SUMMARY
Golgi complexes (Golgi) play important roles in the development and function of neurons [1–3]. Not only are Golgi present in the neuronal soma (somal Golgi) but they also exist in the dendrites as Golgi outposts [4–7]. Previous studies have shown that Golgi outposts serve as local microtubule organizing centers [8] and secretory stations in dendrites [6, 9]. It is unknown whether the structure and function of Golgi outposts differ from those of somal Golgi. Here we show in Drosophila that, unlike somal Golgi, the biochemically distinct cis, medial, and trans compartments of Golgi are often disconnected in dendrites in vivo. The Golgi structural protein GM130 is responsible for connecting distinct Golgi compartments in soma and dendritic branch points, and specific distribution of GM130 determines the compartmental organization of dendritic Golgi in dendritic shafts. We further show that compartmental organization regulates the role of Golgi in acentrosomal microtubule growth in dendrites and in dendritic branching. Our study provides insights into the structure and function of dendritic Golgi outposts as well as the regulation of compartmental organization of Golgi in general.
RESULTS AND DISCUSSIONS
The Golgi complexes in the soma of Drosophila neurons form ring-shaped structures consisting of stacks of cis, medial, and trans compartments
We investigated the architecture of Golgi in soma and dendritic outposts by using the dendritic arborization (da) neurons in Drosophila larva as a model system. These neurons offer an opportunity for combining molecular genetics and high-resolution live imaging to study the Golgi at single-cell resolution in vivo [7, 8, 10, 11]. Different glycosylation enzymes are known to localize to the biochemically distinct Golgi compartments [12, 13]. We previously showed in the da neurons that the medial Golgi marker α-mannosidase II tagged with eGFP (ManII-GFP) is present as multiple large units in the soma and as puncta in dendrites [7]. In order to label additional Golgi compartments, we generated transgenic flies that express HA-tagged α-mannosidase I (HA-ManI, for medial Golgi) [14], TagRFP-T-tagged galactosyltransferase (GalT-TagRFP, for trans-Golgi) [15], and YFP- or TagRFP-T-tagged N-acetylgalactosaminyltransferase 2 (GalNacT2-YFP/TagRFP, for trans-Golgi) [16] (Figure S1). These markers correctly labeled Golgi mini-stacks consisting of cis, medial, and trans compartments in Drosophila epithelial cells (Figure S1A and B, and data not shown) [17, 18].
We used these markers to investigate the in vivo structural organization of Golgi compartments in the da neurons. In the soma of da neurons, the medial and trans markers all labeled several ring-shaped structures that contained endogenous dGMAP, the Drosophila homolog of the cis-localized Golgi membrane associated protein 210 (GMAP-210) [19] (Figures 1A–B, and S1C). These cis, medial, and trans Golgi markers were closely juxtaposed, and in many cases appeared to colocalize with each other due to convoluted three dimensional (3-D) architecture (Figures 1A–C and S1C–E). We also employed reverse tagging strategy by tagging ManII with TagRFP-T and GalT with YFP to ensure that the partial colocalization did not occur due to specific fluorescent proteins. Switching the fluorescent protein tags did not change the localizations of the markers for different Golgi compartments (Figures S1G and S1H). Moreover, Brefeldin A (BFA) treatment dispersed the ring-shaped Golgi in the soma and dendritic Golgi outposts (Figure S1F) [20], which was reversed after washout, further suggesting that the markers faithfully labeled the Golgi.
