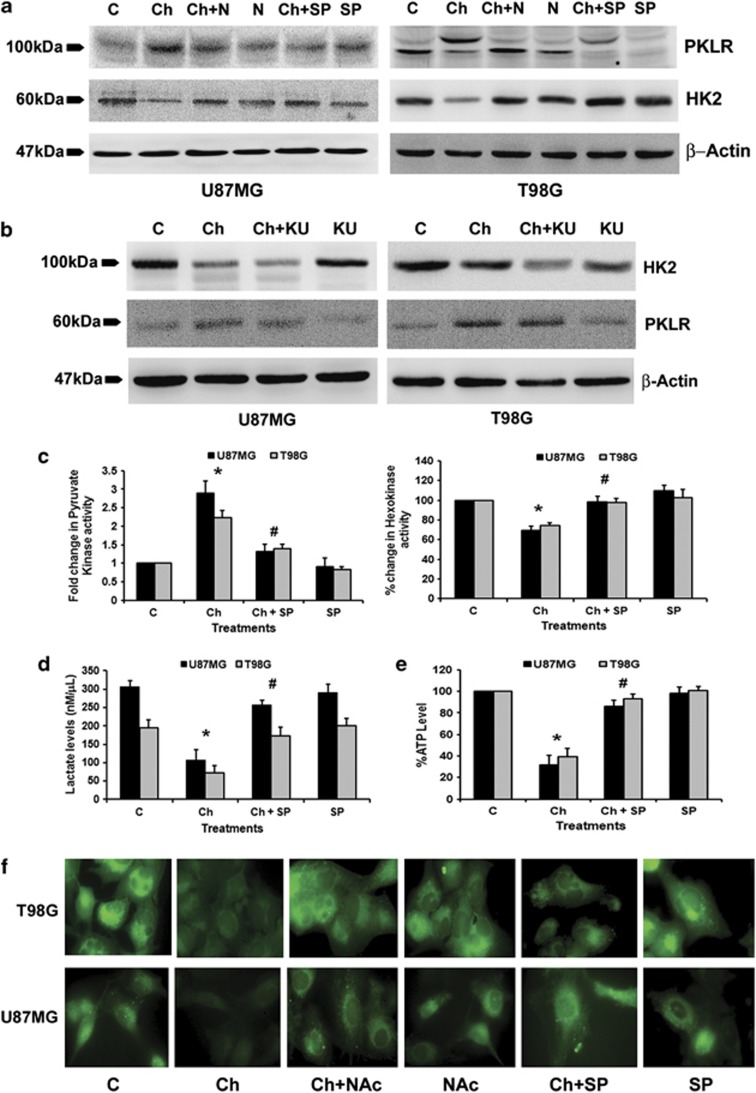
Chaetocin-mediated changes in HK and PK are JNK dependent
As Chaetocin-mediated increase in HK2 and PKLR was concomitant with increased ROS, ATM and JNK levels, we determined the role of these molecules in the regulation of HK2 and PKLR in Chaetocin-treated cells. Western blot analysis revealed a decrease in HK2 and increase in PKLR protein levels upon Chaetocin treatment (Figure 5a). This change in HK2 and PKLR levels was ROS and JNK dependent (Figure 5a) but independent of ATM (Figure 5b). Changes in HK2 and PKLR expression were also reflected in their enzyme activity, as Chaetocin induced ∼30% decrease in HK activity and 2–3-fold increase in PK activity (Figure 5c). This alteration in enzyme activity was JNK dependent, as SP600125 reverted Chaetocin-mediated changes in both HK and PK activity (Figure 5c).
Figure 5.
