Abstract
The immune stimulation induced by short interfering RNAs (siRNAs) has been reported to be quieted or abrogated by methoxy or fluoro modifications of the 2′ position of the ribose sugar. However, variables such as the type of modification, nucleotide preference, and strand bias have not been systematically evaluated. Here, we report the results of a screen of several modified siRNAs via a human peripheral blood monocyte cytokine induction assay. Unlike corresponding modifications of guanosine, cytidine, or uridine, 2′-fluoro modification of adenosine significantly reduced cytokine induction while retaining siRNA knockdown activity. The results of this study suggest adenosine as an optimal target for modification.
Introduction
Unmodified RNAs, including short interfering RNAs (siRNAs), induce an innate immunostimulatory response that is primarily mediated by Toll-like receptors [reviewed in (Judge and MacLachlan, 2008)]. The subsequent immune response and release of inflammatory cytokines represents a major challenge for the development of safe RNA therapeutics. Chemical modifications can be used to improve drug-like characteristics of siRNAs, including increasing stability while reducing toxicity [reviewed in (BEHLKE, 2008; Watts et al., 2008; Bramsen and Kjems, 2011)]. Previously published reports have investigated the immunostimulatory potentials of methoxy (2′OMe), fluoro (2′F), and deoxy (2′H) modified siRNAs (Judge et al., 2005, 2006; Robbins et al., 2007; Sioud et al., 2007; Eberle et al., 2008). Judge et al. (2006) evaluated 2′OMe modifications of the passenger strand of an siRNA targeting apolipoprotein B (ApoB) and found that adenosine (A), guanosine (G), and uridine (U) modifications effectively reduced the levels of tumor necrosis factor alpha (TNFα). However, 2′OMe cytidine (C) was ineffective—a surprising result that was then confirmed in vivo (Judge et al., 2006). Other researchers compared modified uridines in single-strand RNAs and reported that 2′OMe, 2′F, and 2′H modifications reduced TNFα levels (Sioud et al., 2007). Interestingly, only the 2′OMe modification significantly antagonized the TNFα induction by a separate unmodified RNA, indicating a sequence-independent abrogation. A similar study evaluated the effectiveness of 2′OMe modified A, G, and C in single-strand RNAs and reported that all 3 modifications effectively silenced the interferon alpha (IFNα) induction of the RNAs themselves; however, only 2′OMe-A completely antagonized IFNα induction by a separate unmodified RNA (Robbins et al., 2007).
Additional in vivo studies compared siRNAs containing combinations of 2′F and 2′OMe modifications and showed that overall methoxy modifications alone or combined with fluoro pyrimidines were effective in quieting interferon induction, while fluoro-only pyrimidine modifications were less effective (Shin et al., 2007). Cekaite et al. (2007) employed an mRNA biomarker approach that evaluates the effects of 2′F-U and 2′OMe-U modifications on the immunostimulatory potential of single-strand RNAs and found that fluoro and methoxy uridine were equally effective in reducing the induction of immune-related biomarkers). Overall, 2′OMe and 2′F modifications have been described in certain contexts, but none of these studies have systematically compared 2′OMe and 2′F modifications on all 4 nucleotides.
Here, we report the in vitro evaluation of the impact on siRNA-mediated immune stimulation of 2′OMe and 2′F modifications applied in a nucleotide-specific manner to either guide, passenger, or both strands of several siRNAs. This includes the first reported evaluation of the immune stimulation of siRNAs containing 2′F modified purines. Significantly, we discover that adenosine was the only one of the 4 nucleotides that confers immune stealth with both 2′OMe and 2′F ribose modifications. We also confirm previous reports by recapitulating known liabilities of modified cytidine in conferring immune stealth (Judge et al., 2006; Shin et al., 2007; Eberle et al., 2008). These data corroborate that 2′F incorporations within siRNAs are generally more tolerated for siRNA activity than corresponding 2′OMe modifications [reviewed in (BEHLKE, 2008; Watts et al., 2008; Bramsen and Kjems, 2011)]. Overall, 2′F modification of adenosines are recommended for reducing immune stimulation while retaining optimal siRNA knockdown.