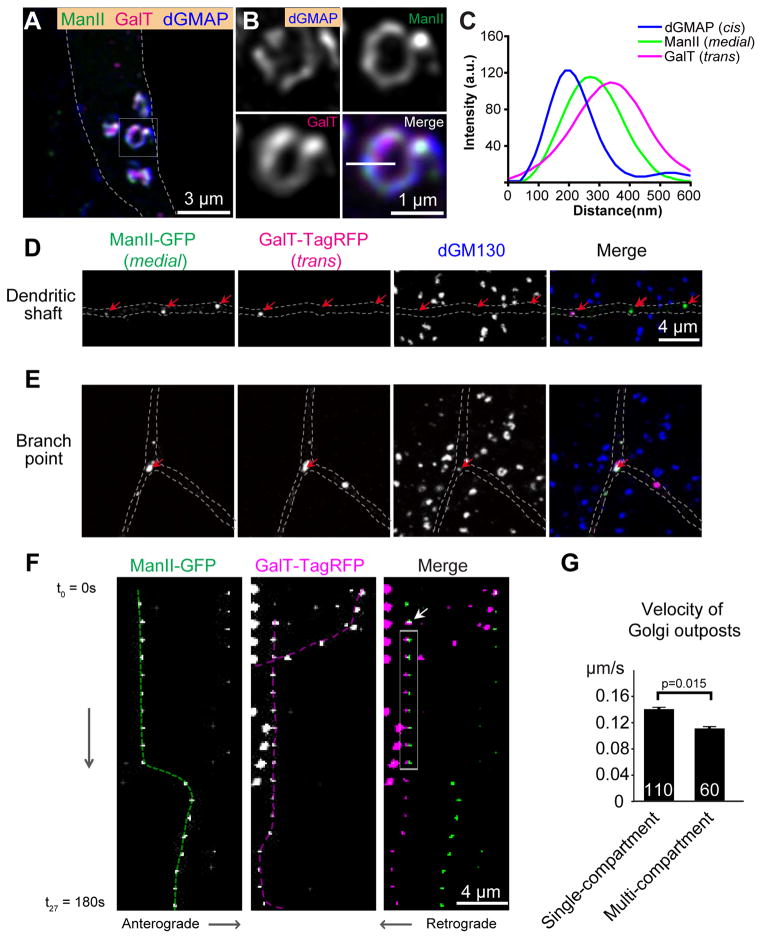
Figure 1. Distinct Golgi architectures in the soma and dendrites of Drosophila da neurons.
(A–C) The cis Golgi marker anti-GMAP (blue), medial Golgi marker ManII-GFP (green), and trans Golgi marker GalT-TagRFP (magenta) label ring-shaped structures in the soma of da neurons. (A) Overview of somal Golgi. (B) Magnified view of a single somal Golgi unit. (C) Line profile shows the fluorescence intensity along the white line in B.
(D) Golgi compartments are often disconnected in dendritic shafts as shown by the separation of the medial Golgi marker ManII-GFP (green) and the trans Golgi marker GalT-TagRFP (magenta). dGM130 protein (blue) is undetectable in dendritic shafts. Red arrows point to the Golgi compartments containing either ManII-GFP or GalT-TagRFP. The dGM130 puncta outside of the dendrites are specific labeling in epithelial cells adjacent to the neuron.
(E) The Golgi outposts in dendritic branch points contain multiple compartments as well as dGM130. Red arrows point to the Golgi compartments containing ManII-GFP, GalT-TagRFP and dGM130.
(F)Dendritic Golgi outposts are dynamic. Kymographs of time-lapse imaging of ManII-GFP and GalT-TagRFP puncta in dendrites. The interval between two images is 10s. The medial and trans compartments exhibit temporary interactions: they became co-colocalized at the time point indicated by the white arrow for 70s (gray box) before separating from each other. The dashed lines indicate the trajectories of the Golgi compartments.
(G) Quantification of the velocity of Golgi outposts.
These results demonstrate that somal Golgi in da neurons form ring-shaped structures consisting of stacks of cis, medial, and trans compartments. Thus, unlike other Drosophila larval cells but similar to mammalian cells [17, 21], larval neurons contain somal Golgi compartments that are not only connected to each other but also form high-order structures.
Disconnected Golgi compartments exist in dendrites
In contrast to the soma, distinct Golgi compartments were often disconnected from one another in the dendrites (Figures 1D and S1E). Whereas 91.6 ± 5.7 % of ManII-GFP and GalT-TagRFP structures (from 7 neurons) colocalized in the soma, only 51.6 ± 4.8 % ManII-GFP puncta (from 29 neurons) colocalized with or contacted GalT-TagRFP puncta in dendritic shafts. This surprising observation represents an animal case of Golgi organization with separated compartments in post-mitotic cells, which is similar to the Golgi in Saccharomyces cerevisiae [22, 23]. Notably, the Golgi outposts in the branch points of proximal dendrites did not follow this pattern. ManII-GFP and GalT-TagRFP were always juxtaposed to each other in these branch points (Figures 1E), similar to the Golgi mini-stacks observed in non-neuronal cells in Drosophila larva (Figures S1A and S1B) [17, 18]. Whereas the Golgi units in the soma were stationary, the Golgi outposts in the dendritic shafts (but not those at the branch points) often moved bi-directionally (Figure 1F). Moreover, in fifty 10-min time-lapse recordings, we observed 3 cases in which the disconnected Golgi compartments became colocalized with each other for a period before moving apart (Figure 1F), which may contribute to the portion of multi-compartment Golgi observed in dendrite shafts. The velocity of moving multi-compartment Golgi outposts was 0.11 ± 0.008 μm/s (n=60), which is slightly slower than that of moving single compartment outposts at 0.14 ± 0.009 μm/s (n = 110)(p = 0.015) (Figure 1G). In the fifty 10-min recordings (6 sec/frame), we observed five events of sudden appearance and disappearance of single compartment outposts. This could be due to either technical reasons (e.g. focal plane) or cisternal maturation of Golgi compartments [23].