Chaetocin-induced changes in glucose metabolism are c-Jun N-terminal kinase (JNK) dependent. (a) Inhibition of reactive oxygen species (ROS) and JNK abrogates Chaetocin-induced changes in PKLR and HK2 levels. Western blots indicating PKLR and HK2 levels in cells treated with Chaetocin in the presence and absence of N-acetyl-cysteine (NAc) or SP. (b) Chaetocin-induced effects on PKLR and HK2 are independent of ATM. Western blots indicating PKLR and HK2 protein levels in Chaetocin-treated glioma cells both in the presence and absence of ATM inhibitor, KU. The blots (a and b) were reprobed with β-actin to establish equal loading. (c) PK and HK enzyme activities were assessed in glioma cells treated with Chaetocin both in the presence or absence of SP. The graph shows that JNK inhibitor SP reverses Chaetocin-induced changes in PK (left panel) and HK activity (right panel). (d) Chaetocin-induced reduction in lactate level and (e) ATP production is JNK dependent. Lactate and ATP levels were measured in glioma cells treated with Chaetocin, in the presence or absence of SP. (f) Chaetocin-mediated reduction of glucose uptake in glioma cells is dependent on JNK and ROS. Glucose uptake was measured in glioma cells treated with different combinations of Chaetocin, SP or NAc using fluorescent glucose (2-NBDG) staining. Western blots (a and b) and fluorescent images (f) are representative of three independent experiments showing similar results. Graphs shown in (c–e) represent data as means±S.E.M. pooled from at least three experiments. *Significant change from control and #significant change from the Chaetocin-treated group (P<0.05)
Chaetocin decreases lactate levels, ATP production and glucose uptake in a ROS- and JNK-dependent manner
Intracellular ATP levels determine chemoresistance in cancer cells, as depletion of ATP by inhibitor of glycolysis sensitizes cells to chemotherapeutics.34 Moreover, lactate is an important contributor to ATP generation in astrocytoma cells.35 We therefore determined lactate levels and ATP production in Chaetocin-treated cells. A significant decrease in lactate production (Figure 5d) and ATP generation (Figure 5e) was observed in Chaetocin-treated cells as compared with the untreated control. These changes in lactate and ATP levels were JNK dependent, as SP600125 significantly reverted Chaetocin-mediated inhibition of lactate and ATP production. The decreased ATP production was accompanied by a ROS/JNK-dependent decrease in cellular glucose uptake by glioma cells in the presence of Chaetocin (Figure 5f). These results indicate that Chaetocin induces an overall inhibition of energy production and glucose metabolism in glioma cells in a ROS- and JNK-dependent manner.
Chaetocin inhibits growth of tumor xenograft in nude mice
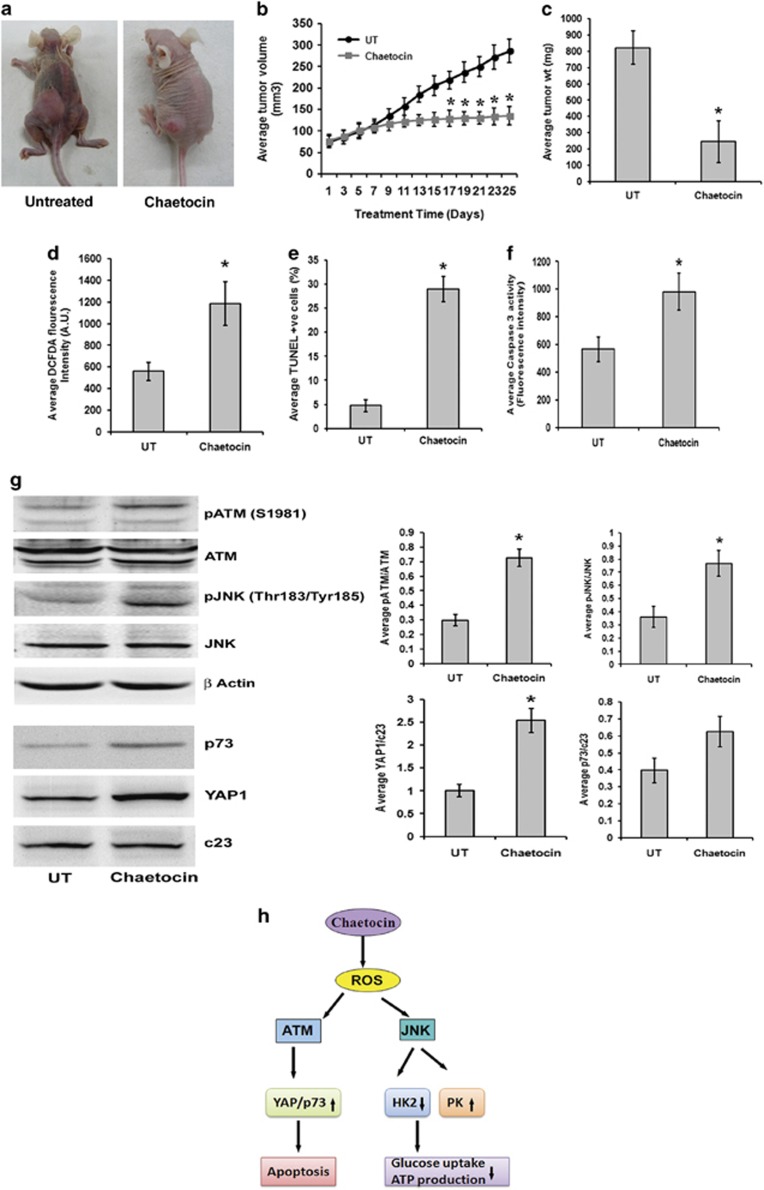
Given that Chaetocin induces potent anti-proliferative effect on glioma cells in vitro, and as the in vivo anti-tumor effect of Chaetocin in ovarian carcinoma has been reported,36 we next investigated the in vivo antiglioma effects of Chaetocin. Compared with vehicle treatment, Chaetocin effectively retarded the tumor growth in the flank of athymic mice (Figure 6a). Inhibition of tumor growth was reflected by a significant decrease in tumor volume (Figure 6b) and weight (Figure 6c) upon Chaetocin treatment. The dosing regimen of Chaetocin was well tolerated, with no significant change in body weight of treated mice (Supplementary Figure 4). In coherence with the in vitro results, Chaetocin-treated tumor mass showed significantly elevated ROS generation (Figure 6d), increased TUNEL-positive cells (Figure 6e) and caspase-3 activity (Figure 6f). Decrease in both PCNA-positive cells and its protein level in Chaetocin-treated tumors indicated inhibition of cell proliferation (Supplementary Figure 5a). Alteration in cell cycle markers p21, p27 and cyclin D1 was also observed in Chaetocin-treated tumors as compared with untreated controls. A significant increase in p21 and p27 levels along with a decrease in cyclin D1 levels (Supplementary Figure 5b) indicated cell cycle inhibition upon Chaetocin treatment in glioma xenografts.
