Abstract
Chromosome instability (CIN), a common feature of solid tumors, promotes tumor evolution and increases drug resistance during therapy. We previously demonstrated that loss of the pRB tumor suppressor causes changes in centromere structure and generates CIN. However, the reason for, and significance of, this change was unclear. Here we show that defects in cohesion are key to the pRB-loss phenotype. pRB loss alters H4K20 methylation, a prerequisite for efficient establishment of cohesion at centromeres. Changes in cohesin regulation are first evident during S-phase where they compromise replication and increase DNA damage. Ultimately such changes compromise mitotic fidelity in pRB-deficient cells. Remarkably, increasing cohesion suppressed all of these phenotypes and dramatically reduced CIN in cancer cells lacking functional pRB. These data explain how loss of pRB undermines genomic integrity. Given the frequent functional inactivation of pRB in cancer, conditions that increase cohesion may provide a general strategy to suppress CIN.
Keywords: Retinoblastoma, CIN, cohesin, Wapl, merotely, nucleoside
Introduction
Whole chromosome instability (CIN), defined as the frequent gains and losses of whole chromosomes (Lengauer et al., 1997), results from defects in mitotic fidelity (Holland and Cleveland, 2009) and contributes to the aberrations in chromosome number common in solid tumors (Albertson et al., 2003; Beroukhim et al., 2000; Hanahan and Weinberg, 2000; Thompson and Compton, 2011). Such changes result in aneuploidy, promote genomic heterogeneity, and have important implications in cancer. It has been demonstrated that the constant “shuffling” of genomic content through CIN facilitates the loss of heterozygosity of tumor suppressors and increases copy number of oncogenes (Baker et al., 2009). The genomic diversity generated by CIN promotes tumor evolution and the development of cancer cells that can adapt to become resistant to therapeutics and more prone to tumor relapse. Consequently, CIN correlates with poor patient prognosis (Choi et al., 2009; Gao et al., 2007; Heilig et al., 2010; Kuukasjarvi et al., 1997; McClelland et al., 2009; Nowell, 1976; Rajagopalan and Lengauer, 2004; Swanton et al., 2009). However, the molecular mechanisms that cause CIN and aneuploidy in tumor cells remain poorly understood and this lack of understanding has limited the development of therapeutics to target this feature of cancer.
Numerous regulators of mitotic fidelity have been shown experimentally to impact whole chromosome instability, including those directly involved in chromosome segregation and the mitotic checkpoint (McGranahan et al., 2012; Thompson and Compton, 2011). However, an understanding of the molecular causes of genome instability in tumor cells is complicated by the fact that mutations in these pathways are rare in cancers and efforts to identify a single mitotic protein or pathway that is commonly altered in sporadic cancers have been unsuccessful (Negrini et al., 2010; Rajagopalan and Lengauer, 2004; Thompson and Compton, 2011).
Recently we demonstrated that depletion of the pRB tumor suppressor from diploid, chromosomally stable human cells causes rates of chromosome missegregation that are comparable with those seen in CIN cells (Manning et al., 2010). As pRB pathway lesions are common in human cancers (Knudsen and Wang, 2010), this may represent a broadly relevant mechanism by which cancer cells become CIN. However, the precise mechanism by which pRB loss causes this type of genomic instability is unclear. Several possibilities have been suggested; including that inactivation of pRB impacts chromosome structure and additionally causes changes in the expression and localization of several mitotic regulators. Such changes, alone or in combination, might explain the high rate of segregation errors present following pRB loss.
Here we show that cohesin function is altered by pRB loss. We demonstrate that pRB depletion alters the distribution of cohesin on chromatin and that changes in histone methyl marks that promote cohesin enrichment at heterochromatin contribute to this difference. Changes in chromosome cohesion were first evident in pRB-depleted cells during S phase (at the time when cohesion is normally established) and persist into mitosis, suggesting that the effects of pRB loss on mitotic progression may be a delayed consequence of changes that occur during S phase or earlier. pRB-depleted cells display several phenotypes that would be expected in cells with reduced cohesin (compromised cohesion, altered DNA replication dynamics, increased DNA damage, and reduced mitotic fidelity) and we show that all of these phenotypes were suppressed when cohesin stability on chromatin was enhanced. Moreover, promoting cohesion was sufficient to reduce CIN in pRB-depleted cells as well as in a panel of cancer cell lines that carry pRB pathway lesions. Together, these results suggest that changes in cohesin function are key to the genesis of CIN in pRB-compromised cells. This work suggests that manipulation of chromosome cohesion may represent a powerful approach to target CIN in cancer cells.
Results
Changes in chromatin structure following pRB loss originate in S phase
Merotelic attachments, where a single chromosome is associated with microtubules from both spindle poles, are a predominant cause of chromosome segregation errors in cancer (Cimini et al., 2001; Thompson and Compton, 2008). Erroneously attached chromosomes lag behind during anaphase segregation and have an increased likelihood of being missegregated. In cells depleted of pRB by treatment with pooled siRNAs, ~25% of cells exhibited lagging chromosomes (Figure S21 & (Manning et al., 2010)). Similar results were obtained using five independent siRNA constructs or, alternatively, following lentiviral infection of one of three shRNA hairpins, each targeting a different pRB-specific sequence. Scrambled siRNA or lentiviral constructs were used as controls respectively (Supplemental Table 1, Supplemental Figure 1). For the experiments described below, pRB depletion was achieved using pooled siRNA constructs, the results were confirmed with independent targeting constructs, and compared to a scrambled control sequence.
Previously, we proposed that the chromosome segregation errors seen in pRB-depleted cells are caused by changes in chromatin structure near the centromere. To quantify such changes in chromosome structure following pRB loss we measured chromatin compaction and cohesion. pRB was depleted from two diploid, genomically stable human cell lines (hTERT-RPE and HCT116) and a dual-colored, Fluorescence in situ hybridization (FISH)-based assay was used to label defined regions on both arms of Chromosome 16. Sister chromatid cohesion (the distance between replicated foci of a single color; inter-chromosome distance) was measured from the time that loci were replicated (S-phase) until M-phase. Gross changes in chromosome compaction (the distance between green and red foci; intra-chromosome distance) were measured throughout the cell cycle. To minimize any secondary effects resulting from continued proliferation in the absence of pRB, the assay was performed at 24 hours after siRNA transfection. Cell cycle staging was based on the replicative state of Chromosome 16 and DAPI staining, enabling cell cycle-dependent changes to be examined in the absence of drug treatment. Comparable results were found in both cell lines (Figures 1 & S2).