Collectively, these results reveal three different organizations of Golgi in Drosophila da neurons: 1) the ring-shaped Golgi units that contain cis, medial, and trans compartments in the soma; 2) the disconnected, punctate Golgi compartments in the dendritic shafts (“single compartment Golgi”); and 3) Golgi mini-stacks in both dendritic branch points and shafts (“multi-compartment Golgi”). The first two organizations have not been characterized previously in Drosophila. These findings also demonstrate that different structural organizations of Golgi may exist in the same post-mitotic cell.
We also observed both single and multi-compartment Golgi in the dendrites of mouse cortical neurons in culture (Figure S1I and S1J), suggesting that these two types of dendritic Golgi organizations are evolutionarily conserved.
dGM130 is required for connecting the cis, medial, and trans compartments in vivo
The Golgi structural protein GM130 has been proposed to organize Golgi into stacks and/or ribbons in mammalian cells in culture [24–27, 28]. We found that, while the Drosophila ortholog of GM130 (dGM130) localized to the ring-shaped Golgi units in the soma (Figures 2C and 2D), it was barely detected in dendritic shafts (Figure 1D). dGM130 was always present at the branch points where ManII-GFP and GalT-TagRFP did form multi-compartment Golgi (Figure 1E). These results raised the possibility that the distribution of dGM130 determines the different organizations of Golgi between the soma and dendrites.
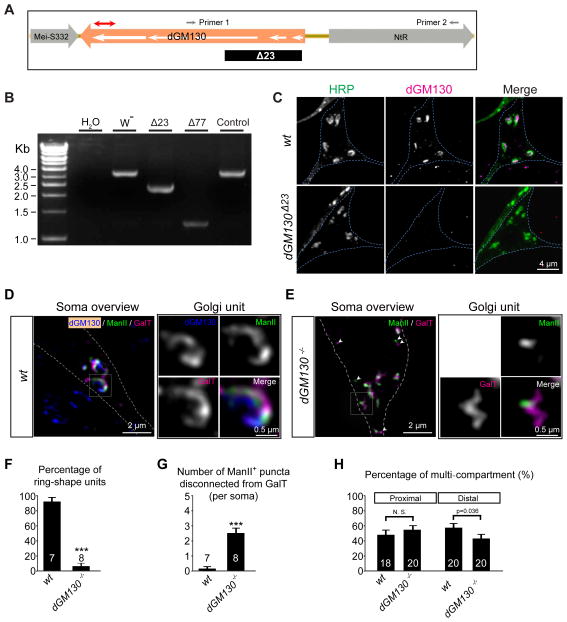
Figure 2. dGM130 is required for connecting the cis, medial, and trans compartments in vivo.
(A–C) Generation and characterization of dGM130 null mutants.(A) A schematic of the genomic region around the dGM130 gene. The white arrows indicate the five exons of the dGM130 gene. The red line with double arrowheads indicates the antigenic region for the anti-dGM130 antibody. The dGM130Δ23 allele carried a deletion that spanned the first two exons and half of the third exon (indicated by the black box) as confirmed by DNA sequencing. Small gray arrows indicate the location of the two primers used to amplify the genomic region for genotyping and DNA sequencing. (B) PCR results confirming genomic deletions caused by imprecise excision in dGM130 mutants. W−: wild-type; Δ77: a deletion that removes part of dGM130 and part of the adjacent gene NtR; Δ23: a deletion that removes the first two exons and half of the third exon; control: a precise excision line of the P-element P{RS3}GM130CB-6408-3. (C) Immunostaining of class III da neurons with an anti-dGM130 antibody that recognizes a C-terminal fragment of dGM130 protein shows that dGM130 protein is undetectable in the dGM130Δ23 mutant, but present in the wild-type.