Figure 6.
Chaetocin inhibits tumor growth in heterotopic xenograft glioma model. (a) Representative photographs depicting the differences between cohorts of tumor-bearing nude mice untreated or treated with Chaetocin. U87MG glioma cell suspension (5 × 106 cells) was injected subcutaneously in the flanks of nude mice. After 15 days of tumor growth, the animals were given either Chaetocin (treated) or vehicle (untreated) on every alternate day for the next 25 days and the parameters depicting tumor growth were scored. The left panel shows representative animal in the untreated group bearing full-grown tumor and the right panel shows the representative animal of the Chaetocin-treated group with relatively regressed tumor. Images clearly demonstrate that Chaetocin restricts the growth of nude mice glioma xenografts. (b) Growth of tumor was assessed by measuring the changes in tumor volume from the first day of Chaetocin treatment in both groups. Average tumor volumes in both treated and untreated groups (n=10) are plotted across the treatment days. Chaetocin significantly restricts the tumor volume on and after 17th day of treatment. (c) Graph denotes the average tumor weights in control and treated groups (n=10). Following 25 days of Chaetocin administration, the tumor mass was dissected out and weighed. Tumors from Chaetocin-treated group weighed significantly less as compared with the untreated controls at the end of treatment schedule. (d) Cells from the dissected tumor masses were analyzed for reactive oxygen species (ROS) levels using DCFDA fluorescence assay. Graph represents the average fluorescent DCFDA intensities in Chaetocin-treated and untreated groups (n=5). Chaetocin-treated tumor cells show increased DCFDA fluorescence as compared with the untreated group. (e) Chaetocin treatment significantly increased terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive cells in the tumor mass. Graph represents TUNEL-positive cells in Chaetocin-treated and untreated groups (n=5). (f) Chaetocin-treated tumors show increased caspase-3 enzyme activity as compared with the untreated tumors. Graph depict the average enzyme activity (raw fluorescence intensity values) in the tumor lysates of Chaetocin-treated and untreated groups (n=5). Values in (b to f) are represented as means±S.E.M. with the indicated number of animals per group. Statistical analysis was carried out using unpaired Student's t-test. Results were considered significant, when P-value was ≤0.05. *Significant change from the untreated group. (g) Western blots depicting an increase in pATM and pJNK, and p73 and YAP1 in the total cell lysates and nuclear extracts prepared from the Chaetocin-treated and untreated tumors. Blots were reprobed for β-actin or c23 to establish equivalent loading. Graph represents average band intensities (raw values) of phosphorylated (p) ATM and pJNK normalized to ATM and JNK, respectively. The average ratio of the band intensities (raw values) of YAP1 or p73 were normalized to c23. Values indicate means±S.E.M. of n=6. *Significant change between the treated and untreated groups (P<0.05). (h) Proposed model for induction of apoptosis and regulation of glucose metabolism by Chaetocin-induced ROS in glioma cells
Elevated pATM, pJNK, YAP1 and p73 levels in chaetocin-treated tumor xenograft tissue
We further validated our in vitro findings in the xenograft tissue obtained from nude mice. Elevated ROS level was accompanied by an increase in pATM and pJNK expression in the tumor tissue of Chaetocin-treated group in comparison with vehicle-treated control (Figure 6g). Similarly, an increase in nuclear YAP1 and p73 levels was also observed in Chaetocin-treated tumors (Figure 6g). Chaetocin treatment also reduced H3K9me3 levels in brain tissue of tumor bearing nude mice as compared with vehicle-treated control (Supplementary Figure 6).
