Abstract
The endosomal sorting complexes required for transport (ESCRT) pathway was initially defined in yeast genetic screens that identified the factors necessary to sort membrane proteins into intraluminal endosomal vesicles. Subsequent studies have revealed that the mammalian ESCRT pathway also functions in a series of other key cellular processes, including formation of extracellular microvesicles, enveloped virus budding, and the abscission stage of cytokinesis. The core ESCRT machinery comprises Bro1 family proteins and ESCRT-I, ESCRT-II, ESCRT-III, and VPS4. Site-specific adaptors recruit these soluble factors to assemble on different cellular membranes, where they carry out membrane fission reactions. ESCRT-III proteins form filaments that draw membranes together from the cytoplasmic face, and mechanistic models have been advanced to explain how ESCRT-III filaments and the VPS4 ATPase can work together to catalyze membrane fission.
Keywords: multivesicular bodies, membrane protein degradation, exosomes, shedding microvesicles, virus budding, cytokinesis, abscission
Overview
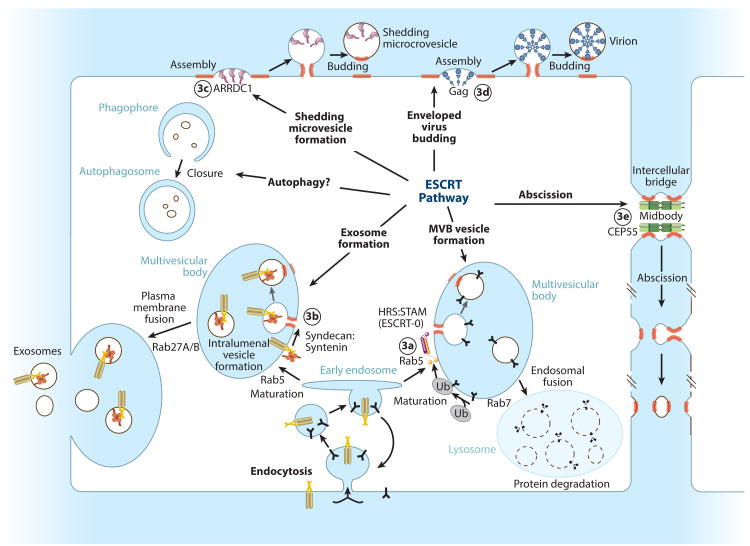
The mammalian endosomal sorting complexes required for transport (ESCRT) pathway comprises more than 30 proteins that catalyze a series of important cellular membrane fission events, including intraluminal vesicle formation at endosomes, virus and vesicle budding from the plasma membrane, and abscission of the intercellular bridge during cytokinesis (Figure 1). In each case, membrane fission is effected from within the cytoplasmic face of thin membrane tubules and therefore differs from vesiculation processes such as endocytosis, where fission is effected from the vesicle neck exterior. Thus, the ESCRT pathway is a mobile machinery that can be recruited to different cellular membranes to catalyze “reverse topology” fission events.
Figure 1.
Membrane fission reactions promoted by the mammalian ESCRT pathway. Adaptor complexes that direct the pathway to specific sites of action are shown schematically, and ESCRT-remodeled membranes are highlighted red. Circled numbers denote the panel in Figure 3 that shows the adaptor structure.
The existence of a reverse topology fission machinery was first implied by the observation that late endosomal compartments, termed multivesicular bodies (MVBs), contained intraluminal vesicles and that integral membrane receptors destined for lysosomal degradation trafficked through these vesicles (reviewed in Reference 1). Core ESCRT components were subsequently identified in seminal yeast genetic studies as factors required for MVB vesicle formation and vacuolar targeting of membrane proteins. The pathway was named when it was shown that ESCRT components could be subdivided into discrete complexes with distinct biochemical functions (1, 2). Yeast studies have continued to illuminate structural and mechanistic aspects of the conserved ESCRT pathway, but metazoan ESCRTs perform a series of additional functions that are best studied in their native contexts.
The ESCRT Machinery
The five different classes of proteins and complexes that constitute the core ESCRT machinery can be subdivided into (a) early acting factors that coordinate ESCRT assembly, membrane deformation, and cargo sorting (Bro1 family proteins, ESCRT-I and ESCRT-II complexes) as well as (b) late-acting factors that catalyze membrane fission and ESCRT disassembly (ESCRT-III and VPS4 complexes) (Figure 2 and see the online Supplemental Table 1 for a summary of ESCRT nomenclature). Follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org. Regulation occurs at multiple steps to ensure that the ESCRT machinery assembles correctly and cooperatively. Control mechanisms include the use of (a) adaptors with specificity for different phosphoinositides and substrates (Figure 3), (b) sequential binding interactions (Figures 2 and 4), (c) specialized subunit isoforms, (d) conformational cycling between “inactive” (soluble) and “active” (assembled, membrane-associated) states, (e) avidity effects that help oligomeric ESCRT subunits to assemble in an “all-or-none” fashion, (f ) regulatory cofactors that promote or inhibit pathway activities, and (g) posttranslational modifications (particularly ubiquitylation) that identify protein cargoes and regulate ESCRT machinery activities (Figure 5).
Figure 2.
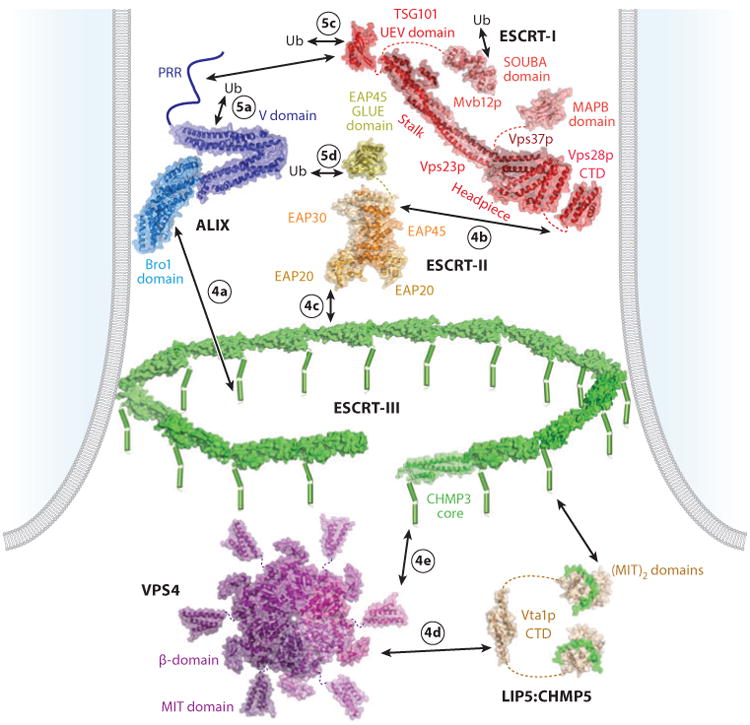
Core ESCRT components and their interactions, shown within a stylized bud neck. Lines denote regions of unknown structure, dashes denote linkers, cylinders denote helices, arrows denote protein-protein interactions, and circled numbers denote the panel in Figure 4 or 5 that shows the relevant interaction. Relevant Protein Data Bank identity numbers are shown in parentheses: human ALIX (20EV); yeast ESCRT-I core (2P22); human TSG101 UEV domain (1S1Q); yeast Vsp28p C-terminal domain (2J9U); MAPB domain (3TOW); SOUBA domain (4AE4), note that the MAPB and SOUBA domains would not be present simultaneously in any single ESCRT-I complex; ESCRT-II core (2ZME); EAP45 GLUE domain (2HTH); human CHMP3 (2GD5), with individual subunits modeled into a circular filament; VPS4 MIT (2JQ9); VPS4 core (1XWI), modeled as a hexamer based on the structure of the related AAA ATPase p97 (1E32); Vta1p C-terminal domain (2RKL); and LIP5:CHMP5 (2LXM). Abbreviations: CTD, C-terminal domain; MIT, microtubule interacting and transport; PRR, proline-rich region; Ub, ubiquitin.
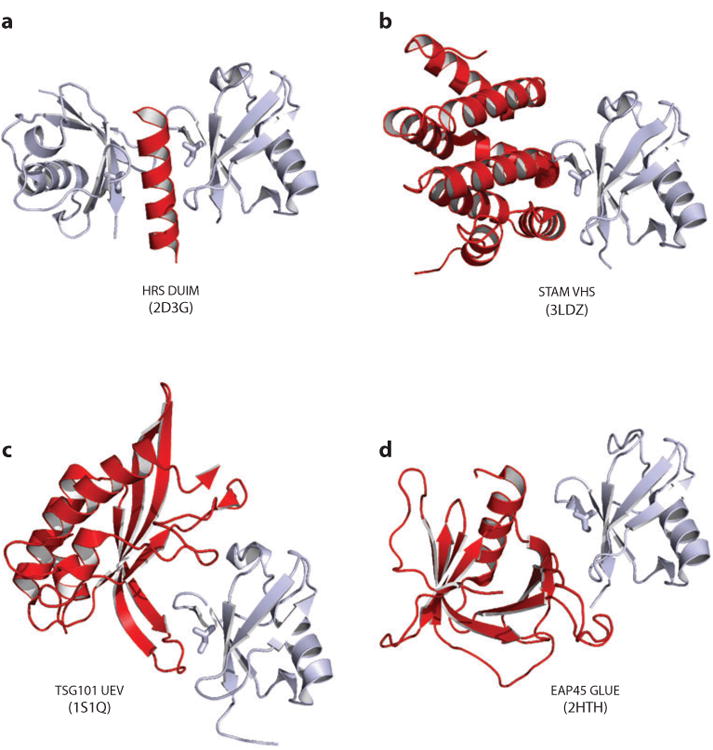
Figure 3.