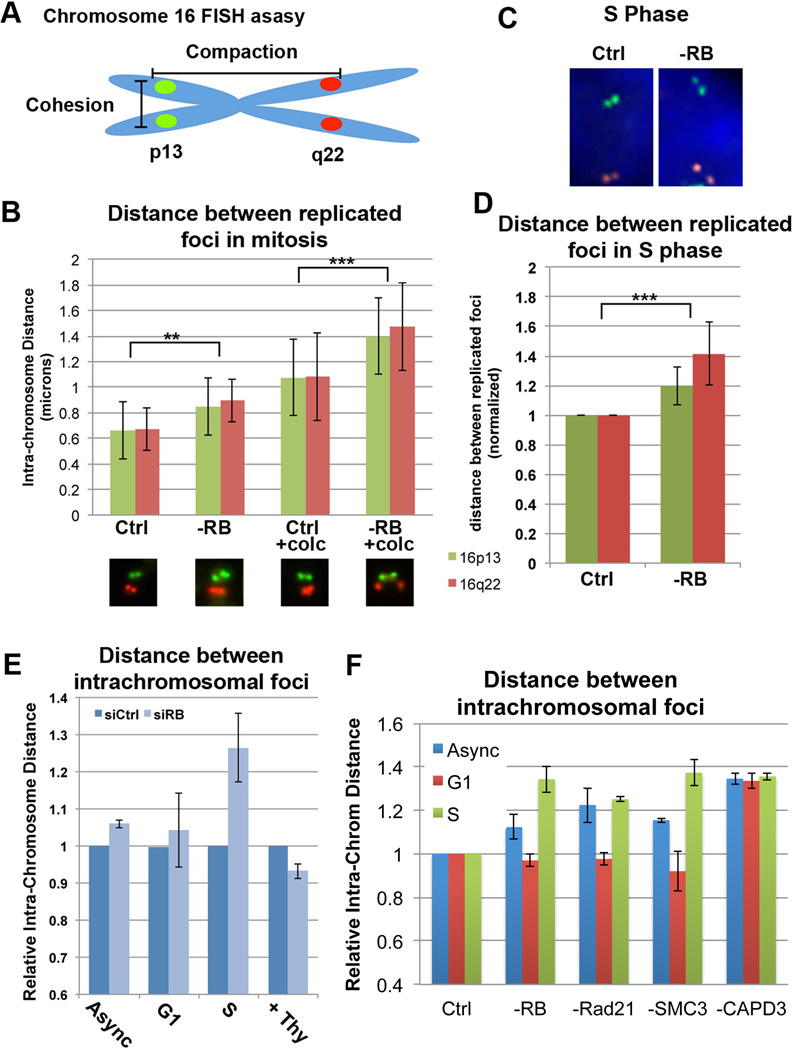
Figure 1. pRB loss compromises chromatid cohesion and compaction during S phase and mitosis.
A&B) Fluorescence in situ hybridization (FISH) with probes specific for 16p13 (green) and 16q22 (red) were used to quantify mitotic cohesion (increased distance between replicated foci of the same color) in control and pRB-depleted cells +/− prolonged mitotic arrest induced by 4 hr colcemid treatment (+ colc). pRB loss decreased mitotic cohesion, a change enhanced by prolonged mitotic delay. C-E) Measures of cohesion and compaction (distance between red and green foci) in interphase cells show significant decreases in chromatin cohesion (D) and compaction (E) as early as S phase following pRB loss. S phase progression is required for decreased compaction following pRB loss (0.2mM Thymidine). F) Cell cycle changes in compaction following depletion of cohesin components (Rad21, SMC3), but not condensin (CAPD3) closely mirrored the effects seen following pRB depletion. **p<0.01; ***p<0.001. See also Figures S1&S2.
FISH-based analysis of chromosome spreads prepared from cycling and nocodazole treated cultures demonstrated that pRB-depleted mitotic cells exhibit a decrease in sister chromatid cohesion that is enhanced upon prolonged mitotic arrest (Figure 1B). This is consistent with changes observed in mitotic spreads prepared from pRB-depleted cells (Figure S1F) and previous reports of pRB-dependent changes in mitotic chromatin structure (Manning et al., 2010; van Harn et al., 2010). Analysis of interphase cells revealed that this decrease in sister chromatid cohesion was first evident in S phase (Figure 1C & D).
Measures of intra-chromosome distance in interphase cells showed that depletion of pRB also increased the distance between foci on the p and q arms (Figure 1C & E). Cell cycle analysis revealed that these changes in compaction were also first evident in S phase cells, indicating that changes in chromosome structure previously noted during mitosis originate much earlier in the cell cycle (Figure 1E & F). The increase in intrachromosome distance observed during S phase in pRB-depleted cells was replication dependent, since inhibition of replication with either thymidine or HU treatment restored distances in pRB-deficient cells to those of controls (Figure 1E and data not shown). Cells depleted of cohesin proteins (Rad21 and SMC3) or condensin II (CAPD3) exhibited similar changes in S phase compaction (Figure 1F). However, changes in chromatid cohesion comparable to pRB-depleted cells were seen exclusively in cohesin-depleted cells (Figure S2A). Together, these results illustrate that reduction of cohesin is sufficient to compromise both chromatin cohesion and compaction.
Lesions in the pRB pathway can lead to the E2F-dependent up-regulation of several mitotic regulators (Chakraborty et al., 2007; Iovino et al., 2006; Ishida et al., 2001; Knudsen and Knudsen, 2008; Ren et al., 2002). These targets include the mitotic checkpoint protein Mad2 (Hernando et al., 2004; Schvartzman et al., 2011; Sotillo et al., 2010), a change that is notable because overexpression of Mad2 can induce CIN (Kabeche and Compton, 2012). Consistent with a previous study showing that Mad2 is co-operatively repressed by p53 and the three pocket proteins (pRB, p107, and p130) and that absence of pRB alone was insufficient to alter the level of Mad2 expression (Schvartzman et al., 2011), we saw no change in Mad2 mRNA or protein levels following pRB depletion (Figure S2D & E). Moreover, overexpression of Mad2 in hTERT-RPE cells was insufficient to alter chromatin compaction or cohesion in either S phase or mitotic cells (Figure S2F–I). Together this suggests that changes in Mad2 expression are unlikely to be significant factor in CIN that results in these pRB-depleted cells.
Collectively these results show that the depletion of pRB causes changes in chromosome cohesion and compaction during interphase. These changes were independent of Mad2, and similar changes could be generated by the depletion of cohesin proteins, but not by the depletion of condensin proteins.
Loss of pRB alters histone methylation and reduces the level of cohesin on pericentric heterochromatin
To understand why pRB-depleted cells have reduced cohesion, we looked for changes in cohesin levels, acetylation, and chromatin association, or for changes in the expression of known cohesin regulators. The cohesin complex is loaded onto DNA concurrent with replication such that the Esco1/2 acetylase, which travels with the replication machinery, is necessary for the stable association of cohesin to chromatin and for functional sister chromatid cohesion (reviewed in (Sherwood et al., 2010)). Cells depleted of pRB did not exhibit significant changes in the abundance of cohesin components or known regulators (Figure 2, Figure S3) but did show decreased SMC3 acetylation (Figure 2A) suggesting that cohesin is less stably associated with chromatin.