Materials and Methods
Oligo sequence and synthesis
β-galactosidase siRNAs are slightly modified versions of those published (Judge et al., 2005) with the addition of 2 nucleotide uridine overhangs on both strands. The β-gal 728 siRNA is as follows: guide strand 5′-3′ (AAAUCGCUGAUU UGUGUAGUU) and passenger strand 5′-3′ (CUACACAAAU CAGCGAUUUUU). The β-gal control siRNA is a nontargeting control sequence: guide strand 5′-3′ (UAGCGACUAAAC ACAUCAAUU) and passenger strand 5′-3′ (UUGAUGUGU UUAGUCGCUAUU). A previously published siRNA targeting ApoB (Judge et al., 2006) was also used: phosphorylated guide strand 5′-3′ (pAUUGGUAUUCAGUGUGAUGA CAC) and passenger strand 5′-3′ (GUCAUCACACUGAAU ACCA AU). Knockdown studies used a nontargeting control siRNA sequence, which contains fluoro 2′F (f ), methoxy 2′OMe (m), deoxy 2′H (d), and ribo 2′OH (r) residues at the indicated positions as well as inverted abasic caps (iB) on the passenger strand: guide (fC;fC;fU;mG;mA;mA;mG;mA;mG;mA;mG;fU;fU;mA;mA;mA;rA;rG;rA;mU;mU) and passenger (iB;fU;fC;fU;fU;fU;fU;dA;dA;fC;fU;fC;fU;fC;fU;fU;fC;dA;dG;dG;dT;dT;iB). siRNAs were synthesized at Merck & Co. using previously described methods (Wincott et al., 1995; Morrissey et al., 2005). See Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/nat) for the listing of specific sequence modifications for all siRNAs used in this study.
siRNA formulation and administration to peripheral blood monocytes
Isolation of human peripheral blood monocytes (PBMCs), formulation of siRNA lipid nanoparticles (LNP), and administration to PBMCs was performed as previously described (Peacock et al., 2011). L201 lipid mixture is a combination of 6% Peg-DMG (Sunbright), 44% Cholesterol (MP Biomedical), and 50% ClinDMA (Merck and Co., Inc.) (Abrams et al., 2010; Kenski et al., 2010). LNP-formulated siRNA at 50uM was diluted 20-fold into freshly plated PBMCs, incubated for 16–20 hours, and conditioned media was then frozen at −80 or directly processed with cytokine ELISAs.
Cytokine ELISA assays
Human TNFα was quantified by a sandwich enzyme-linked immunosorbent assay as previously described (Peacock et al., 2011). Briefly, human TNFα antibody (Thermo Scientific) diluted 1:250 in phosphate buffered saline (PBS) was coated in 96-well plates and incubated overnight. The plate was then blocked with 4% BSA-PBS (blocking buffer) before the PBMC supernatant was transferred to the coated plate. After a 1–2 hour incubation, the plate was washed 3× with PBST; the detection antibody (diluted 1:250) was added and incubated for 2 hours. Strepavidin-HRP conjugate was added and incubated for 30 minutes before reading the plate using Luminol-based horseradish peroxidase detection reagent and SuperSignal West Pico Chemiluminescent Substrate (Pierce). The plate was read on an EnVision plate reader (Perkin Elmer).
Measurement of β-galactosidase activity
A mouse hepatocyte-derived cell line (Hepa1-6) was co-transfected with siRNAs and a β-galactosidase plasmid (pCMV SPORT β-gal, Invitrogen) using Lipofectamine 2000 (Invitrogen). The cells were seeded at 20,000 per well in 96-well PolySorp opaque white plates (Nunc), incubated for 24 hours at 37C, and then transfected with 10 nM siRNA and 0.6 ng/μL of pCMV SPORT β-gal plasmid. The transfected cells were incubated overnight at 37C; then, β-gal enzyme activity was measured using the Gal-Screen luminescence detection system (Applied Biosystems) and SpectraMax plate reader (Molecular Devices). Luminescence values were normalized to a nontargeting control siRNA to calculate percent activity.