(D and E) Ring-shaped Golgi are transformed into irregular shapes in dGM130Δ23 mutant (dGM130−/−) neuron. White arrowheads in (E) point to disconnected ManII- and GalT-puncta.
(F and G) Bar charts showing the percentage of ring-shaped units (F) and the number of ManII-positive puncta that are disconnected from GalT-positive puncta (G) in wild-type (wt) and dGM130−/− neurons.
(H) Quantification of the percentage of multi-compartment Golgi in the proximal and distal dendrites in dGM130−/− mutant neurons.
To test this possibility in vivo, we generated dGM130 null mutants by P-element imprecise excision (Figure 2A–C). The null mutation dGM130Δ23 dramatically perturbed the architecture of somal Golgi in da neurons (Figures 2D and 2E). The ring-shaped Golgi units were rarely detected in dGM130 mutant neurons (Figures 2D–F). Instead, many Golgi compartments of irregular shapes were present. Moreover, puncta that were positive for either ManII-GFP or GalT-TagRFP, but not for both, increased dramatically in the soma (Figures 2D, 2E and 2G), indicating defects in compartmental organization. These results suggest that in da neurons, dGM130 is required for connecting the cis, medial, and trans compartments and for forming the ring-shaped structures.
Loss of dGM130 also led to disconnection of Golgi compartments in the lobula plate tangential cells in the adult fly brain (Figure S2A) and larval epithelial cells (Figure S2B).
We next examined the compartmental organization of Golgi outposts in dendritic shafts of dGM130 null mutant neurons. The percentage of multi-compartment outposts in the proximal 100 μm and distal 100 μm dendrites were quantified separately in control and dGM130 null mutant neurons. Loss of dGM130 decreased the percentage of multi-compartment Golgi in distal dendrites, but did not cause any difference in proximal dendrites between control and dGM130 mutant neurons (Figure 2H and S2D). These results suggest that dGM130 is required for connecting some of the cis, medial, and trans compartments in at least the distal dendrites. This also implies that the dendrites might contain dGM130 at levels that are under detection sensitivity. The reason of the difference between proximal and distal dendrites remains to be determined. It is possible that the dispersion of somal Golgi caused by dGM130 mutation may lead to spillover of multi-compartment Golgi into proximal dendrites.
The role of GM130 in Golgi architecture has been controversial [17, 28–32], which may be caused by the lack of in vivo study carried out with genetically null mutants. Our results show that, although dGM130 null mutant flies were viable and fertile, dGM130 is required for distinct Golgi compartments to join together, establishing GM130 as a key regulator of Golgi architecture in neurons in vivo.
It is known that GM130 functions by forming a protein complex with other proteins, particularly GRASP55 and 65 [33]. The mammalian GRASP65 is required for Golgi cisternal stacking [25]. Our results suggest that dGM130 is necessary for inducing multi-compartment Golgi outposts. Thus, it is possible that GM130 initiates the protein complex for cisternal stacking. We also generated several strains of Drosophila GRASP (dGRASP) null mutant flies by P-element imprecise excision. These dGRASP null flies were viable but the females were sterile. Unlike in dGM130 null neurons, different Golgi compartments were connected in dGRASP null neurons (Figure S2G), raising the possibility of maternally contributed wild-type proteins in the mutant animals. We performed germ-line mosaic analysis and found that the dGRASP mutant clones were sterile, precluding the possibility of removing the potential maternal contribution.
dGM130 is sufficient to induce multi-compartment Golgi outposts in vivo
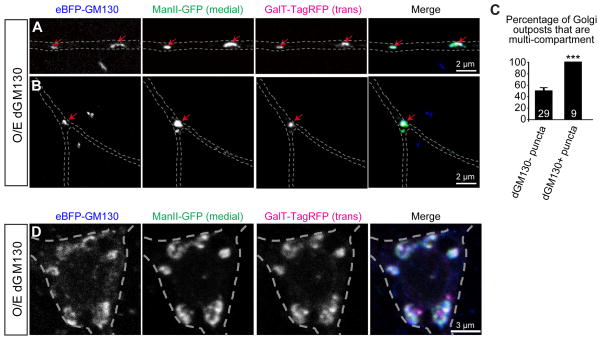
We next tested whether ectopic localization of dGM130 in dendritic shafts is sufficient to connect distinct Golgi compartments. Overexpressing dGM130 led to the appearance of dGM130-positive puncta in the dendrite shafts (Figures 3A and 3C). Strikingly, the cis, medial and trans compartments were juxtaposed to each other at almost every dGM130-postive puncta in the dendritic shafts (Figures 3A and 3C). In contrast to the dendritic shafts, dGM130 overexpression did not significantly affect the compartmental organization of somal Golgi (Figure 3D). Taken together, these results support that notion that dGM130 induces the formation of multi-compartment Golgi outposts in vivo.