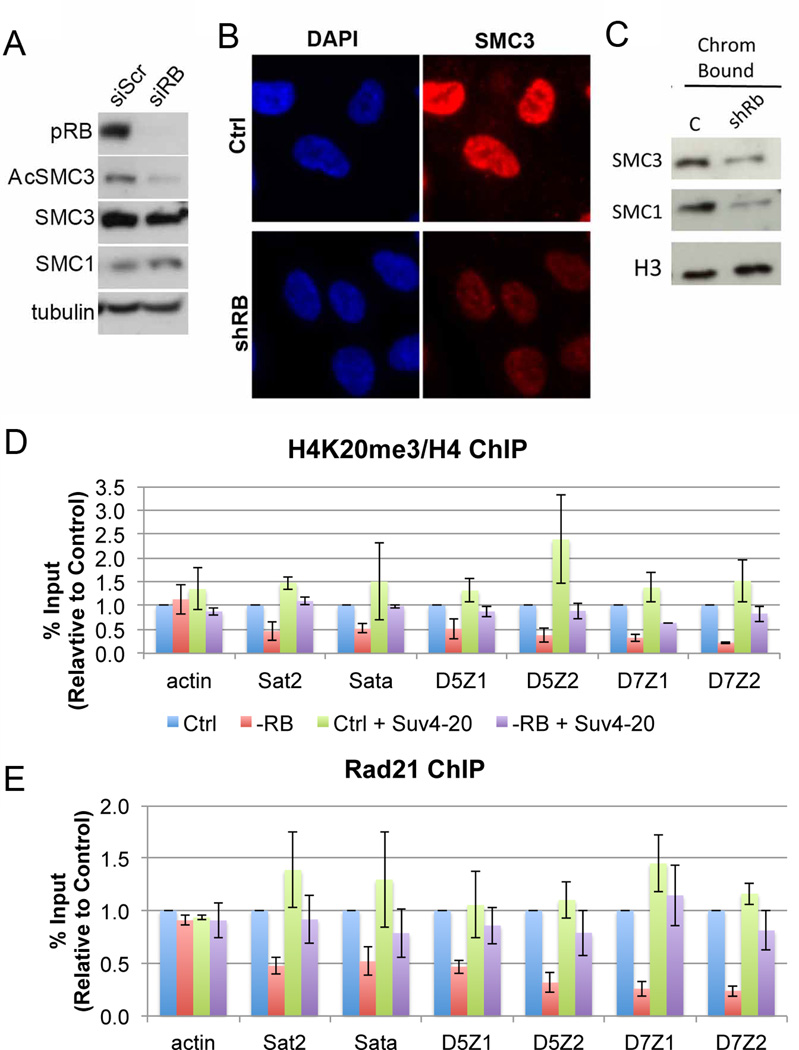
Figure 2. Cohesin association with chromatin is compromised in pRB-depleted cells.
A) Extracts from proliferating cells showed similar total cohesin protein levels (SMC1 and SMC3) between control and pRB depleted conditions, yet RB-depleted cells (-RB) had less acetylated SMC3 (AcSMC3), indicating reduced cohesin stability on chromatin. B) Immunofluorescence and C) cell fractionation analysis of G2 arrested cells indicated a decrease in the amount of cohesin stably associated with chromatin following pRB depletion. D & E) Changes in the methylation status of Lysine 20 of Histone 4 (H4K20) across a panel of pericentromeric heterochromatin regions correspond with decreased cohesin chromatin binding following pRB loss. Overexpression of the H4K20 methyltransferase Suv4-20 is sufficient to promote H4K20 tri methylation and cohesin binding in pRB-depleted cells. Error bars represent the SEM between individual biological replicates. See also Figure S3.
The total amount of cohesin associated with chromatin in an asynchronous population of cells is only moderately reduced when pRB is depleted (Manning et al., 2010). However, our finding that changes in chromatin cohesion and compaction in pRB-depleted cells are most evident at specific times during the cell cycle prompted us to investigate this more carefully. Control and pRB-depleted cells were arrested in G2, a point in the cell cycle when replication has been completed and cohesin is stably associated with chromatin, then analyzed by immunofluorescence and cell fractionation. These assays showed a clear reduction in the level of cohesin stably associated with chromatin in pRB-depleted cells (Figure 2B & C). Consistent with this, and with compromised mitotic cohesion, pRB-depleted cells also exhibit defects in other cohesinlinked processes (Mannini et al., 2010a; Sherwood et al., 2010, Watrin and Peters, 2006, Caron et al., 2012), including reduced rates of DNA replication and increased DNA damage foci (Figure S3).
Although defects in cohesin loading could explain changes in SMC3 acetylation, there is no known link between pRB and the cohesin loading machinery. Instead, pRB has been described to function in the recruitment of chromatin modifying enzymes that may impact the chromatin binding and enrichment of cohesin, upstream of replication-dependent loading and acetylation. In euchromatin, cohesin co-localizes with CTCF and the mediator complex (Kagey et al., 2010; Parelho et al., 2008; Wendt et al., 2008). However, recruitment of cohesin to heterochromatin, including pericentromeric regions where it is essential for accurate chromosome segregation, was recently shown to be regulated by the histone methyltransferase Suv4-20h2 (Hahn et al., 2013). Indeed, loss of Suv4-20h2 reduced Histone 4 Lysine 20 (H4K20) methylation at pericentromeric regions and compromised cohesin enrichment (Hahn et al., 2013).
The link between regulation of H4K20 methylation and cohesin is intriguing as changes in pericentromeric structure are likely to impact CIN. Moreover, pRB has been reported to interact directly with Suv4-20h2 in an E2F-independent manner (Gonzalo et al., 2005) and the mutation of mouse Rb1 dramatically reduces H4K20 methylation (Gonzalo et al., 2005; Isaac et al., 2006). To examine the idea that changes in H4K20 methylation might connect the depletion of pRB to altered accumulation of cohesin, we first asked whether pRB-loss affected the level of H4K20 trimethylation and the amount of cohesin that accumulates at pericentromeric chromatin in human cells. Chromatin immuno-precipitation (ChIP) experiments demonstrated that pRB-depletion decreased H4K20 trimethylation and cohesin association at a panel of pericentromeric regions (Figure 2D & E). In control experiments, the depletion of neither cohesin nor condensin II proteins altered H4K20 trimethylation (Figure S3I).
To test the significance of the changes in H4K20 methylation we examined the effect of increasing H4K20 trimethylation in pRB-depleted cells. Consistent with data in mouse cells (Gonzalo et al., 2005), overexpression of Suv4-20H2 in pRB-depleted cells promoted H4K20 trimethylation and increased the level of cohesin bound at pericentromeric heterochromatin (Figure 2D & E). Together this data suggests that pRB loss alters the pattern of H4K20 methylation and that this reduces the level of cohesin at pericentromeric heterochromatin.
Promoting cohesion suppresses S phase defects
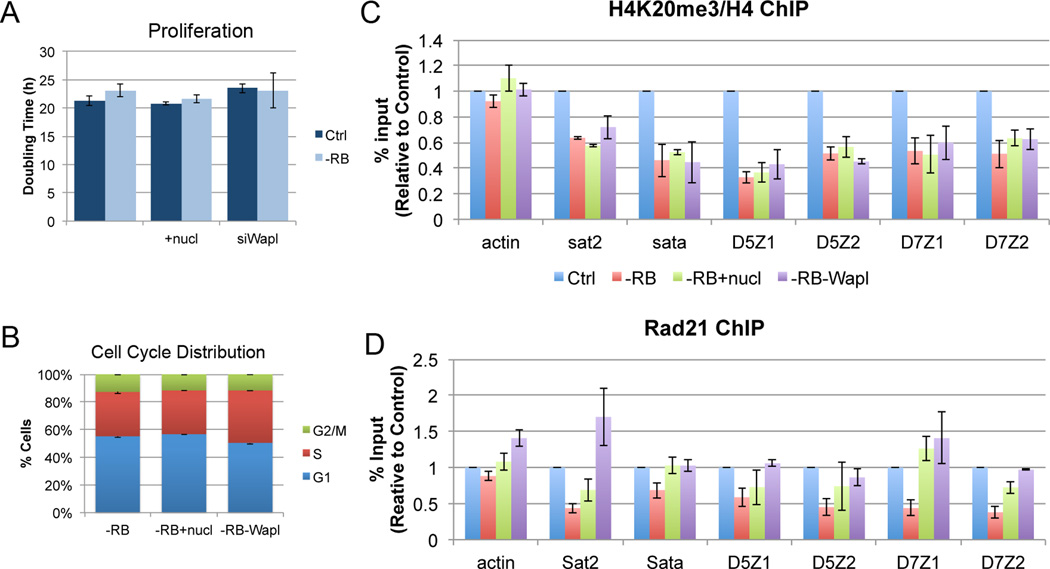
To test whether changes in the levels of chromatin-associated cohesin are functionally significant for any properties of pRB-depleted cells, we experimentally promoted cohesion by depleting Wapl, a negative regulator of cohesin. Wapl depletion increases the stability of the cohesin complex on chromatin in both S phase and mitosis (Peters et al., 2008). Efficient depletion was achieved by transfection of pooled siRNA constructs and all results were confirmed by the transfection of one or more of 5 independent siRNA constructs, each targeting unique Wapl-specific sequences (Supplemental Table 1). To minimize the effect of complete inactivation of Wapl on cell cycle progression, cells were collected 24 to 48 hours following transfection. This time point was chosen because it showed significant targeting of Wapl (>80% reduction in Wapl mRNA) without any significant change in the proliferation or cell cycle distribution of depleted cells (Figure 3A, B & S4).