Discussion
The fungal metabolite chaetocin bearing the epipolythiodioxopiperazine moiety was identified as a specific inhibitor of H3K9 methyltransferase SU(VAR)3–9.37 However, recent findings indicate that the ETP moiety rather than the structure of Chaetocin is critical for its nonspecific histone lysine methyltransferase inhibitory activity.38 Nonetheless, it is evident that the broad specificity of Chaetocin to affect histone methylation also includes H3K9me3. The property of Chaetocin to induce apoptosis in cancer cells has been attributed to its ability to enhance ROS induction through inhibition of thioredoxin reductase.6, 39 The ability of diverse chemotherapeutic agents to induce glioma cell apoptosis through increased intracellular ROS generation2, 3 prompted us to investigate whether Chaetocin could affect glioma cell viability through perturbation of redox homeostasis. Elevated ROS generation in Chaetocin-treated cells was concomitant with the decrease in Trx/TrxR pathway, which is crucial in regulating cellular ROS levels.40 As observed in multiple myeloma and other solid tumors,6, 39 this heightened ROS production is required for the antiglioma effects of Chaetocin. Also, a ROS-dependent decrease in H3K9me3 levels was observed in glioma cells.
In addition to the hippo pathway-mediated classical phosphorylation-dependent regulation of YAP1, other proteins can also regulate YAP1.41 Although YAP phosphorylation by JNK regulates its function in apoptosis,11 Chaetocin-induced JNK had no effect on YAP1 expression. Interestingly, Chaetocin induced YAP1 in a ROS-ATM-dependent manner, without the involvement of the classical hippo pathway. YAP1 knockdown rescued cells from Chaetocin-induced apoptosis, indicating the proapoptotic nature of YAP1 in glioma cells. ATM not only interacted with YAP1 but also regulated its interaction with p73 and p300. As YAP1, p73 and p300 are concomitantly recruited to the regulatory regions of apoptotic target genes,30 it is likely that by enhancing the interaction between these components, ATM possibly contributes to the ability of YAP1 to inhibit glioma cell growth. Akt, a key player in survival pathways, phosphorylates YAP1 to suppress proapoptotic gene expression.25 As aberrant activation of Akt is reported in glioma, it is possible that Akt-mediated phosphorylation of YAP1 suppresses the induction of proapoptotic genes.
The Warburg effect, a metabolic adaptation of cancer cells characterized by enhanced glycolysis and suppressed oxidative phosphorylation (OXPHOS),42, 43 is associated with drug resistance in cancer cells.44 As a consequence, agents targeting glycolysis have shown promising efficacy in reversing drug resistance and are being considered as potential anticancer targets.44, 45 Chaetocin treatment of non-small-cell lung cancer and myeloma altered transcripts of genes associated with inflammatory response, cell death/apoptosis pathways and cellular metabolism such as enolase.36 GBM expresses high levels of HK2 and inhibition of HK2 sensitizes glioma cells to apoptosis.16 Dissociation of HK2 from the mitochondria potentiates Bax-induced cytochrome c release and apoptosis.46 JNK regulates mitochondrial apoptotic pathway,47 and mitochondrial JNK signaling induces ROS production.48 Interestingly, knockdown of HK2 increases ROS levels,17 and excessive ROS causes oxidative mitochondrial damage and massive cell death.1 It is tempting to speculate that Chaetocin-mediated abrogation of HK2 activity could have possibly contributed to both elevated ROS production and caspase-9-mediated apoptosis. Importantly, PK initiates a metabolic loop that regulates redox metabolism,19 and PK also regulates glioma cell proliferation.49 Interestingly, ROS-mediated inhibition of PKM2 induces alterations in glucose metabolism required for cellular antioxidant responses characterized by low PK activity.18 Here, we demonstrate for the first time the role of JNK in the regulation of PK and HK2 activity (Figure 6h).