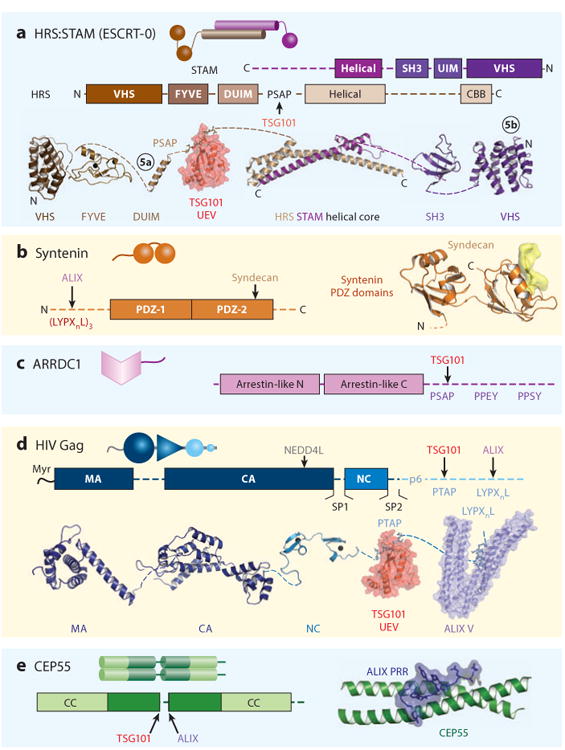
ESCRT adaptors. The functions and cellular locations of the different adaptors are shown in Figure 1. Here, each adaptor is shown as a cartoon representation, as a schematic model showing domain and motif maps, and, where possible, a composite of structurally characterized domains (ribbon diagrams) and intermolecular complexes (surface renderings). Circled numbers in panel a denote ubiquitin complexes that are shown in Figure 5. Relevant Protein Data Bank identity numbers are shown in parentheses. Panel a contains the HRS:STAM helical core (3F1I), HRS tandem VHS-FYVE domain (1DVP), HRS DUIM (2D3G), HRS PSAP:TSG101 UEV domain complex (3OBQ), STAM SH3 domain (1UJ0), and the STAM VHS domain (3LDZ). Panel b contains the syndecan peptide:syntenin PDZ2 domain complex (1YBO). Panel c contains HIV Gag Myr-MA (1UPH), CA (3H47), NC (1MFS), PTAP:TSG101 UEV domain complex (1M4Q), LYPXnL:ALIX (2R05). Panel d contains the CEP55 coiled coil:ALIX proline-rich region (PRR) complex (3E1R). Abbreviation: CBB, clathrin-binding box.
Figure 4.
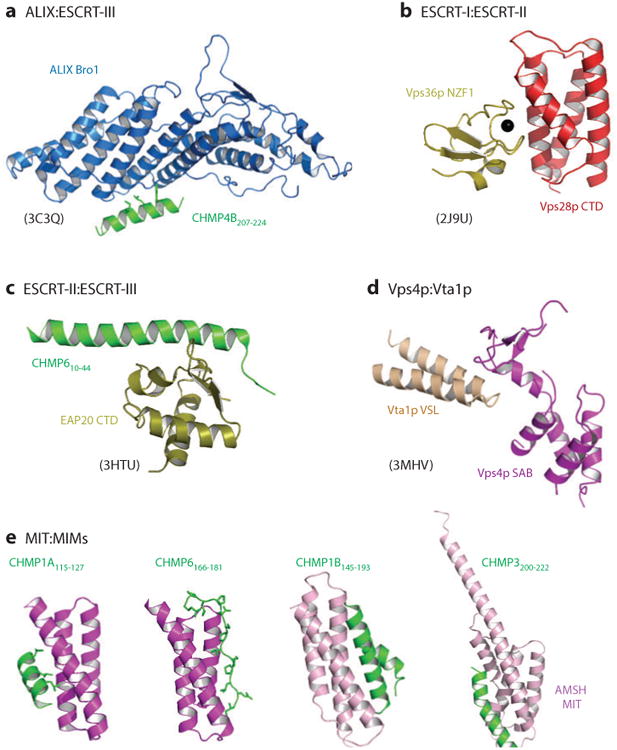
The structural basis for intercomplex interactions within the ESCRT pathway. The relevant interactions are as depicted in Figure 2, except that interacting domains from yeast Vps28p (ESCRT-I) and Vps36p (ESCRT-II) are shown in panel b because the divergent human ESCRT-I:ESCRT-II interaction site has not been structurally characterized and that interacting domains from yeast Vta1p and Vps4p are shown in panel d because the homologous human LIP5:VPS4 interaction has not been structurally characterized. Multiple ESCRT-III MIM:MIT domain interactions have been structurally characterized, and four distinct classes of interactions are shown [and see Figure 2 for the structure of the LIP5(MIT)2:CHMP5 complex]. Relevant Protein Data Bank identity numbers are shown in parentheses. Abbreviations: MIT, microtubule interacting and transport; MIM, MIT interaction motif.
Figure 5.
Structurally characterized ubiquitin interactions within the ESCRT pathway. Ubiquitin (Ub)-binding domains from the HRS:STAM adaptor (ESCRT-0) (see Figure 3a) and from ESCRT-I and ESCRT-II (Figure 2) are shown in red, bound to ubiquitin in gray. The Ub I44 side chains are shown explicitly (stick) to emphasize that the very different ubiquitin-binding domains recognize the same Ub surface. Relevant Protein Data Bank identity numbers are provided within the figure.
ESCRT Pathway Recruitment
At least five different classes of adaptors recruit the ESCRT pathway to different sites of action (Figures 1 and 3):
HRS:STAM (ESCRT-0) and related adaptors bind the TSG101 subunit of ESCRT-I to help create MVB vesicles (3, 4).
Syntenins collaborate with syndecans to bind ALIX and create vesicles destined for extracellular release as exosomes (5, 6).
Arrestin domain-containing proteins, such as ARRDC1, link cargoes to ubiquitin (Ub) ligases and ESCRT machinery during the formation of shedding microvesicles (7–9).
Viral structural proteins like the HIV Gag protein employ late assembly domains to bind TSG101, ALIX, and/or NEDD4family Ub E3 ligases to promote virus budding from the plasma membrane (10, 11).
CEP55 recruits TSG101 and ALIX to intercellular bridges to facilitate abscission (12–14).
Early Acting ESCRT Factors
The early acting ESCRT complexes localize to bud necks where they help facilitate membrane curvature; integrate inputs from site-specific adaptors, membranes, and ubiquitylated protein cargoes; and ultimately recruit the downstream membrane fission machinery (15, 16). They are rich in ubiquitin-binding domains (UBDs), reflecting the prominent role of this posttranslational modification in ESCRT function (Figure 5). UBD-Ub interactions are generally weak and transient but can be enhanced by the concentrating effects of the two-dimensional membrane (17), by avidity and/or cooperativity, and by preferential binding to polymeric Lys63-linked Ub chains (18).
Bro1 proteins
Bro1 protein family members bind membrane-specific adaptors and recruit the downstream ESCRT-III subunits CHMP4 and CHMP5 (19, 20). Humans express at least five Bro1 domain-containing proteins of which ALIX (21) and HD-PTP (22) are the best characterized (see Supplemental Table 1). These two proteins appear to be paralogs, with HDPTP functioning primarily in MVB protein sorting (23) and ALIX functioning in MVB sorting (24), exosome biogenesis (5), virus budding (25), and abscission (12).
ALIX comprises Bro1, V, and proline-rich elements (Figure 2). Short amphipathic helices or β-hairpins from the ESCRT-III proteins bind the concave surface of the banana-shaped Bro1 domain (Figure 4a) (19, 20), whereas the basic convex surface may bind and induce (or sense) negative membrane curvature (26). The V domain has two extended helical “arms,” and the second arm binds both Lys63-linked polyubiquitin chains (27) and YPXnL motifs found within viral and exosomal adaptors and MVB cargoes (Figure 3b,c) (5, 24, 25, 28, 29). The analogous region of HD-PTP binds the ESCRT-I subunit UBAP1, and this interaction helps define an endosome-specific Bro1-ESCRT-I pair (30).
The C-terminal proline-rich region (PRR) of ALIX folds back and autoinhibits ligand binding to the upstream domains (31, 32). The PRR contains binding epitopes for a number of other proteins, including CEP55 (12–14) and TSG101 (25, 33, 34), that presumably capture the “free” PRR conformation, thereby inducing (or sensing) ALIX activation. ALIX can also be phosphorylated, within both the Bro1 domain and the PRR, and both modifications likely play regulatory roles (35, 36). The fully assembled ALIX protein has been modeled as an antiparallel dimer with open, associated V domains. This observation implies that the Bro1 domains can bridge or nucleate two ESCRT-III filaments (37). ALIX also binds other important cellular components, including actin and endocytic, apoptotic, and calcium signaling machinery, although their ESCRT connections are not yet well understood (21, 38).
ESCRT-I
Like Bro1 proteins, ESCRT-I complexes link membrane-specific adaptors and MVB protein cargoes to downstream ESCRT machinery, and ALIX and ESCRT-I can often function interchangeably in supporting virus budding (11, 39). Humans express a complex array of heterotetrameric ESCRT-I complexes, each of which contains a single copy of the unique human TSG101 subunit and single copies of VPS28 (two variants, A and B), VPS37 (four isoforms, A-D), and MVB12 (at least three isoforms, MVB12A, MVB12B, and UBAP1) (30, 40, 41). Several ESCRT-I subunit isoforms have been shown to function in specific applications (30, 42, 43), but the full range of combinatorial complexity and functional diversity of mammalian ESCRT-I complexes remains to be explored.
The core of yeast ESCRT-I is composed of a “stalk” and a “headpiece” that together form a highly extended structure ∼18 nm long, with ligand-binding domains flexibly tethered at either end (Figure 2) (44–46). At one end, the N-terminal UEV domain and adjacent linker of TSG101 bind Ub (Figure 5c) (47, 48); the CEP55 adaptor (Figure 3e) (12–14); and P (T/S)AP peptide motifs within ALIX, the MVB HRS:STAM adaptor, the shedding vesicle ARRDC1 adaptor, and many viral protein adaptors (Figure 3a,c,d) (9, 33, 34, 49, 50). Terminal domains from some VPS37 and MVB12 subunits also have Ub-binding activities (51), including the solenoid UBA domain (SOUBA) of UBAP1, which can bind up to three Ub molecules simultaneously (Figure 2) (42). At the other end of the ESCRT-I core, MVB12A and -B subunits display MVB12-associated β-prism (MABP) domains, which bind membranes (Figure 2) (52). The C-terminal domain of VPS28 forms a four-helix bundle that binds ESCRT-II, albeit via structurally divergent interactions in yeast and mammals (Figure 4b) (44, 45, 53, 54).