Figure 3. Promoting cohesin binding in pRB-depleted cells.
Addition of exogenous nucleosides or co-depletion of Wapl does not alter A) the rate of proliferation, B) the cell cycle distribution, or C) the H4K20 methylation status at pericentromeric regions of cells depleted of pRB. D) However, both addition of exogenous nucleosides and co-depletion of Wapl are sufficient to enhance cohesin binding at pericentromeric heterochromatin in pRB-depleted cells. Error bars represent the SEM between individual biological replicates. See also Figure S4.
Consistent with the described role of Wapl in cohesin regulation, immunofluorescence, cell fractionation, and ChIP experiments demonstrated that Wapl depletion promoted cohesin association with chromatin in both control and pRB-depleted cells. (Figure S4A & B).
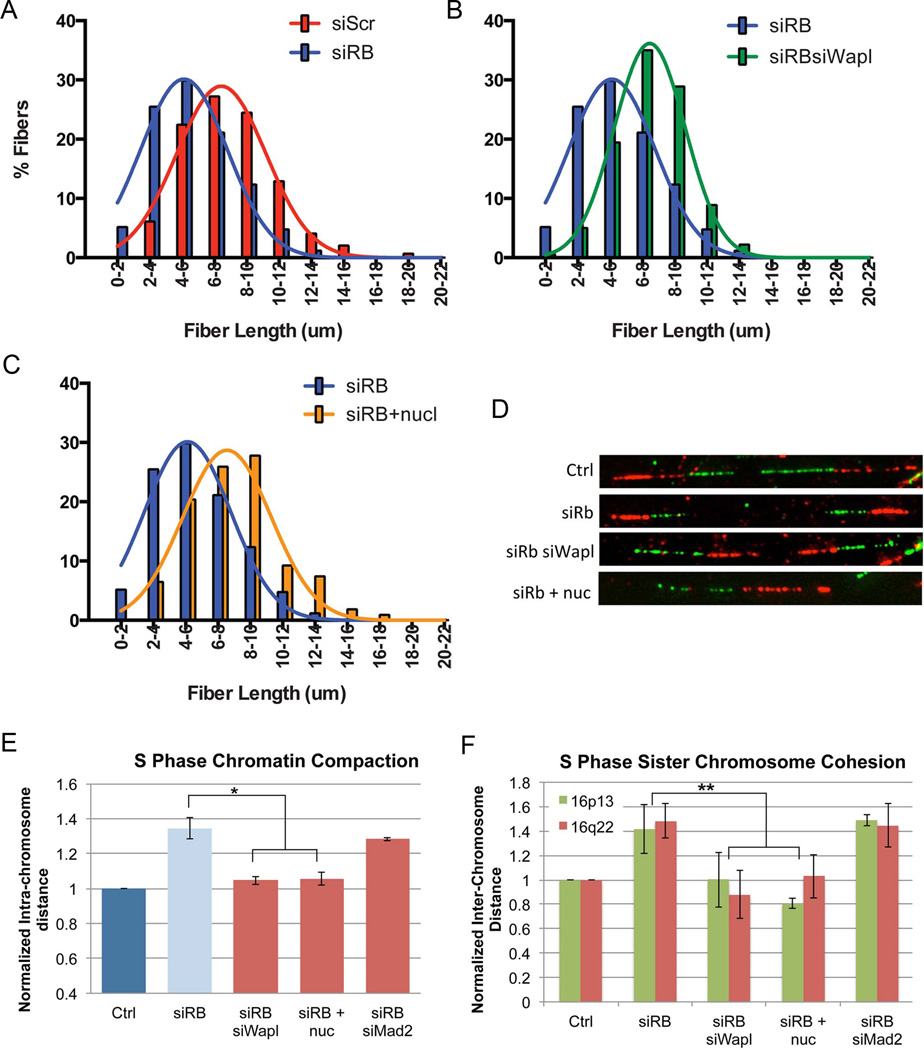
As described in the sections below, we then examined the functional impact of increased cohesion on pRB-depleted cells. We first examined was the effect of Wapl depletion on replication dynamics. Cohesin has been shown to stabilize forks and to promote replication (Guillou et al., 2010; Terret et al., 2009). DNA fiber combing assays demonstrated that pRB-depleted cells have reduced rates of replication at elongating forks ((Bester et al., 2011); Figure 4A). Promoting cohesion by depleting Wapl was sufficient to promote replication in pRB-depleted cells as judged by DNA fiber length (average fiber length in siRB siWapl is 37.5 +/− 7.08; p (v Ctrl)= 0.026; Figure 4B & D; Figure S5).
Figure 4. Promoting cohesion suppresses S phase defects in replication and chromatin structure resulting from pRB loss.
DNA fiber combing assays were performed and fiber lengths measured for control- and pRB- depleted cells alone or following co-depletion of Wapl or addition of nucleosides (+nuc). A) Loss of pRB promoted replication defects and resulted in shorter fibers (p<0.0001). B) Co-depletion of Wapl, or C) supplementation of nucleosides in pRBdepleted cells suppressed replication defects and allowed for longer fiber formation (p<0.0001 v siRB for both). D) Representative fibers from each condition are shown. FISH based assays to measure E) chromatin compaction and F) chromatin cohesion revealed that co-depletion of Wapl and nucleoside addition both suppressed compaction and cohesion defects during S phase in pRB-depleted cells, while Mad2 co-depletion did not. * p<0.05; **p<0.01. See also Figure S5.
Recent studies have reported that replication defects can be suppressed by nucleoside supplementation in the growth media and that this reduces CIN (Bester et al., 2011; Burrell et al., 2013). Therefore, we compared the effects of Wapl depletion with the effects of nucleoside supplementation. As expected, addition of exogenous nucleosides similarly increased the DNA fiber lengths in siRB-treated cells (average fiber length in siRB +nuc cells is 38.24 +/− 5.45; p (v Ctrl) =0.079; Figure 4C & D). Unexpectedly, western blot and ChIP analysis to monitor the association of the cohesin complex with pericentromeric heterochromatin revealed that both Wapl depletion and nucleoside addition promoted SMC3 acetylation and were sufficient to enhance Rad21 binding in the context of pRB depletion (Figures S4B, C & 3D). Importantly, neither of these conditions altered H4K20 trimethylation, proliferation rate, E2F target gene expression, or cell cycle distribution of pRB-depleted cells (Figure 3A–C, S4D), suggesting that both treatments promote cohesin association with chromatin downstream of the epigenetic changes caused by pRB loss. This indicates that increased cohesin loading and acetylation is sufficient to bypass changes that originate from defects in cohesin chromatin binding and enrichment.