GBM still remains one of the most malignant of human cancers, partly because of its refractoriness to current therapies. Studies suggest that diverse chemotherapeutic agents can induce apoptosis in cancer cells by elevating intracellular ROS generation. Chaetocin attenuated the growth of glioma xenografts and this decrease in tumor volume was accompanied by an increase in ROS production. Importantly, the ability of Chaetocin to reduce H3K9me3 levels in brain tissue of tumor bearing nude mice as compared with untreated control (Supplementary Figure 6) indicates that it can cross the blood–brain barrier, making it a potential antiglioma target in clinic. Also, by providing insight into previously uncharacterized mechanisms through which ROS regulates aberrant cellular metabolism and resistance to apoptosis, this study will advance our understanding of glioma biology. The potent efficacy of Chaetocin as an inducer of apoptosis and metabolic modulator in glioma cells may lead to advances in the treatment of GBM and warrants its investigation as a potent antiglioma target.
Materials and Methods
Cell culture and treatment
Glioblastoma cell lines A172, U87MG and T98G were obtained from American Type Culture Collection (Manassas, VA, USA) and ECACC (European Collection of Cell Cultures). Cells were cultured in DMEM supplemented with 10% FBS. On attaining semiconfluence, cells were switched to serum-free media (SFM), and after 6 h, cells were treated with different concentrations of Chaetocin (in dimethyl sulfoxide), in the presence or absence of ROS inhibitor NAc, or ATM inhibitor KU60019 (Tocris Bioscience, Northpoint, UK) or JNK inhibitor SP600125 for different time intervals. All reagents were purchased from Sigma (St. Louis, MO, USA), unless otherwise stated.
Determination of cell viability
Viability of cells treated with Chaetocin, in the presence or absence of NAc, KU60019 or SP600125 for different intervals of time was assessed using the MTS assay (Promega, Madison, WI, USA) as described.3 Values were expressed as percentage change compared to controls.
TUNEL assay
TUNEL assay was performed on glioma cells (104) treated with Chaetocin in the presence and absence of NAc as described previously2 and on tumor tissue sections obtained from glioma xenografts in nude mice. The percentage of cell death was determined from the number of TUNEL-positive cells (red) that colocalized with DAPI (blue) to the total number of cells taken from multiple fields.
Measurement of ROS
Intracellular ROS generation in cells treated with Chaetocin in the presence and absence of NAc was assessed by using 2′, 7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA; Molecular Probes, Eugene, OR, USA) or fluorescent dye dihydroethidium (DHE) as described previously.3 Tumor tissue obtained from glioma xenografts in nude mice immediately after surgical removal was homogenized using Dounce homogenizer to make a single-cell suspension. A total of 105 tumor cells were incubated with 10 μM DCFDA for 45 min and fluorescence intensity was measured at λex=500/λem=530 nm.
Western blot analysis
Western blot analysis was performed on protein lysates isolated from control or Chaetocin-treated cells and tumor tissues as described previously,2 using antibodies against YAP1, pATM, ATM, PKLR, Mst1/2, cyclin D1 (Abcam, Cambridge, UK), pYAP1, pJNK, JNK, p27, p73, pLATS1, HK2 (Cell Signaling, Danvers, MA, USA), p21 (BD Biosciences, San Diego, CA, USA) and YAP1 (Novus Biological, Cambridge Science Park, Cambridge, UK). Antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), unless otherwise mentioned. Secondary antibodies were purchased from Vector Laboratories Inc. (Burlingame, CA, USA). The blots were stripped and reprobed with anti-β-actin (Sigma) or c23 to determine equivalent loading as described.50
Assay for caspase-3 and -9 activities
The Colorimetric Assay Kits for caspase-3 and -9 (Abcam) were used to determine the levels of active caspases in cells treated with different combinations of Chaetocin, NAc or KU as described previously.50
Co-immunoprecipitation
Immunoprecipitation was performed with nuclear extracts (200 μg) obtained from glioma cells treated with different combinations of Chaetocin, NAc or KU. Extracts were incubated with 2 μg of YAP1 antibody (Novus Biological) for 16 h as described previously.13 Interaction of proteins with YAP1 was studied by probing the blots with antibodies against ATM, p300 and p73 as described.13
Transfection
Eighteen hours before transfection, 5 × 103 cells were seeded onto 96-well plates in medium without antibiotics and transfection with 50 nmol/l duplex YAP-1 or non-specific (NS) siRNA (Thermo Fischer Scientific, Lafayette, CO, USA) was carried using Lipofectamine RNAi Max reagent (Life Technologies-Invitrogen, Carlsbad, CA, USA) as described previously,51 and MTS assay was performed. Similarly, transfection with either empty vector (pcDNA) or with pcDNA Flag YAP1 plasmid obtained from Addgene (Cat. no. 18881) and generated in Yosef Shaul laboratory8 was performed on 5 × 103 cells seeded onto 96-well plates as described51 and MTS assay was performed.