ESCRT-II
ESCRT-II is a stable heterotetrameric complex that can bridge the ESCRT-I and ESCRT-III complexes (55). The complex contains single copies of EAP45 and EAP30, as well as two copies of EAP20. Both human and yeast ESCRT-II core complexes are shaped like the letter Y, with EAP20 subunits constituting the arms and EAP45 and EAP30 together constituting the base (Figure 2) (54, 56, 57). All three proteins contain tandem copies of the “winged-helix” fold, suggesting a common molecular ancestry. N-terminal elements from EAP45 and EAP30 are flexibly tethered to the base of the Y, and the EAP45 GLUE domain and adjacent linker bind ESCRT-I, phosphoinositides, and Ub (Figure 5d) (44, 53, 54, 58, 59). The two EAP20 subunits bind the ESCRT-III subunit, CHMP6 (Figure 4c) (53, 56, 60, 61); thus, ESCRT-II can nucleate the assembly of two ESCRT-III filaments (62). The yeast ESCRT-I:ESCRT-II supercomplex can adopt an ensemble of crescent shapes in solution and can stabilize the necks of vesicular membrane invaginations (15, 63).
ESCRT-II activity is essential for endosomal protein sorting and MVB vesicle formation in yeast (55), and appears to perform a similar role at the mammalian MVB (64). Indeed, overexpression of yeast ESCRT-II can bypass the ESCRT-I requirement, suggesting that ESCRT-II activation may be the most important ESCRT-I function during yeast MVB vesicle formation (55).
ESCRT-III Proteins
Humans express eight different subfamilies of ESCRT-III proteins. The conserved ESCRT-III core structure contains a long helical hairpin and two shorter helices that pack against the open end of the hairpin (Figure 2) (65– 67). In their soluble monomeric states, the C-terminal tails of ESCRT-III subunits fold back against the core and autoinhibit protein assembly (66, 68–70). The proteins then “open” when recruited to sites of membrane fission by the early acting factors ALIX and ESCRT-II, thereby freeing the core domains to bind membranes and polymerize. ESCRT-III polymerization also exposes arrays of C-terminal microtubule interacting and transport interaction motif (MIM) elements within their C-terminal tails that can bind microtubule interacting and transport (MIT)-domain containing proteins, including VPS4 (71–73), its activator LIP5 (74– 76), the microtubule severing enzyme spastin (77), and the deubiquitylating enzymes UBPY and AMSH (Figures 2 and 4e) (78, 79).
As discussed below, ESCRT-III proteins coassemble into membrane-bound filaments that play critical roles in membrane fission (80, 81). Despite their structural similarities, different ESCRT-III subunits clearly perform distinct roles, both in membrane fission and in cofactor recruitment. Studies of yeast MVB vesicle formation have revealed that the “core” ESCRT-III proteins are recruited sequentially in the order: CHMP6 (Vps20p), CHMP4 (Snf7p), CHMP3 (Vps24p), and CHMP2 (Did4p), followed by VPS4 (82–85). CHMP4 and CHMP2 subunits appear to constitute particularly important building blocks of ESCRT-III filaments because (a) they are required for all known ESCRT-dependent membrane fission processes (82, 85–87); (b) when overexpressed, both CHMP2 and CHMP4 subunits can form membrane-enclosed helical tubules that extrude from the plasma membrane (88, 89); and (c) CHMP4 subunits are present at higher stoichiometry than the other ESCRT-III proteins (84).
VPS4 ATPase Complexes
VPS4 ATPases power ESCRT-mediated membrane remodeling at the end of the catalytic cycle by using the energy of ATP hydrolysis to disassemble ESCRT-III filaments into their constituent subunits (Figure 2) (90, 91). There are also indications that VPS4 remodeling contributes directly to membrane fission during virus budding and cytokinesis (92–94), although perhaps not during MVB vesicle formation (85). Humans express two closely related VPS4 proteins (VPS4A and -4B) that can function redundantly in at least some contexts (72).
VPS4 contains an N-terminal MIT domain that binds ESCRT-III substrates, a mobile linker, a central ATPase cassette composed of large and small domains that resembles other AAA ATPases, a three-stranded antiparallel β-sheet inserted within the small ATPase domain, and a terminal helix that connects the two ATPase domains (Figure 2) (95, 96). The enzyme cycles through two states: a catalytically inactive, disassembled state and a catalytically active, higher-order assembly (97). Catalytically inactive Saccharomyces cerevisiae Vps4p can form double-ring structures in vitro (reviewed in Reference 90), but more recent analyses indicate that the active enzyme may be a hexamer (Figure 2) (N. Monroe, personal communication).
VPS4 assembly represents an important regulatory node within the ESCRT pathway. Enzyme recruitment, assembly, and activation are promoted by the high concentrations of MIM elements displayed by ESCRT-III filaments (71–73, 98, 99). In addition, two ESCRT-III-containing complexes, LIP5:CHMP5 and IST1:CHMP1B, appear to play particularly important roles in promoting VPS4 localization and activation (Figures 2 and 4d). IST1 and CHMP1B can copolymerize into helical tubes in vitro (J. McCullough & A. Frost, unpublished), and they play particularly important roles in helping to recruit VPS4 and other cofactors during abscission (66, 100–103).
LIP5 and CHMP5 form a soluble cytosolic complex that promotes VPS4 oligomerization and helps link the enzyme to the ESCRT-III lattice (74, 95, 98, 104). The first MIT module within the tandem MIT domain of LIP5 binds the tails of different ESCRT-III proteins, with a preference for CHMP1B, whereas the second MIT module binds tightly to CHMP5 (Figure 2) (74–76). The dimeric C-terminal VSL domain of LIP5 binds the C-terminal domains of VPS4 (Figure 4d) and stimulates VPS4 hexamerization and ATPase activity in vitro (95, 105, 106). This interaction may also link multiple VPS4 hexamers into an extended hexagonal array (106).
VPS4 enzymes apparently disassemble ESCRT-III filaments by first making MIT:MIM interactions with subunit tails (Figure 4e), then making a second contact near the center of the hexameric ring, and ultimately translocating the subunits up into (or through) the ring, thereby disrupting ESCRT-III lattice interactions (74, 90, 95, 99).
ESCRT:Membrane Interactions
In conjunction with the essential functions performed by ESCRT proteins, the lipid membrane also actively participates in ESCRT-mediated, “reverse-topology” membrane fission reactions by (a) targeting ESCRT factors to specific sites of action, (b) providing a two-dimensional surface that concentrates and organizes the ESCRT machinery and restricts MVB cargo diffusion, and (c) helping facilitate the fission reaction (17, 107). In the first case, different phosphoinositides direct the pathway to different cellular membranes. For example, the endosome-specific phospholipid PI(3)P targets the ESCRT pathway to endosomes for MVB vesicle formation through interactions with the HRS FYVE domain (108) and probably also with the GLUE domain of EAP45 (ESCRT-II) (59). By contrast, HIV-1 targets the ESCRT pathway to the cell periphery because the viral Gag adaptor binds specifically to PI(4,5)P2 in the plasma membrane (109). PI(4,5)P2 and PI(3)P also localize to the cleavage furrow (110), but their roles in targeting ESCRT factors to intercellular bridges remain to be determined. More generally, the membrane surface serves as an important organizational platform because all core ESCRT complexes except VPS4 also contain exposed basic/hydrophobic motifs that bind acidic lipids and help orient the complexes on the membrane surface. The topology of the membrane platform also appears to be important, as the dramatic membrane curvature generated prior to fission apparently helps recruit, retain, and organize ESCRT core complexes at the bud neck (15, 63, 111).
In addition to these targeting and organizational functions, specialized lipids such as cholesterol perform important functions in the membrane remodeling and fission reactions. Bilayers can segregate their lipids laterally to form cholesterol-rich microdomains called lipid rafts (112), and both MVB vesicles and HIV virions are cholesterol rich (16, 17, 113). Cholesterol is required for ESCRT-II self-assembly on supported lipid bilayers, and the resulting assemblies induce phase separation into liquid-ordered domains (114). This is potentially important because phase separation between liquid-ordered and -disordered regions can create line tension that promotes membrane scission (17). Two other specialized lipids have also been linked to ESCRT-related vesiculation processes. The phospholipid 2,2′-lysobisphosphatidic acid (LBPA) is enriched at late endosomes, where it can associate with ALIX (115). LBPA can drive spontaneous membrane deformation and intraluminal vesicle formation in vitro (115), apparently by adopting a cone shape under acidic conditions (116). The sphingolipid ceramide also adopts a cone shape and has been implicated in lipid microdomain assembly and exosome formation (117–119).
Biological Functions
Multivesicular Body Vesicle Formation
The ESCRT pathway plays a central role in targeting membrane proteins to the lysosome for degradation. Such proteins are first sorted into regions of the endosomal membrane that bud into the lumen (Figure 6a,b) and can then be degraded when the MVB fuses with the lysosome (Figure 1) (16, 120). Sorting into MVB vesicles requires (a) cargo clustering, (b) membrane invagination, (c) cargo transfer into nascent vesicles, and (d) vesicle neck fission, and ESCRT factors facilitate all of these steps (15, 81). MVB cargos are initially clustered on endosomal membranes by HRS:STAM (ESCRT-0) and related adaptors (e.g., GGA3 and TOM1L1). ESCRT-I:ESCRT-II supercomplexes promote membrane curvature and organize ESCRT assemblies at the vesicle neck, and late-acting ESCRT-III and VPS4 complexes catalyze membrane fission and factor recycling.
Figure 6.