These observations are consistent with the idea that increased cohesion can suppress replication defects in pRB-depleted cells, but they also highlight a series of questions about the effects of exogenous nucleosides. A previous report proposed that nucleoside supplements suppress replication defects by correcting a deficit in nucleotide pools (Bester et al., 2011). However, we found nucleotide pools in pRB-depleted cells to be increased, rather than decreased by the loss of pRB. (Figure S5E & F). In a complementary experiment we found that replication stress caused by Hydroxyurea (HU) treatment is sufficient to cause anaphase defects, but did not cause the changes in chromatin binding or acetylation of cohesin components that we observed in pRB-depleted cells, indicating that these changes are not a simple consequence of replication stalling (Figure S6). The notion that nucleoside supplements suppress phenotypes of pRB-deficient cells by improving rates of replication may be correct, but the cause of shorter replication fibers is unclear. Our data show that Wapl depletion and nucleoside supplements both increase the levels of cohesin on pericentric heterochromatin, a change that may contribute to the improved replication dynamics.
Promoting cohesion suppresses CIN
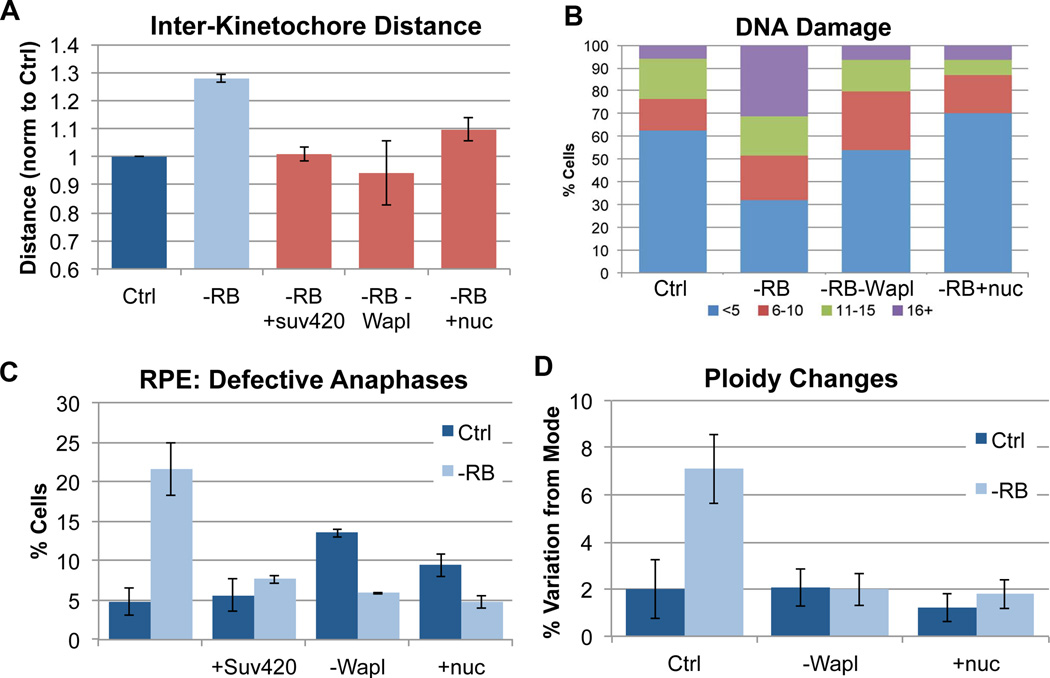
To determine whether increasing the levels of cohesin on chromatin suppress the changes in chromosome structure seen in pRB-depleted cells, siRB treated RPE cells were codepleted of Wapl (or treated with nucleosides) and analyzed for measures of cohesion and compaction in S phase and mitosis. Both Wapl depletion and nucleoside supplements where sufficient to suppress changes in cohesion and compaction during S phase (Figure 4E & F). Measures of inter-kinetochore distance showed that these treatments also restored normal centromeric cohesion in mitotic cells (Figure 5A).
Figure 5. Promoting cohesion in pRB-depleted cells suppresses defects associated with genome instability.
A) The distance between paired sister kinetochores in mitotic cells (marked with ACA & Hec1 antibody) was measured as a readout of functional mitotic cohesion. Overexpression of Suv4-20, co-depletion of Wapl, and nucleoside addition all similarly suppressed centromeric cohesion defects during mitosis in pRB-depleted cells. B) Using γH2AX foci as a readout, pRB depleted cells were seen to exhibit increased DNA damage. Both co-depletion of Wapl and nucleoside addition suppressed the frequency and extent of γH2AX foci. The percent of cells with <5 foci (blue), 6–10 foci (red), 11–15 foci (green), and 16 or more foci (purple) is indicated for each condition. C) Immunofluorescence analysis of DAPI, α-tubulin and ACA staining demonstrate that the presence of lagging chromosomes during anaphase following pRB depletion was significantly decreased by overexpression of Suv4-20, co-depletion of Wapl and nucleoside addition. D) Variation in chromosome copy number determined using FISH analysis indicates that 36 hours after siRNA treatment, pRB-depleted cells exhibit a nearly four-fold increase in cells deviating from the modal copy number (mode = 2 in RPE cells) for each of 3 chromosomes scored. Co-depletion of Wapl and nucleoside addition suppressed the generation of aneuploid cells. See also Figure S6.
To understand how changes in cohesion influence genome stability, we assayed functional readouts of DNA damage and chromosome segregation under conditions where the structural defects resulting from pRB depletion had been suppressed. We examined the influence of changes in chromatin structure on the presence of DNA damage in pRB-depleted cells by scoring the prevalence of γH2AX foci. Conditions that promote cohesion (Wapl depletion and nucleoside addition) suppressed DNA damage in pRB-depleted cells (Figure 5B).
Mitotic cells were then analyzed for evidence of segregation errors. Depletion of Wapl alone delays the loss of sister chromatid cohesion and can lead to paired sisters lagging during anaphase. However, in cohesion-compromised pRB-depleted cells, the increased cohesion provided by Wapl depletion or addition of nucleosides strikingly reduced the incidence of lagging chromosomes near to that seen in control cells (Figure 5C).
Frequent segregation errors result in a heterogeneous aneuploid population where the heterogeneity of chromosome copy number within a population of cells is indicative of the total degree of chromosomal instability. To score for such heterogeneity, copy number changes of individual chromosomes were assessed using centromeric FISH probes specific for each of 3 different chromosomes (2, 6 and 8). Cells depleted of pRB displayed chromosome copy number heterogeneity nearly four-fold higher than control cells. Co-depletion of Wapl or addition of nucleosides suppressed the degree of copy number heterogeneity in cells depleted of pRB (Figure 5D). While pRB-dependent changes in H4K20 methylation had been observed to co-exist with CIN ((Gonzalo et al., 2005); Figure 2), the functional relationship between the two phenomena had yet to be explored. Therefore, we examined the importance of H4K20 methylation for the CIN phenotype. Significantly, overexpression of Suv4-20H2 not only increased H4K20 trimethylation and increased cohesin association with pericentric heterochromatin (Figure 2D & E), but was also sufficient to increase mitotic cohesion and decrease segregation errors in pRB-depleted cells (Figure 5A & C).