Figure 3. Ectopic dGM130 connects distinct Golgi compartments of dendritic Golgi outposts.
(A and B) Dendritic shafts (A) and branch points (B) of da neurons overexpressing dGM130. The medial and trans compartments are labeled by ManII-GFP (green) and GalT-TagRFP (magenta). Red arrows point to dGM130-positive puncta.
(C) Bar chart showing the percentage of Golgi outposts that are multi-compartment in the dendritic shafts of dGM130-overexpressing neurons. The multi-compartment Golgi outposts were identified by the presence of medial and trans Golgi markers.
(D) Soma of da neurons overexpressing dGM130.
The compartmental organization of Golgi outposts regulates acentrosomal microtubule growth
Dendritic Golgi outposts have been previously shown to participate in the patterning of dendritic branches of Drosophila da neurons [7]. To test whether Golgi compartmental organization might contribute to dendritic branching, we examined Golgi compartmental organization in several mutants with dendritic branching defects. Loss-of-function mutations of the transcription factor dar1, which reduces dendritic branching in class III da (C3da) neurons [34], led to a decrease in the percentage of multi-compartmental Golgi in dendrites without affecting the Golgi in the soma and branch points (Figure S3A and B, and data not shown). In contrast, overexpression of Knot, a transcription factor known to increase dendritic growth [35, 36], did not change the compartmental organization in dendrites (Figure S3C and D). These results suggest that certain regulators of dendritic branching and growth may act by regulating the compartmental organization of dendritic Golgi outposts.
A recent study suggests that dendritic Golgi outposts regulate dendritic branching by functioning as acentrosomal nucleation sites for microtubules [8]. In light of our findings that the Golgi outposts comprise two populations, one with single compartments and the other with multiple compartments, we asked whether the structural organization of Golgi outposts regulates microtubule growth. We examined microtubule growth in da neurons in vivo by time-lapse imaging of EB1-GFP [8, 10, 37]. EB1 binds to growing microtubule plus ends and moves in a way that resembles comets (hence termed “EB1-GFP comets”) as microtubules grow [37]. We compared the association of microtubule growth initiation with multi-compartment outposts to that with single-compartment outpost by live-imaging the presence of ManII-eBFP and GalT-TagRFP together with EB1-GFP in wild type da neurons. The number of microtubule initiation events associated with the Golgi outposts containing both medial and trans Golgi compartments was significantly greater than the events associated with single-compartment outposts (Figures 4A and 4B). Consistent with the result that Golgi outposts at branch points contain multiple compartments, branch points also initiated more microtubule growth than the single-compartment outposts in dendritic shafts (Figure 4B). In the dendritic shafts, 57.8% of microtubule growth initiation sites were associated with Golgi outposts containing both medial and trans compartments, compared to 8.9% for medial-only and 13.3% for trans-only outposts (Figure 4C). Twenty percent of dendritic microtubule growth initiation sites were either not associated with dendritic Golgi outposts or associated with Golgi outposts that were below detection sensitivity (Figure 4C). These results raised the possibility that connecting multiple Golgi compartments promotes microtubule growth in vivo.
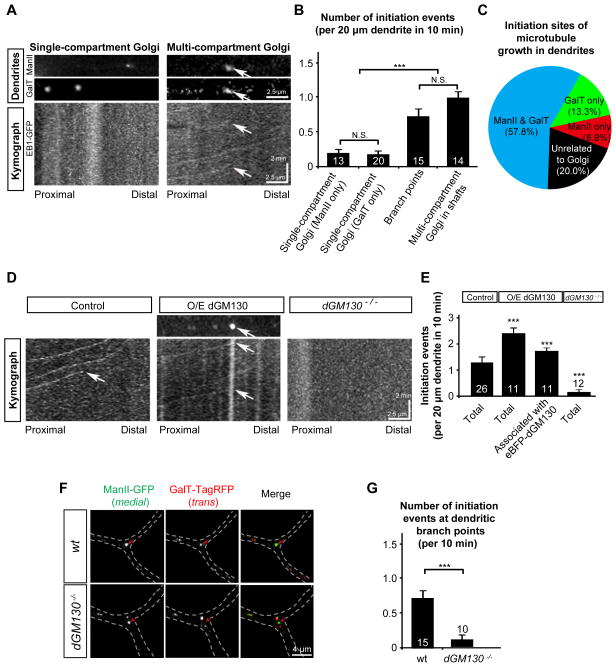
Figure 4. Multi-compartment Golgi outposts are the main initiation sites for microtubule growth in dendrites.