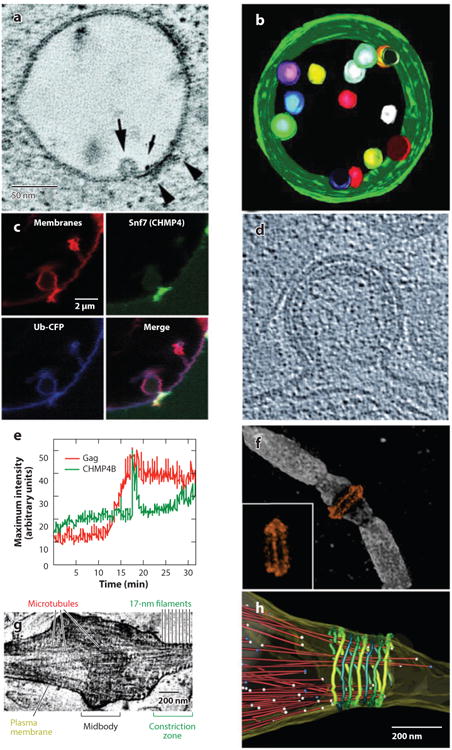
(a-c) ESCRT pathway functions in multivesicular body (MVB) vesicle formation, (d-e) enveloped virus budding, and (f-h) the abscission stage of cytokinesis. (a) Slice from an electron microscopy (EM) tomogram showing a vesicle budding into an endosome (large arrow) at a site adjacent to the HRS:STAM:clathrin coat (arrowheads). The small arrow shows a gold particle used to label the endosome. (b) EM tomographic reconstruction of an MVB, pseudocolored to show the internal vesicles and limiting membrane (green). Panels a and b were reprinted with permission from Reference 193, copyright 2003, National Academy of Sciences, U.S.A. (c) Fluorescence micrographs showing that Snf7p (CHMP4, green) concentrates at the neck of an MVB-like vesicle budding into a giant unilamellar vesicle in a reconstituted system. Ubiquitin cargos (blue) and membranes (red) are shown to define the vesicle. Reprinted with permission from Reference 15. (d) Cryoelectron microscopy (cryo-EM) image of a budding HIV-1 viral particle. The virion is ∼100 nm in diameter, the Gag protein lattice is visible as a protein-dense layer inside the plasma membrane, and the open neck of the virion is the site where ESCRT-mediated membrane fission occurs. Reprinted with permission from Reference 109, copyright 2012, Cold Spring Harbor Laboratory Press. (e) Graph showing the time course of CHMP4B (ESCRT-III) recruitment (green) as HIV Gag molecules (red) assemble into a single budding virion on a HeLa cell plasma membrane. Note that Gag assembles gradually (over ∼7 min), whereas CHMP4B appears in a sharp burst immediately prior to virion release. Reprinted with permission from Reference 92. (f) Structured illumination microscopy fluorescence image of an intercellular bridge prior to abscission, showing TSG101 (orange) forming two rings on either side of the midbody (with an alternate view inset) and microtubules (white). Reprinted with permission from Reference 94 (g) Cryo-EM tomographic image of an intercellular bridge showing the midbody, microtubules, and 17-nm filaments within constriction zones that undergo microtubule severing and abscission. Panels g and h were modified and reprinted with permission from Reference 171. (h) Pseudocolored EM reconstructions showing an intercellular bridge late in cytokinesis. Microtubules are shown in red with balls denoting their ends, and 17-nm filaments are shown in green shades. Abscission occurs at the narrow, microtubule-free constriction zone to the right of the filaments.
The heterodimeric HRS:STAM adaptor is shown schematically in Figure 3a (121). Important motifs include an HRSPSAP motif that helps recruit TSG101(ESCRT-I) (50), an HRS clathrin-binding box that connects the adaptor to a flat clathrin coat (122, 123), a STAM SH3 domain that recruits deubiquitinating enzymes (124), and four different UBDs that help concentrate ubiquitylated cargoes (Figures 3a and 5a,b) (125). Ub is the major signal for cargo sorting, and Lys63-linked polyubiquitin chains serve as the sorting signals for at least some cargoes, such as the epidermal growth factor receptor (18, 126).
NEDD4 proteins (Rsp5p in yeast) are the most important class of ESCRT-associated Ub ligases. NEDD4 ligases can either bind directly to PPXY motifs within cargoes (127) or can be recruited by arrestin-related adaptor proteins (Figure 3c) (7). Ubiquitylation is a highly dynamic process, and Ub is ultimately removed from cargoes prior to MVB vesicle incorporation by the deubiquitylating ESCRT enzymes, UBPY or AMSH, which interact with both the HRS:STAM adaptor and ESCRT-III subunits (124, 128). Cargoes can also interact directly with early acting ESCRT factors, as when the G protein–coupled receptor PAR1 uses a YPXnL motif to bind ALIX (24).
Intraluminal vesicles appear to form just beyond the edge of the HRS:STAM:clathrin coat (Figure 6a), which begs the question of how cargo proteins are transferred from the adapters into the nascent vesicles (15). One possibility is that the crescent-shaped ESCRT-I:ESCRT-II supercomplexes open and close to extract cargoes from the HRS:STAM:clathrin adaptor and transfer them into the nascent vesicles (63). Coassembly of ESCRT factors at the nascent bud neck would then provide a barrier against cargo diffusion from the vesicle prior to fission (Figure 6c).
Intriguing future issues surrounding MVB biology include (a) the relative importance of endosomal membranes and associated ESCRT protein networks as platforms for signaling (129) and small interfering RNA (siRNA) generation (130), and (b) the mechanistic basis for the remarkable observation that direct contacts between multivesicular endosomes and the endoplasmic reticulum stimulate ESCRT-mediated sorting of epidermal growth factor receptors into MVB vesicles (131).
Exosomes and Shedding Microvesicles
Cells can release different types of extracellular microvesicles, including two that have ESCRT connections: exosomes and shedding microvesicles (ectosomes). Exosomes likely constitute a major pathway for intercellular communication, with reported functions in antigen presentation and T-cell activation; immune tolerance; immune evasion; maternal immunosuppression and transplant tolerance; intercellular transport of proteins, DNA, and RNA; tissue repair; and neural communication (including roles in the spread of prion/plaque diseases) (reviewed in References 132 and 133). However, although exosome production and physiological responses have been demonstrated in tissue culture systems, exosomes are challenging to purify and assay, and clear examples of in vivo functions are rare.
ESCRT pathway involvement is expected because exosomes originate as MVB vesicles, and proteomic analyses of purified exosomes have identified subunits from all core ESCRT complexes (134, 135). Exosome production from cells requires the dimeric cytoplasmic protein syntenin, which acts as an adaptor that recruits ALIX to sites of intraluminal vesicle formation via interactions between the ALIX V domain and three syntenin LYPXnL motifs (Figures 1 and 3b) (5). Syntenin binds the cytoplasmic tails of the transmembrane protein syndecan (136). Syndecan clustering stimulates exosome production and the process also requires ALIX, EAP30 (ESCRT-II), CHMP2A, CHMP4A/B/C, and VPS4 proteins, implying ESCRT dependence. MVB trafficking away from the lysosome and to the plasma membrane for fusion remains to be understood in detail but is reportedly regulated by Rab27A/B GTPases (132).
Shedding microvesicle formation is less well characterized but also appears to involve the ESCRT pathway. For example, ESCRT-I is required for early stages of shedding microvesicle formation in Caenorhabditis elegans (137) and the human arrestin domain-containing protein ARRDC1 uses a PSAP motif to recruit TSG101 (ESCRT-I) for shedding microvesicle release from the plasma membrane (Figure 3c) (9).
In summary, exosomes and shedding microvesicles have an exciting research future, particularly if they prove to be major pathways for intercellular communication. Future goals include developing robust functional assays, learning how specific proteins and RNA cargoes are packaged, determining how MVBs traffic and fuse with the plasma membrane, and understanding how target cells receive signals from microvesicles.
Enveloped Virus Budding
Viruses rely upon host cellular machinery for many functions, and many enveloped viruses usurp the ESCRT pathway to bud from cells. ESCRT-mediated virus release is best characterized for retroviruses, particularly HIV-1 (11, 39), but it is also well documented for arena-, filo-, paramyxo-, orbi-, and rhabdoviruses. There is also a recent report that Epstein-Barr virus, a herpesvirus, uses ESCRT machinery for nuclear egress (138; and see References 139 and 140). Many other classes of enveloped viruses have additional intriguing ESCRT links, although some encode their own membrane fission machinery, e.g., influenza A, (141), whereas others form external protein coats that apparently alleviate the need for ESCRT-catalyzed membrane fission, e.g., alphaviruses (142).
ESCRT-dependent viruses employ late assembly domains, often in multiple copies, to recruit early acting factors of the ESCRT pathway. The three best-characterized late assembly domains are P(T/S)AP (e.g., as in HIV-1 Gag), which binds directly to the UEV domain of TSG101(ESCRT-I) (Figure 3d); YPXnL [e.g., equine infectious anemia virus (EIAV) Gag], which binds the V domain of ALIX; and PPXY [e.g., Rous sarcoma virus (RSV) Gag], which binds the WW domains of NEDD4 family Ub ligases (11, 39). All three of these viral late domains mimic interactions used during cellular MVB vesicle and shedding microvesicle formation (see above). Viruses can also connect to the ESCRT pathway in additional ways, and elucidating these connections will likely reveal new mechanisms by which cellular factors can link into the pathway (143–147). Ub contributes to the budding of many retroviruses, including HIV-1 where Lys63-linked chains have been implicated. Gag ubiquitylation likely contributes to ALIX and ESCRT-I recruitment in some cases, but ubiquitylation of host ESCRT machinery may also be important because budding of the prototypic foamy virus remains Ub dependent, even when the Gag protein is mutated so that it lacks any lysines (148–151).
Viruses apparently require only a subset of the downstream ESCRT machinery for efficient budding (39). This is presumably because (a) viral structural proteins, unlike MVB cargoes, are capable of targeting to specific membranes, self-assembling, and generating membrane curvature (Figure 6d), and because (b) viruses do not require all of the biochemical activities that are needed during abscission, such as microtubule severing.
The ESCRT protein network is least well understood in the PPXY-dependent viruses, but the arrestin-related trafficking proteins appear to participate (8), which is consistent with known connections between yeast arrestins and Rsp5p (the sole yeast NEDD4 homolog) (7). There is also a report that ESCRT-II is required for RSV budding (152).
Viruses like EIAV that bud via YPXnL motifs use ALIX to recruit CHMP4 family members, particularly CHMP4B (19, 37, 86, 153, 154). ALIX builds up steadily throughout the entire EIAV assembly process (92), whereas ESCRT-III and VPS4 levels spike immediately prior to the membrane fission event (Figure 6e) (92, 93). This implies that physical mechanisms (such as threshold effects), generation of proper membrane curvature, or Ub transfer must induce ESCRT-III recruitment late in the process (92). ALIX functions are augmented by V domain Ub binding (27) and by additional unidentified Bro1 domain interactions (154–156).