Collectively, these experiments support a model whereby changes in methylation at pericentromeric heterochromatin following pRB loss lead to reduced cohesin association, cohesion defects and CIN. Conditions that increase cohesin stability on chromatin suppress the changes in chromosome compaction and cohesion seen during Sphase in pRB-depleted cells and restore normal kinetochore structure during mitosis. Improved cohesion also suppressed DNA damage in pRB-depleted cells, prevented the appearance of lagging chromosomes, and suppressed aneuploidy. That so many of the defects seen in pRB-deficient cells could be similarly suppressed suggests cohesin function is integral to the changes that occur following pRB loss.
Suppressing CIN in Cancer Cells
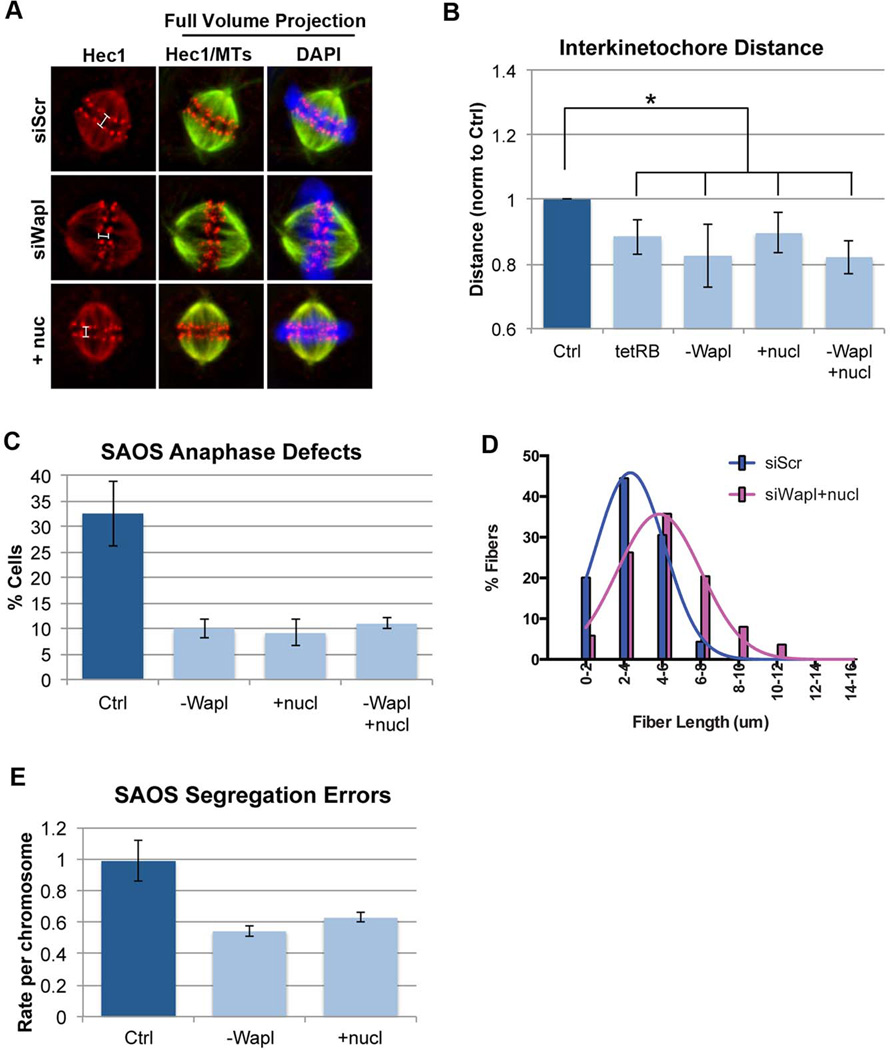
If the inactivation of pRB is a significant source of CIN in cancer, and increased cohesion suppresses CIN resulting from pRB-loss, then increased cohesion would be expected to suppress CIN in human cancer cells. To test this idea we first examined an osteosarcoma cell line (SAOS-2). Like many osteosarcomas, SAOS-2 cells are mutant for RB1 and display high genomic heterogeneity. Previously, we showed that re-expression of pRB from an inducible promoter is sufficient to promote mitotic centromeric cohesion in these cells (Manning et al., 2013). Similarly, we found that depletion of Wapl, or addition of nucleosides promoted cohesion between mitotic centromeres (Figure 6A & B). As seen when cohesion defects were suppressed in pRB-depleted RPE cells, SAOS-2 cells depleted of Wapl or supplemented with nucleosides, alone or in combination, displayed improved rates of replication during S-phase and fewer lagging chromosomes during mitosis (Figure 6C & D).
Figure 6. Promoting cohesion suppresses CIN in cancer cells lacking pRB.
A & B) RB1 null osteosarcoma (SAOS2) cells were fixed and stained for tubulin, Hec1, and DAPI following re-expression of pRB with a tetracycline regulated construct (tetRB), depletion of Wapl, and/or addition of nucleosides. All three conditions induced a moderate, but significant increase in mitotic sister chromatid cohesion, C) reduced anaphase defects and suppressed replication defects (Figure S5). Combined depletion of Wapl with nucleoside addition provided no additional suppression of cohesion or anaphase defects, but does enhance D) suppression of replication defects (siScr: 3.22µm; siWapl+nucl: 4.969µm; p<0.0001; and Figure S5). E) FISH-based measures of chromosome segregation errors in recently divided daughter cells show that depletion of Wapl and addition of nucleosides decreased CIN in SAOS2 cells. *p<0.05
To understand how the suppression of these defects impacts CIN, we quantified the frequency of segregation errors in recently divided SAOS-2 daughter cells, either with or without Wapl depletion or nucleoside treatment. Cells were plated at extremely low density, fixed within one cell cycle and analyzed for copy number of chromosomes 6 and 8 using centromeric FISH probes. SAOS-2 cells exhibit high chromosome instability and on average mis-segregate a chromosome about once every other division. Like many cancer cells, SAOS-2 cells have a complex genotype and contain many abnormalities that could contribute to the generation of segregation errors, including a high incidence of extra centrosomes and multipolar spindle structures. Despite this, both the transient depletion of Wapl, and nucleoside supplementation, was sufficient to suppress the rate of segregation errors in these cells by half (Figure 6E). Combined Wapl depletion and nucleoside addition was more efficient at promoting replication in SAOS2 cells than either treatment alone (Figure 6D, Figure S6). However the combination did not further decrease the frequency of anaphase defects compared to each individual condition (Figure 6C), suggesting that cohesion defects, rather than the replication defects, represent the major contributing factor to CIN in these cells.