Immunohistochemistry
Immunohistochemistry was performed on tumor tissue samples obtained from either vehicle or Chaetocin-treated mice as described previously.52
Thioredoxin reductase assay
Thioredoxin reductase activity in cells treated with Chaetocin for 12 h was measured with Thioredoxin Reductase Assay Kit (Abcam), according to the manufacturer's instructions.
Measurement of mitochondrial ROS generation
Intracellular ROS generation was determined using fluorescent dye MitoSOX Red Mitochondrial Superoxide Indicator (Life Technologies-Invitrogen). Briefly, cells seeded in 4-well chamber slides and treated with Chaetocin in the presence or absence of NAc, SP600125 or KU60019, were incubated with 2 μM MitoSOX for 20 min. Images of fluorescently labeled cells were captured by Zeiss ApoTome Imager fluorescence microscope (Carl Zeiss, Oberkochen, Germany), using a Rhodamine filter.
HK and PK activity assay
HK and PK activity in cells treated with Chaetocin in the presence and absence of SP600125 for 12 h was measured with Hexokinase Colorimetric Assay Kit (Biovision Inc., Milpitas, CA, USA) and PK Activity Assay Kit (Sigma), respectively, according to the manufacturer's instructions.
Lactate and ATP measurement
Lactate released in the supernatant collected from cells treated with Chaetocin, either alone or in the presence of NAc or SP600125 for 12 h, was measured with a Lactate Colorimetric Assay Kit (Biovision Inc.), according to the manufacturer's instructions. Intracellular ATP levels were measured by luminometric assay using the ATP lite, Luminescence ATP detection assay system (Perkin Elmer, Waltham, MA, USA) as described previously.12 Values were expressed as a percentage relative to those obtained in controls.
Human metabolism qRT-PCR array
qRT-PCR was performed using Human Glucose Metabolism RT2 Profiler comprising of 84 metabolism-related genes (SuperArray Biosciences, Hilden, Germany), as described previously.12 Five housekeeping genes were included on the array (B2M, HPRT1, RPL13A, GAPDH and ACTB) to normalize the transcript levels. Results were analyzed as per user manual guidelines using integrated web-based software package for the PCR Array System (RT2 Profiler PCR Array Human Glucose Metabolism PAHS-006Z).
Generation of heterotopic glioma xenografts
U87MG cells (5 × 106 cells suspended in 100 μl SFM) were injected subcutaneously in the flank of the anesthetized nude mice. The tumor growth was observed once every alternate day. After 15 days of inoculation, when measurable tumors (5-6 mm diameter) were formed, animals were divided randomly into two groups of 10 animals each and were administered either with vehicle or Chaetocin (0.5 mg/kg body weight), intraperitoneally on alternate days for 25 days. Body weight of the animals and tumor dimensions were measured using a caliper on the days of injection. Tumor volume (V) was calculated using the formula V=length × width × height/2. The animals were killed and tumor mass from each mouse was excised, weighed and processed for further experiments. Athymic nude mice were handled within a specific pathogen-free facility and all the experimental manipulations were undertaken in accordance with the guidelines of the institutional animal ethics committee.
Statistical analysis
In in vitro experiments, comparisons between groups were performed using paired Student's t-test. In in vivo experiments, statistical analysis was carried out using unpaired Student's t-test. All values of P <0.05 were taken as significant.