HIV-1 budding requires CHMP4, CHMP2, and VPS4 (72, 86). These proteins can interact directly in vitro and apparently represent the minimal set of required late-acting ESCRT proteins, although CHMP3 and CHMP1 proteins can also contribute to budding efficiency (86, 157, 158). The protein-protein interactions that lead to ESCRT-I-mediated recruitment of ESCRT-III subunits are not yet fully defined, however, because several groups have reported that siRNA depletion of ESCRT-II and CHMP6 subunits does not significantly reduce virus budding efficiency (86, 159, 160). This result implies either that the ESCRT-I:ESCRT-II:CHMP6:CHMP4 relay system, which is used in MVB cargo sorting (1, 15), may be nonessential (or redundant) in virus budding or that siRNA-treated cells retain sufficient quantities of ESCRT-II and CHMP6 subunits to support budding. Conceivably, ESCRT-II functions may be most critical when bud neck deformation and ESCRT-III recruitment are difficult, as in MVB vesicle formation, but less important during virus budding and abscission when viral structural proteins or the contractile ring helps deform the membrane and stabilize the narrow membrane tubules. Alternatively, viral adaptors may recruit additional ESCRT-associated factors, such as ALIX and NEDD4 family Ub ligases, that provide additional links to ESCRT-III and reduce the ESCRT-II requirement. This issue requires further study, however, because ESCRT-II does play an essential role in recruiting ESCRT-III subunits in a reconstituted HIV-1 assembly system (158) and the complex also reportedly functions in HIV-1 RNA trafficking (161).
The position of the membrane fission site relative to the ESCRT assemblies is of considerable interest. If fission occurs on the virion-proximal side of the ESCRT-III assembly (i.e., as in traditional models for MVB vesicle formation), then there must be a mechanism for disrupting Gag:ALIX or ALIX:ESCRT-III linkages during budding. If fission occurs on the virion-distal side of the ESCRT-III assembly (i.e., as in abscission), then ESCRT factors should end up within the virion. Thus far, only ALIX has been shown to accumulate to appreciable levels within HIV-1 and EIAV virions (25, 33, 92), whereas the ESCRT-III and VPS4 proteins appear to be recycled back into the cytoplasm following membrane fission (92). However, the fate of ESCRT subunits during virus budding merits further study. There is also much to be learned about how (a) ESCRT-dependent budding functions can be negatively regulated or even bypassed in some contexts (162, 163); (b) the roles of candidate accessory factors such as actin, ALG-2 (164), sprouty 2 (165), and CC2D1 proteins (166, 167); and (c) the contribution of ESCRT pathway regulation to antiviral innate immunity (168).
The Abscission Stage of Cytokinesis
The process of cytokinesis separates daughter cells and completes cell division. Early in mammalian cell cytokinesis, the actomyosin contractile ring constricts the cleavage furrow into an intercellular bridge that contains antiparallel microtubule bundles and a midbody (Figure 6f–b). The intercellular bridge is subsequently resolved through the complex membrane fission process of abscission, which can occur on either (or both) side (s) of the midbody (reviewed in Reference 169). ALIX, ESCRT-I, both VPS4 isoforms, and nearly all of the different ESCRT-III proteins participate in abscission, likely catalyzing the membrane fission step (12, 13, 87, 94, 100, 101, 169–171). Cytokinesis appears to have been the original ancestral function of the ESCRT pathway because some crenarchaeal genera like Sulfolobus that diverged from eukaryotes several billion years ago use a minimal set of ESCRT factors for cell division but lack endosomes (172–174). Furthermore, although the ESCRT pathway is not essential for cell division in S. cerevisiae, ESCRT depletions synthetically enhance other cytokinesis defects (175).
The early acting ESCRT-I and ALIX proteins form large membrane-associated rings on either side of the mammalian midbody (Figure 6f). Both are recruited through direct interactions with the central hinge region of CEP55, which forms an unusual coiled coil that binds proline-rich motifs in ALIX and TSG101(Figure 3e) (12, 13, 77). ESCRT-III proteins then form more distal rings that appear to polymerize outward, apparently forming the large (∼17-nm) spiraling filaments visualized within intercellular bridges (Figure 6f–h) (94, 170, 171). These 17-nm filaments require CHMP2A to form and likely contain multiple ESCRT-III subunits, but the spatial organization of different subunits within the filaments remains to be determined. ESCRT-III factors, particularly IST1, then recruit the VPS4 enzymes prior to membrane fission (94, 100, 101). Membrane fission occurs ∼1 μm away from the midbody at constriction zones where the microtubules are cut (Figure 6g,h) (94, 171). ESCRT-III assemblies localize to both midbodies and cut sites but appear to be discontinuous (170).
ESCRT proteins also recruit a subset of the many other factors that localize to intercellular bridges (176). The complexity of abscission appears to explain why mammals have evolved so many different ESCRT-III subunits, and all proteins that contain MIT domains must be considered candidates for ESCRT-regulated roles in cytokinesis. For example, the ESCRT-III protein CHMP1B binds the MIT domains of the AAA ATPase spastin (Figure 4e), which severs microtubules immediately prior to abscission (77). Additional MIT domain-containing proteins recruited by ESCRT-III subunits to function in abscission include the phospholipase D–like protein, MITD1 (CHMP1A, CHMP1B, CHMP2A, and IST1) (177, 178), spartin (SPG20) (IST1) (179), and the protease calpain 7 (IST1:CHMP1B complexes) (180).
Multiple signaling and cell cycling pathways presumably help coordinate ESCRT activities during abscission, and their roles are now emerging. For example, Wnt5a signaling was recently shown to position ESCRT-III at functional sites within intercellular bridges (181). The role(s) of Ub in cytokinesis is (are) not yet well understood, but Ub concentrates at midbodies prior to abscission, and the Ub ligase BRUCE as well as the deubiquitylating enzymes UBPY and AMSH are required for abscission (182, 183). ESCRT-III proteins also help regulate abscission timing via the Aurora B-mediated abscission checkpoint (NoCut) (184). CHMP4C phosphorylation within a unique region that is absent in CHMP4A and CHMP4B promotes binding to the Borealin subunit of the chromosomal passenger complex, which activates the abscission checkpoint and thereby delays abscission until any lagging chromosomes have cleared the intercellular bridge. Thus, this system links proper chromosome segregation to ESCRT-III abscission activity via the phosphorylation state of CHMP4C.
In summary, the mammalian ESCRT pathway catalyzes a series of critical membrane fission reactions and is integrated into many important cellular processes resulting in rich biology and physiology: See the sidebar ESCRTs in Cell Biology, Development, and Disease and Table 1.
ESCRTs in Cell Biology, Development, and Disease.
The ESCRT pathway impacts developmental biology and physiology in important ways, reflecting the central roles of cellular ESCRT functions. Mammalian ESCRT factor knockouts are typically embryonically lethal (189), but partial loss of function mutations have been linked to a variety of different pathologies, including neurodegeneration and cancer, whose underlying mechanisms are often unclear (reviewed in References 130 and 189–192). ESCRT factors have also been implicated in other less well-characterized cellular functions, including autophagy, cell cycle regulation, RNA localization, and cell polarity and migration. In some cases, these functions do not obviously involve membranes and may instead reflect ways in which the ESCRT pathway is integrated with other cellular pathways. Table 1 highlights Such “noncanonical” ESCRT pathway functions and important ESCRT connections to developmental biology and disease.
Table 1. ESCRTs in cell biology, development, and disease.
| Biological phenomenon | ESCRT connection | Key references |
|---|---|---|
|
| ||
| Autophagy | ||
|
| ||
| Protein aggregate clearance | CHMP2B | 194 |
|
| ||
| Autophagosome accumulation | CHMP2B | 195 |
|
| ||
| Endosomal microautophagy | TSG101, VPS4 | 196 |
|
| ||
| Cell cycle regulation | ||
|
| ||
| Abscission checkpoint/chromosomal passenger complex | CHMP4C | 184 |
|
| ||
| Chromatin structure | CHMP1A | 197 |
|
| ||
| Centrosome/spindle maintenance | TSG101 | 198 |
| Arabidopsis Tsg101 | 199 | |
| Schizosaccharomyces pombe, Vps22 | 200 | |
| ESCRT-III, VPS4 | 87 | |
| Drosophila CHMP5 | 201 | |
|
| ||
| RNA localization | ||
|
| ||
| Maternal RNA localization | Drosophila ESCRT-II | 202 |
|
| ||
| siRNA spreading | Drosophila Vps25 | 203 |
| Drosophila Hrs | 203 | |
| HRS, TSG101 | 204 | |
| Drosophila Hrs, VPS36, Alix | 205 | |
|
| ||
| Cell polarity and migration | ||
|
| ||
| Apicobasal polarity, cell migration | ESCRT pathway | 206 |
|
| ||
| Cancer | ||
|
| ||
| Tumorigenesis and metastatic potential | HRS | 207 |
|
| ||
| Benign brain tumors | HRS | 208, 209 |
|
| ||
| Ovarian cancer | TSG101 | 210, 211 |
|
| ||
| Mammary cancer | TSG101 | 212 |
|
| ||
| Gastrointestinal stromal tumors | TSG101 | 213 |
|
| ||
| Papillary thyroid cancer | TSG101 | 214 |
|
| ||
| Metastatic tumors | TSG101 | 215 |
|
| ||
| Neoplastic transformation | Drosophila Tsg101 | 203, 216 |
|
| ||
| Hepatocellular carcinoma | VPS37A | 217 |
|
| ||
| Ovarian cancer | VPS37A | 218 |
|
| ||
| Neoplastic transformation | Drosophila Vps25 | 219 |
|
| ||
| Prostate cancer | CHMP3 | 220 |
|
| ||
| Non-small cell lung cancer | CHMP3 | 221 |
|
| ||
| Ductal pancreatic cancer | CHMP1A | 222 |
|
| ||
| Renal cell carcinoma | CHMP1A | 223 |
|
| ||
| Hyperactive epidermal growth factor receptor signaling | Drosophila Hrs | 224 |
| TSG101 | 225 | |
| CHMP6, VPS4A | 226 | |
|
| ||
| Neurodegeneration/other diseases | ||
|
| ||
| Frontotemporal dementia | UBAP1 | 227 |
| CHMP2B | 228 | |
|
| ||
| Amyotrophic lateral sclerosis | CHMP2B | 229 |
|
| ||
| Lewy body disease | CHMP2B | 230, 231 |
|
| ||
| Spongiform neurodegeneration | TSG101 | 232 |
|
| ||
| Charcot-Marie-Tooth disease | TSG101 | 233 |
| HRS:STAM | 234 | |
|
| ||
| Hereditary spastic paraplegia | CHMP1B | 235 |
|
| ||
| Autosomal dominant cataracts | CHMP4B | 236 |
Models for Membrane Constriction and Fission
It is of fundamental importance to understand how the ESCRT machinery draws opposing membranes together and mediates fission. ESCRT-III proteins perform central roles in these processes, apparently by forming filaments within the necks of budding vesicles, viruses, and intercellular bridges (16, 80, 81, 169). Many ESCRT-III proteins can form filaments in vitro (37, 66, 91, 111, 185, 186) and in cells (88, 89, 157, 171), and these filaments often wrap up into helical tubes or spirals. The role of VPS4 in fission is less clear because the enzyme is not required for single rounds of vesicle formation in a reconstituted yeast MVB system (85), but appears immediately prior to virus budding (Figure 6e) (92, 93) and abscission (94), where it seems to be required for fission (92, 93).