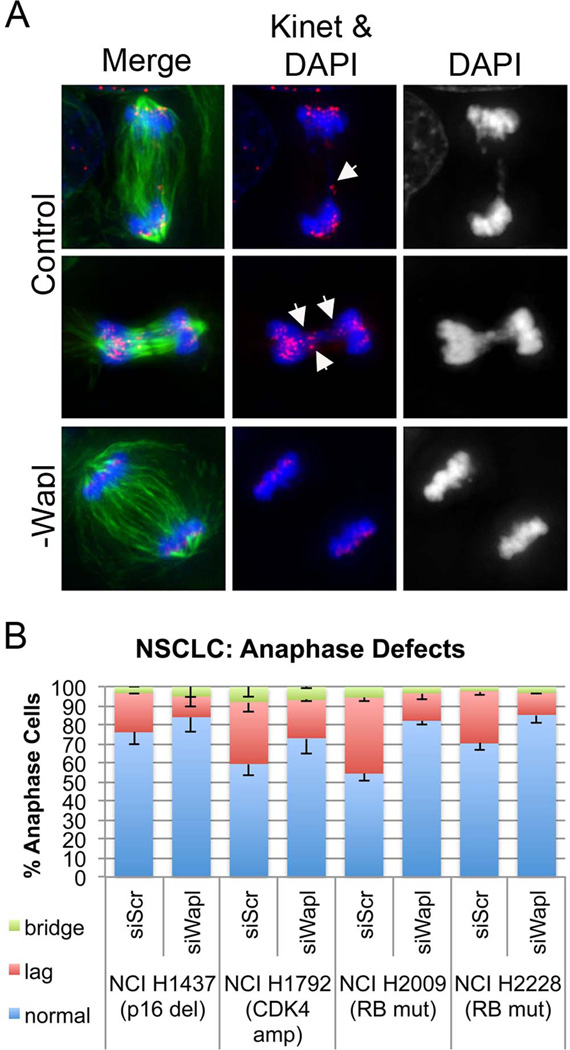
To test the generality of these effects, we next assessed a panel of non-small cell lung cancer (NSCLC) lines in which the pRB pathway is compromised. We examined two cell lines with mutation of RB1 (NCI H2009 and NCI H2228), one cell line with homozygous deletion of p16INK4A (NCI H1437), and one cell line with copy gains of cdk4 (NCI H1792). In each line, promoting cohesion suppressed anaphase defects (Figure 7).
Figure 7. Promoting Cohesion is sufficient to suppress CIN in a panel of non small cell lung cancer (NSCLC) lines.
A) NSCLC lines in which pRB function is compromised were treated with siRNA constructs targeting Wapl, or alternatively with a scrambled control for 36 h, fixed, stained (tubulin, ACA (kinet), and DAPI) and assessed for anaphase defects. Each NSCLC line analyzed exhibited massive segregation defects, including the presence of single and numerous lagging chromosomes and DNA bridges (representative images in panel A; arrow heads indicate kinetochores of lagging chromosomes). B) Wapl depletion suppressed the incidence of lagging chromosomes in each cell line.
Discussion
While most solid tumors exhibit CIN (Beroukhim et al., 2000; Hanahan and Weinberg, 2000), the potential therapeutic benefit of targeting CIN has received little attention, in large part because it has been unclear what changes cause CIN, or how to prevent them from occurring. Here we demonstrate that a connection between pRB and cohesion is an important contributor to genomic stability. These results provide unexpected mechanistic insight into how CIN can be manipulated in cancer cells. We propose a model in which the loss of pRB causes epigenetic changes that compromise cohesin levels, particularly at pericentromeric heterochromatin. These changes have a series of consequences: reduced centromeric cohesion, increased DNA damage, changes in DNA replication, and CIN. The importance of cohesion for the properties of pRB-deficient cells is illustrated by experiments showing that increasing the amount of cohesin on chromatin is sufficient to correct all of these defects. Importantly, promoting cohesion suppressed CIN in cell lines acutely depleted of pRB, and in cancer cells with a variety of pRB pathway lesions.
Chromatin compaction, cohesion establishment, and CIN
The initial characterization of segregation errors following pRB loss by us and others noted that changes in mitotic centromere structure and defects in chromosome segregation are associated with changes in both Condensin II and cohesin complexes (Manning et al., 2010; Coshi et al., 2010; Van harn et al., 2010). Evidence that Condensin II depletion can compromise centromeric structure and promote segregation errors (Hirota et al., 2004; Ono et al., 2004) led to the suggestion that loss of pRB-dependent regulation of Condensin II may underlie CIN. (Coschi et al., 2010; Longworth et al., 2008; Manning et al., 2010). Currently it is not possible to specifically alter pRB’s regulation of Condensin II, or to directly alter its recruitment independent of pRB, and this has precluded any direct test of the functional significance of the changes in Condensin II levels in pRB-deficient cells.
There are several reasons to think that changes in Condensin II levels cannot fully explain the chromosomal changes resulting from pRB loss. First, loss of the Condensin II complex has been shown to cause a moderate increase in cohesion (Hirota et al., 2004; Ono et al., 2013). In addition, the anaphase defects seen following Condensin II depletion typically consist of chromatin bridges and paired lagging chromosomes (Coschi et al., 2010; Green et al., 2012; Ono et al., 2013). These types of defects are substantially different from the profile of defects seen in pRB depleted human cells, where mitotic cohesion is decreased and the predominant anaphase defect is the presence of individual lagging chromosomes.
Here we show that depletion of pRB decreases the enrichment of cohesin at pericentromeric regions. This change has functional consequences for mitotic fidelity. The fact that suppression of the cohesion defects is sufficient to suppress CIN does not exclude the possibility that decreased Condensin II recruitment may also contribute to CIN following pRB loss. However, while Wapl depletion corrected the effects of pRBloss, it failed to correct mitotic defects resulting from depletion of Condensin II (CAPD3), and failed to suppress the effects of combined pRB and Cap-D3 depletion (Figure S6). Together, this data strongly suggest that the effects of pRB loss on cohesin function play a dominant role in the CIN phenotype.
Mis-regulation of pericentromeric heterochromatin: the origin of CIN
pRB can promote the formation of both facultative and constitutive heterochromatin by recruiting chromatin binding and modifying proteins (Gonzalo and Blasco, 2005; Isaac et al., 2006; Siddiqui et al., 2007). Epigenetic regulators that physically interact with pRB include regulators of H3K9 methylation (KDM4A and Suv3-9) and the methyltransferase responsible for di and tri methylation of H4K20 (Suv4-20H2). These methyl marks are significant because they are common in pericentromeric chromatin, have been shown to influence cohesin enrichment, and are expected to impact centromeric structure and cohesion (Lehnertz et al., 2003; Schotta et al., 2004). Our results demonstrate that loss of pRB decreases H4K20 trimethylation and reduces cohesin enrichment at pericentromeric heterochromatin. While cohesin depletion alone does not alter H4K20 trimethylation, overexpression of the methyltransferase responsible for regulation of this key residue was sufficient to promote cohesin association and to suppress mitotic defects. This suggests that changes in methylation are the root cause of the centromeric defects seen in pRB-depleted cells. Although H4K20 trimethylation is enriched at pericentric heterochromatin, it is also distributed throughout the genome, and changes in this mark following pRB loss may contribute to global effects on cohesion, replication and DNA damage. While CIN is generally thought to result from defects in mitosis, the presence of both pRB and Suv4-20 activity during G1, together with the ability of Wapl depletion and nucleoside addition to suppress both S phase and mitotic defects of pRB deficient cells, suggests that the changes causing the mitotic errors likely originate much earlier in the cell cycle.