Acknowledgments
The work was supported by a research grant from the Department of Biotechnology (DBT, Government of India No. BT/PR5818/Med/30/839/2012) to ES. DD is supported by a research fellowship from Council of Scientific and Industrial Research (CSIR, Government of India). We acknowledge Shanker Dutt Joshi for technical assistance with the animal experiments. We acknowledge Dr. Inderjeet Yadav for his help and support with animal facility.
Author contributions
Conception and design: DD and ES; acquisition of data: DD, RG, NPA; analysis and interpretation of data (e.g., statistical analysis): DD, RG and ES; writing, review and/or revision of the manuscript: DD, RG, ES; study supervision: ES.
Glossary
- GBM
glioblastoma multiforme
- ROS
reactive oxygen species
- ATM
ataxia telangiectasia mutated
- YAP1
Yes-Associated Protein 1
- JNK
c-Jun N-terminal kinase
- HK
hexokinase
- PK
pyruvate kinase
- NS siRNA
non-specific (scrambled) siRNA
- DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- DHE
dihydroethidium
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- Trx1
thioredoxin 1
- IP
immunoprecipitation
- NAc
N-acetyl-L-cysteine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Finazzi-Agrò
Supplementary Material
References
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Sharma V, Joseph C, Ghosh S, Agarwal A, Mishra MK, Sen E. Kaempferol induces apoptosis in glioblastoma cells through oxidative stress. Mol Cancer Ther. 2007;6:2544–2553. doi: 10.1158/1535-7163.MCT-06-0788. [DOI] [PubMed] [Google Scholar]
- Dixit D, Sharma V, Ghosh S, Koul N, Mishra PK, Sen E. Manumycin inhibits STAT3, telomerase activity, and growth of glioma cells by elevating intracellular reactive oxygen species generation. Free Radic Biol Med. 2009;47:364–374. doi: 10.1016/j.freeradbiomed.2009.04.031. [DOI] [PubMed] [Google Scholar]
- Chervona Y, Costa M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic Biol Med. 2012;53:1041–1047. doi: 10.1016/j.freeradbiomed.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D, Huse JT, et al. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol. 2013;72:298–306. doi: 10.1097/NEN.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib H, Nebbioso A, Prebet T, Castellano R, Garbit S, Restouin A, et al. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase SUV39H1. Leukemia. 2012;26:662–674. doi: 10.1038/leu.2011.271. [DOI] [PubMed] [Google Scholar]
- Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG. Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J Neuropathol Exp Neurol. 2011;70:568–577. doi: 10.1097/NEN.0b013e31821ff8d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Chiu WH, Luo SJ, Chen CL, Cheng JH, Hsieh CY, Wang CY, et al. Vinca alkaloids cause aberrant ROS-mediated JNK activation, Mcl-1 downregulation, DNA damage, mitochondrial dysfunction, and apoptosis in lung adenocarcinoma cells. Biochem Pharmacol. 2012;83:1159–1171. doi: 10.1016/j.bcp.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Schneider JG, Finck BN, Ren J, Standley KN, Takagi M, Maclean KH, et al. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell metabolism. 2006;4:377–389. doi: 10.1016/j.cmet.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Tomlinson V, Gudmundsdottir K, Luong P, Leung KY, Knebel A, Basu S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010;1:e29. doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Ghildiyal R, Mehta VS, Sen E. ATM-NFkappaB axis-driven TIGAR regulates sensitivity of glioma cells to radiomimetics in the presence of TNFalpha. Cell Death Dis. 2013;4:e615. doi: 10.1038/cddis.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Dixit D, Sen E. Oncrasin targets the JNK-NF-kappaB axis to sensitize glioma cells to TNFalpha-induced apoptosis. Carcinogenesis. 2013;34:388–396. doi: 10.1093/carcin/bgs352. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Kageji T, Kuwayama K, Kitazato KT, Mure H, Hara K, et al. Up-regulation of endogenous PML induced by a combination of interferon-beta and temozolomide enhances p73/YAP-mediated apoptosis in glioblastoma. Cancer Lett. 2012;323:199–207. doi: 10.1016/j.canlet.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wyatt E, Chawla K, Tran M, Ghanefar M, Laakso M, et al. Hexokinase II knockdown results in exaggerated cardiac hypertrophy via increased ROS production. EMBO Mol Med. 2012;4:633–646. doi: 10.1002/emmm.201200240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J, Phillips JJ, Zheng S, Wiencke J, Ronen SM, Pieper RO. Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma. PLoS One. 2013;8:e57610. doi: 10.1371/journal.pone.0057610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MM, Lehrach H, et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011;14:415–427. doi: 10.1016/j.cmet.