ESCRT-III filaments likely function in pairs (or contain paired strands) because both known ESCRT-III-recruiting complexes, ESCRT-II and ALIX, contain two ESCRT-III-binding sites (37, 54, 56, 57, 62) and because yeast Snf7p (CHMP4) can form paired helical filaments on supported lipid monolayers (187). One recurring assembly is a 3–4-nm wide filament, which has been observed in several ESCRT-III assemblies in vitro, including helical CHMP2A:CHMP3 copolymers (157), CHMP4B and Snf7p filaments (37, 187), and lipidated IST1 tubes (J. McCullough & A. Frost unpublished findings), as well as in cellular CHMP2B and CHMP4A assemblies (88, 89). These filaments can further associate into single, paired, or multistart helical tubes with diameters of 40–400 nm. However, distinct ESCRT-III filaments and tubes of differing widths and morphologies have been reported in other cases (111, 171, 185, 186), and a clear consensus structure has therefore yet to emerge. In particular, the 17-nm ESCRT-III filaments that spiral toward the cut sites within the intercellular bridge (171) must either contain multiple copies of smaller ESCRT-III filaments or be different structures entirely.
Another key issue is how ESCRT-III filaments can promote membrane constriction and fission. As illustrated in Figure 7a, a leading model holds that ESCRT-III filaments spiral inward and promote membrane constriction by pulling the opposing membranes together over the resulting “domes” (80, 188). Spiraling would require ESCRT-III subunits to make nonidentical contacts between consecutive coils, which is unusual behavior because protein:protein interfaces typically do not accommodate the required continuum of different contacts. Nevertheless, ESCRT-III tubes can taper into terminal dome-like structures in vitro, demonstrating that such constricting spirals are physically possible (66, 91). Furthermore, cryoelectron microscopy tomograms of the ESCRT coats that mediate cleavage furrow ingression and cell division in Sulfolobus suggest an “hourglass” model in which two opposing domes of spiraling ESCRT-III filaments create a symmetric cleavage furrow, pulling the membranes on either side of the furrow down into a single fission point (G. Jensen, personal communication). Spiraling dome models are attractive because they nicely explain how ESCRT-III assemblies can mediate membrane constriction and fission across very different size scales, ranging from the necks of budding MVB vesicles (∼25 nm) and retroviruses (∼50 nm) to mammalian intercellular bridges and crenarchaeal cells (each ∼1 μm). In all cases, ESCRT-III assemblies could initially form rings that match the starting diameter and then spiral closed toward the fission point. In principle, VPS4 could help promote ESCRT-III filament constriction (188) or remodel the dome to promote the hemifission to fission transition (93).
Figure 7.
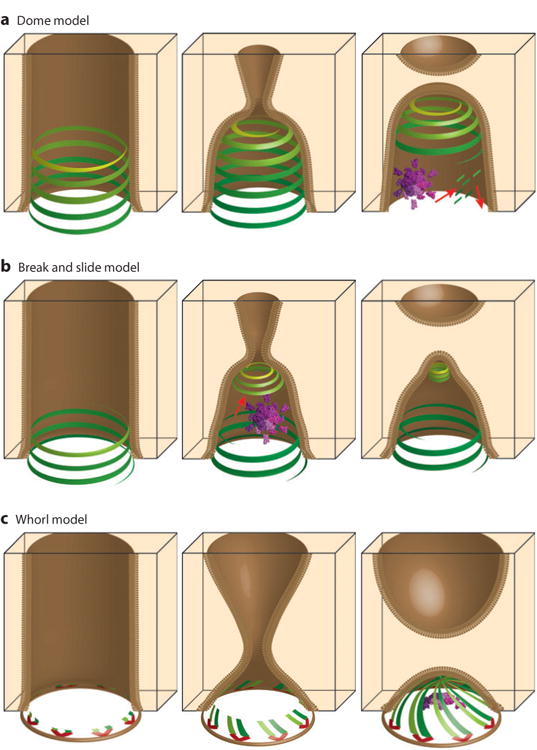
Models for ESCRT-mediated membrane fission reactions. (a) The “dome” model for multivesicular body (MVB) vesicle budding (188), (b) the “break and slide” model for intercellular bridge constriction and abscission (94, 170), and (c) the “whorl” model for MVB vesicle budding (63). ESCRT-I:ESCRT-II supercomplexes are shown in red; ESCRT-III filaments are shown in shades of green and yellow; VPS4 enzyme complexes are shown in purple; and red arrows denote motion. Panels in the left column show ESCRT factors assembling within membrane tubules (brown). Panels in the central column show models for membrane constriction. Panels in the right column show models for the membrane fission step (see text for details).
A variation on the dome model (Figure 7b) was proposed to explain the observation that ESCRT-III proteins form discontinuous structures within intercellular bridges, first appearing as wide rings that are proximal to the midbody and later concentrating at the distal abscission sites (94, 170). This “break and slide” model holds that (a) ESCRT-III filaments nucleate near the midbody and initially begin to polymerize as wide spirals; (b) these wide spirals are unstable owing to their expanded diameters and eventually break, apparently assisted by VPS4 activity; (c) the unanchored spirals then slide along the membrane and constrict into lower-energy helices that narrow the intercellular bridge to the point where a fission dome can form. This model nicely explains the observation that ESCRT-III components concentrate at two different sites, and the model is supported by computational analyses, but ESCRT-III filament sliding has yet to be visualized directly.
Finally, Hurley and colleagues have recently proposed another significant variation on the dome model, termed the “whorl” model (Figure 7c). They propose that the necks of MVB vesicles are ringed with 6–10 copies of the ESCRT-I:ESCRT-II supercomplex that align with their long axes parallel to the neck and that each nucleate two ESCRT-III filaments (63). The filaments then grow toward the center of the neck like spokes, forming a multistart whorl that constricts the membrane along one face. VPS4 assemblies could then help organize the central site, where the growing filaments ultimately meet. This model is consistent with a large body of biophysical and structural data on MVB vesicle formation (63) but is more difficult to envision in the cases of mammalian abscission and crenarchaeal cytokinesis because the whorling ESCRT-III filaments would need to extend ∼500 nm to meet at the center of the midbody. Hence, a hybrid model seems more attractive in those cases.
In summary, ESCRT-III filaments play central roles in membrane fission reactions, and it is critical to define (a) their molecular structure(s); (b) how the different ESCRT-III subunits copolymerize and why so many different subunits are required, particularly during cytokinesis; (c) the mechanism by which ESCRT-III filaments constrict membranes; and (d) whether VPS4 enzymes play an active role in this process or instead function primarily to disassemble the ESCRT-III filaments following fission.
Supplementary Material
Acknowledgments
We apologize to our many colleagues whose primary references we were unable to cite owing to space limitations. We thank Matt Lalonde for help with artwork. Our ESCRT research is supported by National Institute of Health grants AI051174 and GM082545 (W.I.S.) and American Cancer Society grant PF-11-279-01-CCG (L.A.C.).