We found no significant change in the levels of known cohesin regulators in pRB-depleted cells, suggesting that the primary link between pRB and cohesin at pericentromeric regions is through changes in epigenetic marks. We note that Wapl depletion did not suppress the increased expression of E2F-regulated genes in pRB-depleted cells, and did not affect the altered H4K20 methylation at pericentric heterochromatin. However, given that cohesin associates with enhancer elements and the complete inactivation of Wapl affects gene expression, we cannot formally exclude the possibility that changes in cohesin also affect the expression of an unknown gene(s) that indirectly impacts pericentric heterochromatin and influences the levels of merotely in pRB-deficient cells.
CIN and Cancer
Our findings suggest that changes in cohesion are likely the major contributing factor to CIN in pRB-deficient cells. Interestingly, cohesion defects have similarly been implicated in the generation of CIN in colorectal cancer (Pino and Chung, 2010) where sequencing revealed eleven conserved somatic mutations, ten of which were in confirmed or suspected cohesin regulatory genes (Barber et al., 2008). Consistent with the described cohesin mutations in this cancer type and the functional relationship between replication and cohesion, it has recently been demonstrated that CIN colon cancer cell lines exhibit defects in replication (Burrell et al., 2013). It is noteworthy that RB1 mutations are rare in colon cancer. We hypothesize that defects in chromatid cohesion and replication may be a common mechanism to promote CIN. In tumors such as colorectal cancers, that do not mutate RB1, mutations in key components of the cohesin regulatory pathway may be needed to promote CIN. In cancers where RB1 is frequently mutated, disruption of pRB function alone may corrupt the normal control of cohesion, promoting replication defects, DNA damage, and CIN.
The results described here are significant for two reasons. First, they illuminate the mechanism by which pRB inactivation promotes CIN. Second, they demonstrate that CIN can be altered by manipulating chromatin structure. Both very high and very low CIN has been correlated with better patient outcome in various cancers. This is consistent with the view that while moderate levels of segregation errors contribute a beneficial degree of genomic instability, high rates of segregation errors are catastrophic and incompatible with cancer cell viability. Following this line of reasoning, two therapeutic approaches have been proposed: to promote segregation errors and decrease cell viability, or alternatively, to suppress segregation errors and decrease genetic heterogeneity. A caveat of the former approach is that the potential for collateral damage is high, as all dividing cells will be sensitive to perturbations that increase segregation errors. However, the alternative approach, to suppress CIN and stabilize the cancer genome, could render cancer cells more sensitive to existing therapeutics, thereby reducing relapse and improving patient outcome.
NSCLC represent one specific context where CIN has been well documented and the pRB pathway is frequently compromised. Advances in targeted therapies have shown some success, but heterogeneity within the tumor often promotes acquired drug resistance and relapse. The observation that promoting cohesion prevents CIN resulting from pRB loss, and the evidence that this suppressive mechanism works in a panel of NSCLC lines, highlights the idea that the manipulation of cohesin regulation may have therapeutic benefit. However, the complete inactivation of Wapl in normal cells causes problems in S phase (Tedeschi et al., 2013), while high dNTP concentrations are known to increase mutation rates (Chabes et al., 2003; Sabouri et al., 2008). These limitations underscore the continued need to identify ways to improve cohesion in tumor cells that do not have unwanted side effects in normal cells.
Materials and Methods
Cell culture and RNAi
pRB depletion was achieved by transient transfection of one of 5 individual or a pool of four siRNA constructs using RNAi MAX transfection reagent (Invitrogen), per manufacturer’s directions. Alternatively depletion was obtained by infection with one of four shRNA lentiviral constructs followed by puromycin selection to induce constitutive (Manning et al., 2010), or doxycyline inducible (Meerbrey et al., 2011) knockdown. 2 µg/mL of Doxycycline was used to induce expression of the RB1-targeting shRNA or overexpression constructs. Wapl was similarly depleted using one of 5 individual, or a pool of four siRNA constructs. All experiments were confirmed with at least two independent means of knockdown. Supplementation with exogenous nucleosides was performed as previously described (Bester et al., 2011). Samples were treated with nucleosides, siRNA, and/or induced to express shRNA for 36–48 hours before collection or fixation of samples for each experiment. When indicated, cells were treated with 10 µM of the CDK1 inhibitor RO3306 for 20 hours to induce a G2 arrest. Construct sequences, antibody information and detailed methods appear in the Supplemental Material.
Supplementary Material
Highlights.
pRB loss compromises centromeric cohesion and promotes whole chromosome instability
pRB-dependent epigenetic regulation of centromeres promotes cohesin enrichment
CIN is suppressed by promoting centromeric H4K20 methylation or enhancing cohesion
Improved cohesion suppresses altered replication and DNA damage following pRB loss
Acknowledgements
We thank Katsuhiko Shirahige for the generous gift of AcSMC3 antibodies, Duane Compton for sharing the mCherry Mad2 construct, and Andreas Heilman for technical support. ALM is supported by an MGH ECOR Tosteson postdoctoral fellowship; SAY is supported by a fellowship from the Department of Defense (BC120504); LZ is a Jim and Ann Orr MGH Research Scholar and NJD is a James and Shirley Curvey MGH Research Scholar. This work was supported by NIH grants to NJD (CA155202) and LZ (GM076388).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2000;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Khare S, Dorairaj SK, Prabhakaran VC, Prakash DR, Kumar A. Identification of genes associated with tumorigenesis of retinoblastoma by microarray analysis. Genomics. 2007;90:344–353. doi: 10.1016/j.ygeno.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Choi CM, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, Lee DS, Lee SD. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, Werts A, Haak P, Vande Woude GF. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Blasco MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 2005;4:752–755. doi: 10.4161/cc.4.6.1720. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Green LC, Kalitsis P, Chang TM, Cipetic M, Kim JH, Marshall O, Turnbull L, Whitchurch CB, Vagnarelli P, Samejima K, et al. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. Journal of cell science. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J. Cohesin organizes chromatin loops at DNA replication factories. Genes & development. 2010;24:2812–2822. doi: 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Worz S, Sadic D, Schulte M, Mallm JP, Maiser A, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes & development. 2013;27:859–872. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Heilig CE, Loffler H, Mahlknecht U, Janssen JW, Ho AD, Jauch A, Kramer A. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2010;14:895–902. doi: 10.1111/j.1582-4934.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. Journal of cell science. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino F, Lentini L, Amato A, Di Leonardo A. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Molecular and cellular biology. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Current biology : CB. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nature reviews. Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY. Targeting the RB-pathway in cancer therapy. Clin Cancer Res. 2010;16:1094–1099. doi: 10.1158/1078-0432.CCR-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Current biology : CB. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Benes C, Dyson NJ. Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation. Oncogene. 2013 doi: 10.1038/onc.2013.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes & development. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Yamashita D, Hirano T. Condensin II initiates sister chromatid resolution during S phase. The Journal of cell biology. 2013;200:429–441. doi: 10.1083/jcb.201208008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes & development. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & development. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic acids research. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & development. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 Is a Critical Mediator of the Chromosome Instability Observed upon Rb and p53 Pathway Inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood R, Takahashi TS, Jallepalli PV. Sister acts: coordinating DNA replication and cohesion establishment. Genes & development. 2010;24:2723–2731. doi: 10.1101/gad.1976710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui H, Fox SR, Gunawardena RW, Knudsen ES. Loss of RB compromises specific heterochromatin modifications and modulates HP1alpha dynamics. J Cell Physiol. 2007;211:131–137. doi: 10.1002/jcp.20913. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013 doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:433–444. doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.