Results
We quantified siRNA-mediated immune stimulation using a recently described in vitro method that measures the induction of TNFα, resulting from the administration of LNP-formulated siRNAs to human PBMC (Peacock et al., 2011). The siRNAs were formulated in LNP that approximate those used for in vivo studies and the development of therapeutic siRNAs (Abrams et al., 2010). Two published siRNAs with known immunostimulatory potentials (Judge et al., 2005) were selected for the systematic evaluation of methoxy (2′OMe) and fluoro (2′F) ribose modifications of the 4 nucleotides of the passenger and guide strands. One of these siRNAs targets β-galactosidase (β-gal 728), while the other is a related but nontargeting control (β-gal control). Additionally, we conducted a limited 2′OMe analysis of an ApoB siRNA, which also has a previously described immune response (Judge et al., 2006).
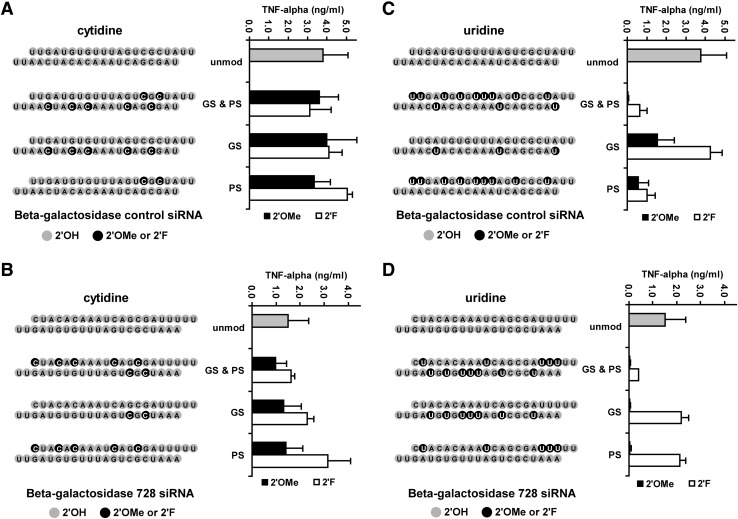
Cytidine modifications do not abrogate immune stimulation
Methoxy modification (2′OMe) of cytidines within the 2 β-gal siRNAs was largely ineffective in reducing siRNA-mediated immune stimulation (Fig. 1A, B), and similar results were seen with the ApoB siRNA (Supplementary Fig. S1A). This recapitulates published reports finding that 2′OMe modification of cytidine was not effective in reducing immune stimulation (Judge et al., 2006; Shin et al., 2007; Eberle et al., 2008). We extend these observations to fluoro modifications (2′F), which also did not quiet siRNA-mediated TNFα induction (Fig. 1A, B). Lastly, since our study employs an LNP delivery vehicle that is distinct from the vehicles used in previous studies, the concordance of immune stimulation results highlights the fact that differing lipid delivery platforms do not impact siRNA-mediated immune stimulation in vitro.
FIG. 1.
Comparison of the effect of pyrimidine modifications on TNFα levels induced in human PBMC cultures after overnight incubation with lipid nanoparticle- formulated siRNAs. 2′OMe and 2′F ribose modifications in both guide and passenger (GS & PS), guide strand only (GS), or passenger strand only (PS) are compared with an unmodified siRNA (unmod). Distribution of modifications with the siRNA strands are represented in the schematic duplexes as black dots with unmodified positions depicted as gray. Modified cytidines present in control siRNA (A) or 728 siRNA (B) are compared with modified uridines in control siRNA (C) or 728 siRNA (D). TNFα levels are shown for unmodified siRNA (gray bars), 2′OMe modified (black bars), and 2′F modified (white bars). TNFα levels for each siRNA are listed in Supplementary Tables S2 and S3. PBMC, peripheral blood monocyte; siRNA, short interfering RNA; TNFα, tumor necrosis factor alpha; 2′OMe, methoxy; 2′F, fluoro.