Glossary
- ESCRT
endosomal sorting complexes required for transport
- Intercellular bridge
a thin membrane tubule ∼1 μm in diameter that connects daughter cells during cytokinesis
- Multivesicular body (MVB)
a late endosome filled with small intraluminal vesicles
- Phosphoinositides
phospholipids that help define different cellular membranes, including endosomes [PI(3)P] and plasma membranes [PI(4,5)P2]
- Exosomes
small extracellular vesicles (50–100 nm in diameter) that are released when multivesicular bodies fuse with the plasma membrane
- Shedding microvesicles
small extracellular vesicles (50–80 nm in, diameter) that bud directly from the plasma embrane
- Late assembly domains
short viral protein motifs that recruit ESCRT factors, including YPXnL:ALIX, P (T/S)AP:TSG101 (ESCRT-I), and PPXY:NEDD4 proteins
- NEDD4 proteins
a family of nine human ubiquitin E3 ligases, many of which are associated with the ESCRT pathway
- Ubiquitin-binding domains (UBDs)
these domains bind ubiquitin and are common in adaptors and early acting ESCRT factors
- ESCRT-III proteins
a homologous family of 12 small filament-forming human proteins named CHMP (charged multivesicular body protein) or IST1
- Microtubule interacting and transport interaction motifs (MIMs)
motifs within the C-terminal tails of ESCRT-III proteins that are bound by MIT domains
- Microtubule interacting and transport (MIT)
domains found in more than a dozen human proteins that bind ESCRT-III filaments
- AAA ATPases
ATPases associated with diverse cellular activities that form ring-like assemblies that remodel cellular macromolecules
- Midbody
a protein-dense structure in the center of the intercellular bridge
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–47. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raiborg C, Stenmark H. Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell Struct Funct. 2002;27:403–8. doi: 10.1247/csf.27.403. [DOI] [PubMed] [Google Scholar]
- 4.Clague MJ, Urbé S. Hrs function: Viruses provide the clue. Trends Cell Biol. 2003;13:603–6. doi: 10.1016/j.tcb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–85. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14:654–55. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- 7.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–25. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol. 2011;85:3546–56. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1–mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–51. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsch S, Müller B, Krausslich HG. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–97. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–31. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 12.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–12. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 13.MoritaE, Sandrin V, Chung HY, Morham SG, Gygi SP, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–27. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–80. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–9. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:1–26. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell. 2010;143:875–87. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 19.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA. 2008;105:7687–91. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mu R, Dussupt V, Jiang J, Sette P, Rudd V, et al. Two distinct binding modes define the interaction of Brox with the C-terminal tails of CHMP5 and CHMP4B. Structure. 2012;20:887–98. doi: 10.1016/j.str.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadoul R. Do Alix and ALG-2 really control endosomes for better or for worse? Biol Cell. 2006;98:69–77. doi: 10.1042/BC20050007. [DOI] [PubMed] [Google Scholar]
- 22.Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, et al. HD-PTP: a novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun. 2000;278:671–78. doi: 10.1006/bbrc.2000.3870. [DOI] [PubMed] [Google Scholar]
- 23.Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci USA. 2008;105:6308–13. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dores MR, Chen B, Lin H, Soh UJ, Paing MM, et al. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol. 2012;197:407–19. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–99. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–47. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, et al. ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. 2012;23:1247–54. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 29.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol. 2011;85:632–37. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefani F, Zhang L, Taylor S, Donovan J, Rollinson S, et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol. 2011;21:1245–50. doi: 10.1016/j.cub.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J. 2010;432:525–34. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, et al. Activation of the retroviral budding factor ALIX. J Virol. 2011;85:9222–26. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, et al. The protein network of HIV budding. Cell. 2003;114:701–13. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Serrano J, Yaravoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12414–19. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt MH, Dikic I, Bogler O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J Biol Chem. 2005;280:3414–25. doi: 10.1074/jbc.M409839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dejournett RE, Kobayashi R, Pan S, Wu C, Etkin LD, et al. Phosphorylation of the proline-rich domain of Xp95 modulates Xp95 interaction with partner proteins. Biochem J. 2007;401:521–31. doi: 10.1042/BJ20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–56. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattei S, Klein G, Satre M, Aubry L. Trafficking and developmental signaling: Alix at the cross-roads. Eur J Cell Biol. 2006;85:925–36. doi: 10.1016/j.ejcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Weiss ER, Göttlinger H. The role of cellular factors in promoting HIV budding. J Mol Biol. 2011;410:525–33. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Souza RF, Aravind L. UMA and MABP domains throw light on receptor endocytosis and selection of endosomal cargoes. Bioinformatics. 2010;26:1477–80. doi: 10.1093/bioinformatics/btq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agromayor M, Soler N, Caballe A, Kueck T, Freund SM, et al. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–28. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsunematsu T, Yamauchi E, Shibata H, Maki M, Ohta T, Konishi H. Distinct functions of human MVB12A and MVB12B in the ESCRT-I dependent on their posttranslational modifications. Biochem Biophys Res Commun. 2010;399:232–37. doi: 10.1016/j.bbrc.2010.07.060. [DOI] [PubMed] [Google Scholar]
- 44.Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 45.Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, et al. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–26. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kostelansky MS, Schluter C, Tam YY, Lee S, Ghirlando R, et al. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–98. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–89. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 48.Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J Biol Chem. 2004;279:28689–96. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- 49.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc Natl Acad Sci USA. 2001;98:7724–29. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, et al. Crystallographic and functional analysis of the ESCRT-I /HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–47. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–17. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boura E, Hurley JH. Structural basis for membrane targeting by the MVB12-associated β-prism domain of the human ESCRT-I MVB12 subunit. Proc Natl Acad Sci USA. 2012;109:1901–6. doi: 10.1073/pnas.1117597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–13. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–89. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 56.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–69. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Hierro A, Sun J, Rusnak AS, Kim J, Prag G, et al. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–25. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- 58.Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–32. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- 59.Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–6. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 60.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, et al. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Im YJ, Wollert T, Boura E, Hurley JH. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev Cell. 2009;17:234–43. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–83. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, et al. Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure. 2012;20:874–86. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malerod L, Stuffers S, Brech A, Stenmark H. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 2007;8:1617–29. doi: 10.1111/j.1600-0854.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 65.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–30. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 66.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, et al. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–62. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514–24. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–79. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 69.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 70.Lata S, Roessle M, Solomons J, Jamin M, Göttlinger HG, et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:816–25. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–44. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 72.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 73.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–39. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 74.Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19:2661–72. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, et al. Interactions of the hu-man LIP5 regulatory protein with endosomal sorting complexes required for transport. J Biol Chem. 2012;287:43910–26. doi: 10.1074/jbc.M112.417899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Z, Vild C, Ju J, Zhang X, Liu J, et al. Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA–ATPase regulator Vta1 in the multivesicular body pathway. J Biol Chem. 2012;287:43899–908. doi: 10.1074/jbc.M112.390724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–86. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282:30929–37. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 79.Solomons J, Sabin C, Poudevigne E, Usami Y, Hulsik DL, et al. Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure. 2011;19:1149–59. doi: 10.1016/j.str.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–66. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–82. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 83.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–89. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 85.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–77. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–42. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107:12889–94. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–86. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hill CP, Babst M. Structure and function of the membrane deformation AAA ATPase Vps4. Biochim Biophys Acta. 2012;1823:172–81. doi: 10.1016/j.bbamcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lata S, Schoehn G, Jain A, Pires R, Piehler J, et al. Helical structures of ESCRT-III are disassem-bled by VPS4. Science. 2008;321:1354–57. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–74. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 94.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–51. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–69. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci USA. 2005;102:13813–18. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377:364–77. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 99.Merrill SA, Hanson PI. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem. 2010;285:35428–38. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–87. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20:1360–73. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol Biol Cell. 2008;19:465–74. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19:475–84. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, et al. Structural basis of Vta1 function in the multi-vesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, et al. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–17. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang D, Hurley JH. Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure. 2010;18:976–84. doi: 10.1016/j.str.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol. 2011;23:452–57. doi: 10.1016/j.ceb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–66. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 109.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;7:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schiel JA, Prekeris R. Making the final cut—mechanisms mediating the abscission step of cytokinesis. Sci World J. 2010;10:1424–34. doi: 10.1100/tsw.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fyfe I, Schuh AL, Edwardson JM, Audhya A. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J Biol Chem. 2011;286:34262–70. doi: 10.1074/jbc.M111.266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103:2641–46. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boura E, Ivanov V, Carlson LA, Mizuuchi K, Hurley JH. Endosomal sorting complex required for transport (ESCRT) complexes induce phase-separated microdomains in supported lipid bilayers. J Biol Chem. 2012;287:28144–51. doi: 10.1074/jbc.M112.378646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–34. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 116.Goursot A, Mineva T, Bissig C, Gruenberg J, Salahub DR. Structure, dynamics, and energetics of lysobisphosphatidic acid (LBPA) isomers. J Phys Chem B. 2010;114:15712–20. doi: 10.1021/jp108361d. [DOI] [PubMed] [Google Scholar]
- 117.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–47. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 118.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.RaiborgC, Stenmark H. The ESCRT machinery in endosomal sortingofubiquitylated membrane proteins. Nature. 2009;458:445–52. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 121.Ren X, Kloer DP, Kim YC, Ghirlando R, Saidi LF, et al. Hybrid structural model of the complete human ESCRT-0 complex. Structure. 2009;17:406–16. doi: 10.1016/j.str.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sachse M, Urbé S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–28. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degrada-tive protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci. 2006;119:2414–24. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- 124.Wright MH, Berlin I, Nash PD. Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem Biophys. 2011;60:39–46. doi: 10.1007/s12013-011-9181-9. [DOI] [PubMed] [Google Scholar]
- 125.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–48. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 127.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–84. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 128.Clague MJ, Coulson JM, Urbeé S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–86. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 129.von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–45. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat Cell Biol. 2012;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- 131.Eden ER, Burgoyne T, Edgar JR, Sorkin A, Futter CE. The relationship between ER-multivesicular body membrane contacts and the ESCRT machinery. Biochem Soc Trans. 2012;40:464–68. doi: 10.1042/BST20110774. [DOI] [PubMed] [Google Scholar]