(A and B) Dendritic Golgi outposts with multiple compartments, but not those with single compartments, co-localize with the initiation sites of microtubule growth in dendrites. (A) Kymographs showing the trajectory of EB1-GFP comets in dendritic shafts of wild-type neurons. The arrows point to the position of a multi-compartment Golgi outpost that contains both ManII-eBFP (medial) and GalT-TagRFP (trans). (B) Quantification of the initiation events from dendritic Golgi outposts with different compartmental organizations: single-compartment Golgi outposts (medial or trans only), Golgi outposts at branch points, multi-compartment Golgi outposts (with both medial and trans).
(C) Pie-chart of the percentage of microtubule initiation sites associated with Golgi outposts with different compartmental organizations.
(D and E) Reconstitution of multi-compartment Golgi in dendritic shafts increases, while loss of dGM130 decreases, the number of microtubule growth events in dendrites. (D) Kymographs showing the trajectory of EB1-GFP comets in dendrites of control (mKate2/mRFP-overexpressing), dGM130-overexpressing, and dGM130 null mutant neurons. The arrows indicate the initiation events of microtubule growth. The comets that bypass the imaged dendritic segment during the indicated period are visible in the kymographs but are not counted as initiation events. (E) Quantification of the initiation events in control and dGM130-overexpressing neurons.
(F) Golgi compartments are disconnected in the dendritic branch points of dGM130 mutant neurons;
(G) Quantification of the initiation events at the branch points in wild type and dGM130 mutant neurons.
Because introducing dGM130 into dendrites connects the cis, medial, and trans compartments of dendritic Golgi (Figures 3A and C), we compared the number of initiation events of microtubule growth in the dendrites between control and dGM130-overexpressing neurons. The number of microtubule growth initiation was significantly increased in the dendritic shafts of neurons overexpressing dGM130, compared to control (Figures 4D and 4E). The increase was largely due to events associated with dGM130-containing multi-compartment Golgi outposts (Figure 4E).
In dGM130 null mutant neurons, microtubule growth initiation events were reduced in distal dendrites (Figure 4D and E). Since Golgi compartments in dGM130-deficient neurons were dispersed in branch points (Figure 4F), we examined microtubule growth initiation at these branch points. Consistently, microtubule growth initiation at dendritic branch points was dramatically suppressed by dGM130 mutations (Figure 4G). In contrast, loss of dGMAP, another Golgi structural protein, did not affect microtubule growth initiation (Figures S4A and S4B). Taken together, these results suggest that the dGM130-mediated compartmental organization of dendritic Golgi outposts regulates microtubule growth in dendrites.
We also assessed the role of dGM130 in dendritic branching. The total number of dendritic branch points as well as the number of higher order branches (4th order and up) was significantly reduced in dGM130 mutant class III da neurons but increased in dGM130-overexpressing neurons of the same type (Figures S4C–E). These results suggest that dGM130, and possibly compartmental organization of Golgi outposts, is a factor that determines the number of higher order dendritic branches.
The mechanism underlying the microtubule growth regulated by dGM130 is currently unclear. dGM130 might regulate microtubule growth through three different mechanisms. First, different Golgi compartments may each serve a unique role in microtubule nucleation. Thus, multi-compartment Golgi serve as a functional scaffold for the microtubule nucleation machinery. Second, GM130, rather than multi-compartment Golgi, may be responsible for initiating microtubule growth. Third, it is also possible that dGM130 and compartmental organization of Golgi regulates microtubule growth indirectly through other Golgi functions such as membrane trafficking.
Previous studies on mammalian hippocampal neurons have shown that ribbon-like Golgi stacks that are disconnected from somal Golgi and positive for GM130 are only located in the soma and proximal dendrites [1]. This has led to the speculation that Golgi outposts might only exist in proximal dendrites. However, this speculation contradict the proposal that membrane proteins are synthesized locally at synapses in distal dendrites [38] and the ultrastructural and immunofluorescence studies showing the presence of membranous organelles positive for Golgi markers [5, 39]. The findings described in this study reconcile this contradiction by showing that Golgi in the soma and those in the dendrites assume different compartmental organizations.