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science (New York, NY) 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1:552–562. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxidants Redox Signal. 2009;11:1097–1106. doi: 10.1089/ars.2008.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swa HL, Blackstock WP, Lim LH, Gunaratne J. Quantitative proteomics profiling of murine mammary gland cells unravels impact of annexin-1 on DNA damage response, cell adhesion, and migration. Mol Cell Proteomics. 2012;11:381–393. doi: 10.1074/mcp.M111.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Lai JM, Chang JT, Wen CL, Hsu SL. Emodin induces a reactive oxygen species-dependent and ATM-p53-Bax mediated cytotoxicity in lung cancer cells. Eur J Pharmacol. 2009;623:1–9. doi: 10.1016/j.ejphar.2009.08.031. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Kang MA, So EY, Simons AL, Spitz DR, Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausti F, Di Agostino S, Cioce M, Bielli P, Sette C, Pandolfi PP, et al. ATM kinase enables the functional axis of YAP, PML and p53 to ameliorate loss of Werner protein-mediated oncogenic senescence. Cell Death Differ. 2013;20:1498–1509. doi: 10.1038/cdd.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Zagurovskaya M, Shareef MM, Das A, Reeves A, Gupta S, Sudol M, et al. EGR-1 forms a complex with YAP-1 and upregulates Bax expression in irradiated prostate carcinoma cells. Oncogene. 2009;28:1121–1131. doi: 10.1038/onc.2008.461. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang M, Kar S, Carr BI. Involvement of ATM-mediated Chk1/2 and JNK kinase signaling activation in HKH40A-induced cell growth inhibition. J Cell Physiol. 2009;221:213–220. doi: 10.1002/jcp.21844. [DOI] [PubMed] [Google Scholar]
- Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tozzi F, Chen J, Fan F, Xia L, Wang J, et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012;72:304–314. doi: 10.1158/0008-5472.CAN-11-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire J, Mailloux RJ, Appanna VD. Mitochondrial lactate dehydrogenase is involved in oxidative-energy metabolism in human astrocytoma cells (CCF-STTG1) PLoS One. 2008;3:e1550. doi: 10.1371/journal.pone.0001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isham CR, Tibodeau JD, Bossou AR, Merchan JR, Bible KC. The anticancer effects of chaetocin are independent of programmed cell death and hypoxia, and are associated with inhibition of endothelial cell proliferation. Br J Cancer. 2012;106:314–323. doi: 10.1038/bjc.2011.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Cherblanc FL, Chapman KL, Brown R, Fuchter MJ. Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases. Nat Chem Biol. 2013;9:136–137. doi: 10.1038/nchembio.1187. [DOI] [PubMed] [Google Scholar]
- Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007;109:2579–2588. doi: 10.1182/blood-2006-07-027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlof AK, Arner ES. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxidants Redox Signal. 2004;6:41–52. doi: 10.1089/152308604771978336. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2012;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res. 2011;71:4585–4597. doi: 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Mancuso A, Bui TV, Tong X, Gruber JJ, Swider CR, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene. 2010;29:2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science (New York, NY) 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- Chambers JW, LoGrasso PV. Mitochondrial c-Jun N-terminal kinase (JNK) signaling initiates physiological changes resulting in amplification of reactive oxygen species generation. J Biol Chem. 2011;286:16052–16062. doi: 10.1074/jbc.M111.223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit D, Ghildiyal R, Anto NP, Ghosh S, Sharma V, Sen E. Guggulsterone sensitizes glioblastoma cells to Sonic hedgehog inhibitor SANT-1 induced apoptosis in a Ras/NFkappaB dependent manner. Cancer Lett. 2013;336:347–358. doi: 10.1016/j.canlet.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Dixit D, Sharma V, Ghosh S, Mehta VS, Sen E. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor (TNFalpha)-induced apoptosis through SIRT1 inhibition. Cell Death Dis. 2011;3:e271. doi: 10.1038/cddis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Choudhury SR, Ghosh S, Mehta VS, Sen E. Involvement of TNFalpha-induced TLR4-NF-kappaB and TLR4-HIF-1alpha feed-forward loops in the regulation of inflammatory responses in glioma. J Mol Med (Berl) 2012;90:67–80. doi: 10.1007/s00109-011-0807-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.