- 132.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 133.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80:1948–57. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 135.Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 136.Grembecka J, Cierpicki T, Devedjiev Y, Derewenda U, Kang BS, et al. The binding of the PDZ tandem of syntenin to target proteins. Biochemistry. 2006;45:3674–83. doi: 10.1021/bi052225y. [DOI] [PubMed] [Google Scholar]
- 137.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–59. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee CP, Liu PT, Kung HN, Su MT, Chua HH, et al. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mettenleiter TC, Mu¨ller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 2013;15:170–78. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 140.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–46. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting Alphavirus budding. J Virol. 2007;81:13631–39. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Popov S, Popova E, Inoue M, Göttlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83:7185–93. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, et al. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Usami Y, Popov S, Popova E, Göttlinger HG. Efficient and specific rescue of human immunode-ficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chung HY, Morita E, von Schwedler U, Müller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–97. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harrison MS, Schmitt PT, Pei Z, Schmitt AP. Role of ubiquitin in parainfluenza virus 5 particle formation. J Virol. 2012;86:3474–85. doi: 10.1128/JVI.06021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vogt VM. Ubiquitin in retrovirus assembly: actor or bystander? Proc Natl Acad Sci USA. 2000;97:12945–47. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 150.Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Göttlinger H. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–84. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- 151.Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci USA. 2007;104:20031–36. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pincetic A, Medina G, Carter C, Leis J. Avian sarcoma virus and human immunodeficiency virus, type 1use different subsets of ESCRT proteins tofacilitate the budding process. J Biol Chem. 2008;283:29822–30. doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–52. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 154.Usami Y, Popov S, Göttlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–22. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sette P, Mu R, Dussupt V, Jiang J, Snyder G, et al. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure. 2011;19:1485–95. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS ONE. 2011;6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Effantin G, Dordor A, Sandrin V, Martinelli C, Sundquist WI, et al. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell Microbiol. 2013;15:213–26. doi: 10.1111/cmi.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci USA. 2012;109:16928–33. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhadina M, Bieniasz PD. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 2010;6:e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ghoujal B, Milev MP, Ajamian L, Abel K, Mouland AJ. ESCRT-II's involvement in HIV-1 genomic RNA trafficking and assembly. Biol Cell. 2012;104:706–21. doi: 10.1111/boc.201200021. [DOI] [PubMed] [Google Scholar]
- 162.Gan X, Gould SJ. Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol Biol Cell. 2011;22:817–30. doi: 10.1091/mbc.E10-07-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Popova E, Popov S, Göttlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol. 2010;84:6590–97. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maki M, Suzuki H, Shibata H. Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Sci China Life Sci. 2011;54:770–79. doi: 10.1007/s11427-011-4204-8. [DOI] [PubMed] [Google Scholar]
- 165.Medina GN, Ehrlich LS, Chen MH, Khan MB, Powell MD, Carter CA. Sprouty 2binds ESCRT-II factor Eap20 and facilitates HIV-1 Gag release. J Virol. 2011;85:7353–62. doi: 10.1128/JVI.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Göttlinger HG. Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A. J Virol. 2012;86:3746–56. doi: 10.1128/JVI.06539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, et al. CC2D1A is a regulator of ESCRT-III CHMP4B. J Mol Biol. 2012;419:75–88. doi: 10.1016/j.jmb.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kuang Z, Seo EJ, Leis J. The mechanism of inhibition of retrovirus release from cells by interferon induced gene ISG15. J Virol. 2011;85:7153–61. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–40. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 170.Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys J. 2012;102:2309–20. doi: 10.1016/j.bpj.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–20. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 172.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–46. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–13. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle forma-tion systems in Archaea. Nat Rev Microbiol. 2010;8:731–41. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McMurray MA, Stefan CJ, Wemmer M, Odorizzi G, Emr SD, Thorner J. Genetic interactions with mutations affecting septin assembly reveal ESCRT functions in budding yeast cytokinesis. Biol Chem. 2011;392:699–712. doi: 10.1515/BC.2011.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–47. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 177.Hadders MA, Agromayor M, Obita T, Perisic O, Caballe A, et al. ESCRT-III binding protein MITD1 is involved in cytokinesis and has an unanticipated PLD fold that binds membranes. Proc Natl Acad Sci USA. 2012;109:17424–29. doi: 10.1073/pnas.1206839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee S, Chang J, Renvoisé B, Tipirneni A, Yang S, Blackstone C. MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol Biol Cell. 2012;23:4347–61. doi: 10.1091/mbc.E12-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Renvoisé B, Parker RL, Yang D, Bakowska JC, Hurley JH, Blackstone C. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol Biol Cell. 2010;21:3293–303. doi: 10.1091/mbc.E09-10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Maemoto Y, Osako Y, Goto E, Nozawa E, Shibata H, Maki M. Calpain-7 binds to CHMP1B at its second α-helical region and forms a ternary complex with IST1. J Biochem. 2011;150:411–21. doi: 10.1093/jb/mvr071. [DOI] [PubMed] [Google Scholar]
- 181.Fumoto K, Kikuchi K, Gon H, Kikuchi A. Wnt5a signaling controls cytokinesis by positioning ESCRT-III to the proper site at the midbody. J Cell Sci. 2012;125:4822–32. doi: 10.1242/jcs.108142. [DOI] [PubMed] [Google Scholar]
- 182.Mukai A, Mizuno E, Kobayashi K, Matsumoto M, Nakayama KI, et al. Dynamic regulation of ubiquitylation and deubiquitylation at the central spindle during cytokinesis. J Cell Sci. 2008;121:1325–33. doi: 10.1242/jcs.027417. [DOI] [PubMed] [Google Scholar]
- 183.Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–45. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 184.Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–25. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, et al. Structure and disas-sembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–56. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 186.Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS ONE. 2011;6:e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–71. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 188.Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Michelet X, Djeddi A, Legouis R. Developmental and cellular functions of the ESCRT machinery in pluricellular organisms. Biol Cell. 2010;102:191–202. doi: 10.1042/BC20090145. [DOI] [PubMed] [Google Scholar]
- 190.Stuffers S, Brech A, Stenmark H. ESCRT proteins in physiology and disease. Exp Cell Res. 2009;315:1619–26. doi: 10.1016/j.yexcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 191.Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–72. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- 192.Rusten TE, Simonsen A. ESCRT functions in autophagy and associated disease. Cell Cycle. 2008;7:1166–72. doi: 10.4161/cc.7.9.5784. [DOI] [PubMed] [Google Scholar]
- 193.Murk JL, Humbel BM, Ziese U, Griffith JM, Posthuma G, et al. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc Natl Acad Sci USA. 2003;100:13332–37. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophago-some accumulation and neurodegeneration. Curr Biol. 2007;17:1561–67. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 196.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–39. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–93. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 198.Xie W, Li L, Cohen SN. Cell cycle–dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci USA. 1998;95:1595–600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, et al. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–89. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- 200.Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell. 2005;9:63–73. doi: 10.1016/j.devcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 201.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Irion U, St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445:554–58. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overpro-liferation by regulating notch trafficking. Dev Cell. 2005;9:687–98. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 204.Lee YS, Pressman S, Andress AP, Kim K, White JL, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–56. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–49. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 206.Lobert VH, Stenmark H. Cell polarity and migration: emerging role for the endosomal sorting machinery. Physiology. 2011;26:171–80. doi: 10.1152/physiol.00054.2010. [DOI] [PubMed] [Google Scholar]
- 207.Tamai K, Toyoshima M, Tanaka N, Yamamoto N, Owada Y, et al. Loss of Hrs in the central nervous system causes accumulation of ubiquitinated proteins and neurodegeneration. Am J Pathol. 2008;173:1806–17. doi: 10.2353/ajpath.2008.080684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gutmann DH, Haipek CA, Burke SP, Sun CX, Scoles DR, Pulst SM. The NF2 interactor, hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), associates with merlin in the ‘open’ conformation and suppresses cell growth and motility. Hum Mol Genet. 2001;10:825–34. doi: 10.1093/hmg/10.8.825. [DOI] [PubMed] [Google Scholar]
- 209.Scoles DR, Nguyen VD, Qin Y, Sun CX, Morrison H, et al. Neurofibromatosis 2 (NF2) tumor suppressor schwannomin and its interacting protein HRS regulate STAT signaling. Hum Mol Genet. 2002;11:3179–89. doi: 10.1093/hmg/11.25.3179. [DOI] [PubMed] [Google Scholar]
- 210.Young TW, Mei FC, Rosen DG, Yang G, Li N, et al. Up-regulation of tumor susceptibility gene 101 protein in ovarian carcinomas revealed by proteomics analyses. Mol Cell Proteomics. 2007;6:294–304. doi: 10.1074/mcp.M600305-MCP200. [DOI] [PubMed] [Google Scholar]
- 211.Young TW, Rosen DG, Mei FC, Li N, Liu J, et al. Up-regulation of tumor susceptibility gene 101 conveys poor prognosis through suppression of p21 expression in ovarian cancer. Clin Cancer Res. 2007;13:3848–54. doi: 10.1158/1078-0432.CCR-07-0337. [DOI] [PubMed] [Google Scholar]
- 212.Oh KB, Stanton MJ, West WW, Todd GL, Wagner KU. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene. 2007;26:5950–59. doi: 10.1038/sj.onc.1210401. [DOI] [PubMed] [Google Scholar]
- 213.Koon N, Schneider-Stock R, Sarlomo-Rikala M, Lasota J, Smolkin M, et al. Molecular targets for tumour progression in gastrointestinal stromal tumours. Gut. 2004;53:235–40. doi: 10.1136/gut.2003.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu RT, Huang CC, You HL, Chou FF, Hu CC, et al. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene. 2002;21:4830–37. doi: 10.1038/sj.onc.1205612. [DOI] [PubMed] [Google Scholar]
- 215.Li L, Cohen SN. tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–29. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 216.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 217.Xu Z, Liang L, Wang H, Li T, Zhao M. HCRP1, a novel gene that is downregulated in hepato-cellular carcinoma, encodes a growth-inhibitory protein. Biochem Biophys Res Commun. 2003;311:1057–66. doi: 10.1016/j.bbrc.2003.10.109. [DOI] [PubMed] [Google Scholar]
- 218.Wittinger M, Vanhara P, El-Gazzar A, Savarese-Brenner B, Pils D, et al. hVps37A status affects prognosis and cetuximab sensitivity in ovarian cancer. Clin Cancer Res. 2011;17:7816–27. doi: 10.1158/1078-0432.CCR-11-0408. [DOI] [PubMed] [Google Scholar]
- 219.Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–20. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 220.Wilson EM, Oh Y, Hwa V, Rosenfeld RG. Interaction of IGF-binding protein-related protein 1 with a novel protein, neuroendocrine differentiation factor, results in neuroendocrine differentiation of prostate cancer cells. J Clin Endocrinol Metab. 2001;86:4504–11. doi: 10.1210/jcem.86.9.7845. [DOI] [PubMed] [Google Scholar]
- 221.Walker GE, Antoniono RJ, Ross HJ, Paisley TE, Oh Y. Neuroendocrine-like differentiation of non-small cell lung carcinoma cells: regulation by cAMP and the interaction of mac25/IGFBP-rP1 and 25.1. Oncogene. 2006;25:1943–54. doi: 10.1038/sj.onc.1209213. [DOI] [PubMed] [Google Scholar]
- 222.Li J, Belogortseva N, Porter D, Park M. Chmp1A functions as a novel tumor suppressor gene in human embryonic kidney and ductal pancreatic tumor cells. Cell Cycle. 2008;7:2886–93. doi: 10.4161/cc.7.18.6677. [DOI] [PubMed] [Google Scholar]
- 223.You Z, Xin Y, Liu Y, Sun J, Zhou G, et al. Chmp1A acts as a tumor suppressor gene that inhibits proliferation of renal cell carcinoma. Cancer Lett. 2012;319:190–96. doi: 10.1016/j.canlet.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 224.Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–69. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 225.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–58. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 226.Zheng ZY, Cheng CM, Fu XR, Chen LY, Xu L, et al. CHMP6 and VPS4A mediate the recycling of Ras to the plasma membrane to promote growth factor signaling. Oncogene. 2012;31:4630–38. doi: 10.1038/onc.2011.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rollinson S, Rizzu P, Sikkink S, Baker M, Halliwell N, et al. Ubiquitin associated protein 1 is a risk factor for frontotemporal lobar degeneration. Neurobiol Aging. 2009;30:656–65. doi: 10.1016/j.neurobiolaging.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 229.Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–77. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 230.Tanikawa S, Mori F, Tanji K, Kakita A, Takahashi H, Wakabayashi K. Endosomal sorting related protein CHMP2B is localized in Lewy bodies and glial cytoplasmic inclusions in α-synucleinopathy. Neurosci Lett. 2012;527:16–21. doi: 10.1016/j.neulet.2012.08.035. [DOI] [PubMed] [Google Scholar]
- 231.Kurashige T, Takahashi T, Yamazaki Y, Hiji M, Izumi Y, et al. Localization of CHMP2B-immunoreactivity in the brainstem of Lewy body disease. Neuropathology. 2012 doi: 10.1111/j.1440-1789.2012.01346.x. [DOI] [PubMed] [Google Scholar]
- 232.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18:1129–42. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL. SIMPLE interacts with NEDD4 and TSG101: evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J Neurosci Res. 2005;82:43–50. doi: 10.1002/jnr.20628. [DOI] [PubMed] [Google Scholar]
- 234.Lee SM, Chin LS, Li L. Charcot-Marie-Tooth disease–linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J Cell Biol. 2012;199:799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 236.Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, et al. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.