EXPERIMENTAL PROCEDURES
Fly stocks and cDNA constructs
The following fly stocks published before were used in this study: P{RS3}GM130CB-6408-3 (Drosophila Genetic Resource Center), GAL419–12 [40], UAS-EB1-GFP [37], UAS-ManII-GFP [7], dar13232 [34], and UAS-Knot [36].
To generate the UAS-GalT-TagRFP and UAS-ManII-eBFP2 or TagRFP transgenic lines, we replaced EGFP/YFP with either eBFP2 or TagRFP-T in the pUAST-ManII-eGFP and pUAST-GalT-YFP constructs [7]. To make UAS-eBFP-dGM130 transgenic line, we fused dGM130 cDNA to the C-terminus of eBFP and inserted it into the pUAST vector. To make the UAS-HA-dManI transgenic line, full length cDNA of Drosophila ManI (dManI) was amplified from the EST clone RE43942 by PCR and inserted in-frame into the C-terminus of the HA-tag in the vector pAttB-HA-UAST. To make the UAS-GalNacT2-YFP/TagRFP transgenic lines, we fused the full length human GalNacT2 cDNA (gift from Graham Warren and Ayano Satoh) with YFP or TagRFP-T. The transformation constructs were injected into w1118 embryos to generate transgenic flies, with the exception of UAS-HA-ManI, which was injected into P{CaryP}attP40 embryos.
Immunostaining and Imaging of Golgi markers in larval da neurons
To image the Golgi structures labeled by fluorescent protein-tagged markers, third instar larvae were dissected in insect saline, fixed with 4% formaldehyde for 40 min, and imaged without staining. To image endogenous dGM130 and dGMAP, the fixed samples were immunostained with rabbit anti-dGM130 (1:1000; Abcam, Cambridge, MA) and rabbit anti-dGMAP (1:1000; gift from Drs. Pascal Therond and Florence Friggi-Grelin).
Three dimensional fluorescence images were acquired using a Leica SP5 AOBS spectral laser scanning confocal microscope (Leica Microsystems). Both somal Golgi and dendritic Golgi outposts images were acquired using a 63x oil objective lens (N.A. = 1.41) in XYZ mode at a z step of 113 nm. Image stacks were then deconvolved with the deconvolution function included in the Leica confocal software.
Live imaging and analysis of EB1-GFP in dendrites
Third-instar larvae expressing EB1-GFP driven by the Gal419–12 driver were anesthetized by ether, mounted in halocarbon oil, and live-imaged with a Leica SP5, an Olympus FV1000/1200, or a Zeiss 710/780 confocal system. The class III da neurons in the dorsal cluster (ddaA and ddaF) were imaged. The control larvae carried UAS -EB1-GFP, UAS-GalT-TagRFP, UAS-ManII-eBFP2, and Gal419–12. The experiments for dGM130 overexpression were done on larvae carrying UAS-EB1-GFP, UAS-dGM130, Uas-eBFP2-dGM130, and Gal419–12. Multi-channel z-stacks of time-lapse images were collected in distal (~ 100 μm) dendrites at 6 sec interval for 10 min. We imaged either two channels for eBFP -dGM130 and EB1-GFP or three channels for ManII-eBFP, GalT-TagRFP and EB1-GFP. Time-lapse images with at least one EB1-GFP comet, regardless of the origin, were analyzed. Kymographs were generated with ImageJ (NIH) and used to count the total number of microtubule growth initiation events and the number of initiation sites.
Supplementary Material
Highlights.
Disconnected Golgi compartments exist in dendrites but not in soma;
The lack of dGM130 in dendritic shafts leads to disconnected Golgi compartments;
Golgi outposts with multiple compartments promote microtubule growth.
Acknowledgments
We thank Drs. Graham Warren, Ayano Satoh, Florence Friggi-Grelin, Pascal Therond, Hans Bakker, Hugo Maccioni, Catherine Rabouille, Yanzhuang Wang, Yang Xiang, and Tadashi Uemura for sharing reagents. We also thank Drs. Jingshi Shen, Yukiko Yamashita, Haoxing Xu, Xiaowei Chen, Ting Han, and Gabriella Sterne for critical reading of an earlier draft of the manuscript, and the Optical Bioimaging Core Facility of WNLO-HUST for support of imaging systems. This work was supported by grants from China Postdoctoral Science Foundation (No. 2013M542010) to J.C., the Fundamental Research Funds for the Central Universities/HUST (No. 2012TS029) and Major Research Program of National Natural Science Foundation of China (No. 91232306) to W. Z., NIH (R01MH091186 and R21AA021204), Protein Folding Disease Initiative of the University of Michigan, and the Pew Scholars Program in the Biological Sciences to B.Y.
Footnotes
The authors declare no conflict of financial interests.
AUTHOR CONTRIBUTIONS: W.Z. and B.Y. designed the experiments. W.Z., J.C., X.W., Y.Z., S.K., and M.S. conducted the experiments. W.Z. and J.C. analyzed the data. W.Z. and B.Y. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Hanus C, Ehlers MD. Secretory Outposts for the Local Processing of Membrane Cargo in Neuronal Dendrites. Traffic. 2008;9:1437–1445. doi: 10.1111/j.1600-0854.2008.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thayer DA, Jan YN, Jan LY. Increased neuronal activity fragments the Golgi complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1482–1487. doi: 10.1073/pnas.1220978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Camilli P, Moretti M, Donini SD, Walter U, Lohmann SM. Heterogeneous distribution of the cAMP receptor protein RII in the nervous system: evidence for its intracellular accumulation on microtubules, microtubule-organizing centers, and in the area of the Golgi complex. J Cell Biol. 1986;103:189–203. doi: 10.1083/jcb.103.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardiol A, Racca C, Triller A. Dendritic and postsynaptic protein synthetic machinery. J Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. The Journal of neuroscience. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeyifous O, Waites CL, Specht CG, Fujisawa S, Schubert M, Lin EI, Marshall J, Aoki C, de Silva T, Montgomery JM, et al. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12:1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM. Directed Microtubule Growth, +TIPs, and Kinesin-2 Are Required for Uniform Microtubule Polarity in Dendrites. Current Biology. 2010;20:2169–2177. doi: 10.1016/j.cub.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann J, Jokitalo E, Pypaert M, Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 2000;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- 13.Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 14.Velasco A, Hendricks L, Moremen KW, Tulsiani DR, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. The Journal of cell biology. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Molecular biology of the cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- 17.Kondylis V, Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yano H, Yamamoto-Hino M, Abe M, Kuwahara R, Haraguchi S, Kusaka I, Awano W, Kinoshita-Toyoda A, Toyoda H, Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proceedings of the National Academy of Sciences. 2005;102:13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friggi-Grelin F, Rabouille C, Therond P. The cis-Golgi Drosophila GMAP has a role in anterograde transport and Golgi organization in vivo, similar to its mammalian ortholog in tissue culture cells. European journal of cell biology. 2006;85:1155–1166. doi: 10.1016/j.ejcb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. The Journal of cell biology. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klumperman J. Architecture of the mammalian Golgi. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Molecular biology of the cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. The Journal of cell biology. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 26.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. The EMBO journal. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 Recruits [gamma]-Tubulin Complexes to cis-Golgi Membranes and Is Required for Golgi Ribbon Formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 29.Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. The Journal of cell biology. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marra P, Salvatore L, Mironov A, Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Molecular biology of the cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Seminars in cell & developmental biology. 2009;20:770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. The Biochemical journal. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- 34.Ye B, Kim JH, Yang L, McLachlan I, Younger S, Jan LY, Jan YN. Differential regulation of dendritic and axonal development by the novel kruppel-like factor dar1. J Neurosci. 2011;31:3309–3319. doi: 10.1523/JNEUROSCI.6307-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and Cut Control Different Aspects of Dendrite Cytoskeleton and Synergize to Define Final Arbor Shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 36.Hattori Y, Sugimura K, Uemura T. Selective expression of Knot/Collier, a transcriptional regulator of the EBF/Olf-1 family, endows the Drosophila sensory system with neuronal class-specific elaborated dendritic patterns. Genes to Cells. 2007;12:1011–1022. doi: 10.1111/j.1365-2443.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 37.Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ. Polarity and intracellular compartmentalization of Drosophila neurons. Neural development. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanus C, Schuman EM. Proteostasis in complex dendrites. Nat Rev Neurosci. 2013;14:638–648. doi: 10.1038/nrn3546. [DOI] [PubMed] [Google Scholar]
- 39.Pierce JP, Mayer T, McCarthy JB. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol. 2001;11:351–355. doi: 10.1016/s0960-9822(01)00077-x. [DOI] [PubMed] [Google Scholar]
- 40.Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.