Methoxy modified uridines can reduce immune stimulation
Uridine modification with 2′OMe is markedly more effective in reducing siRNA-mediated immune stimulation of both β-gal (Fig. 1C, D) siRNAs. Compared with the 7- to 77-fold reduction observed with 2′OMe uridine, 2′F modifications resulted in a less pronounced 4- to 6-fold reduction of TNFα levels (Supplementary Fig. S2, Tables S2– S3). Significant reductions in TNFα levels were also observed for the ApoB siRNA modified with 2′OMe uridine (Supplementary Fig. S1B). Interestingly, when the guide strand of the β-gal control siRNA was modified at only 3 positions, 2′F and 2′OMe conferred immune stealth was significantly compromised (Fig. 1C and Supplementary Fig. S2). This suggests that a threshold number of uridines should be modified to significantly abrogate immune stimulation. However, a more comprehensive titration of 2′OMe and 2′F modifications will be required for more definitive proof.
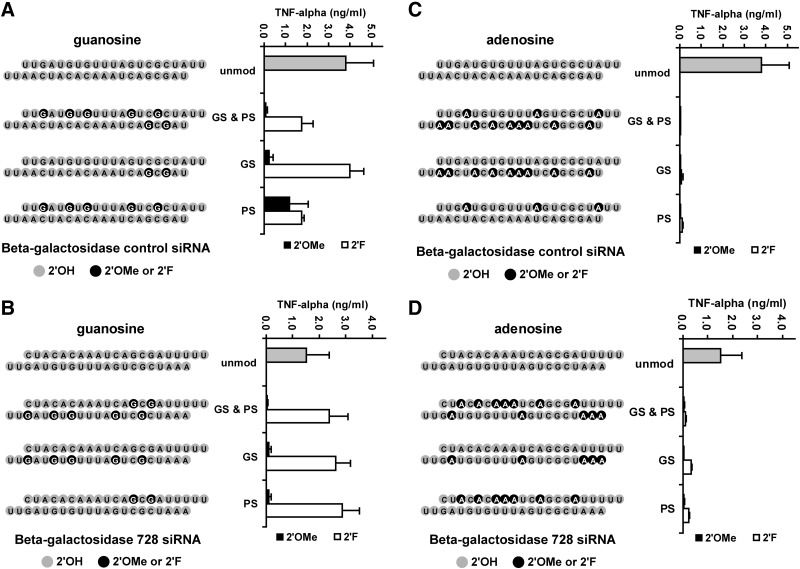
Small numbers of methoxy modified guanosines can reduce immune stimulation
Methoxy guanosine modifications are effective in reducing TNFα induction in β-gal (Fig. 2A, B) and ApoB siRNAs (Supplementary Fig. S1C). Unlike uridine modifications, small numbers of guanosine 2′OMe modifications largely abrogated siRNA-mediated immune stimulation. As few as 2 methoxy modified guanosines in either the passenger strand or the guide strand conferred a 15- to 17-fold reduction in TNFα levels (Supplementary Tables S2, S3, and S4). Similar to uridine, 2′F modifications were not effective in significantly reducing TNFα levels on a consistent basis.
FIG. 2.
Evaluation of the impact of purine modifications on siRNA-mediated immune stimulation in human PBMC cultures. Design of the experiment and schematic depiction of siRNAs is discussed in the Fig. 1 legend. Modified guanosines present in control siRNA (A) or 728 siRNA (B) are compared with modified adenosines present in control siRNA (C) or 728 siRNA (D). TNFα levels for each siRNA are listed in Supplementary Tables S2 and S3.
Adenosine modification with either 2′OMe or 2′F confers immune stealth
Strikingly, either methoxy or fluoro modifications of adenosine significantly reduced immune stimulation by the β-gal siRNAs in PBMCs (Fig. 2C, D). Overall, 2′OMe modified adenosines reduced TNFα levels ∼50-fold, while 2′F modifications reduced levels 4- to 13-fold (Supplementary Fig. S2, Tables S2–S3). While the 2′F-mediated reduction of TNFα levels was less pronounced than corresponding 2′OMe modifications, 2′F adenosine modifications were still far more effective than 2′F modified guanosine, uridine, or cytidine. As few as 3 modified adenosines in the passenger strand of β-gal control siRNA can reduce TNFα levels 39-fold (for 2′F modifications) or 110-fold (for 2′OMe) (Supplementary Fig. S2, Tables S2–S3). Methoxy modified adenosines were also effective in reducing TNFα induction for the ApoB siRNA (Supplementary Fig. S1D).
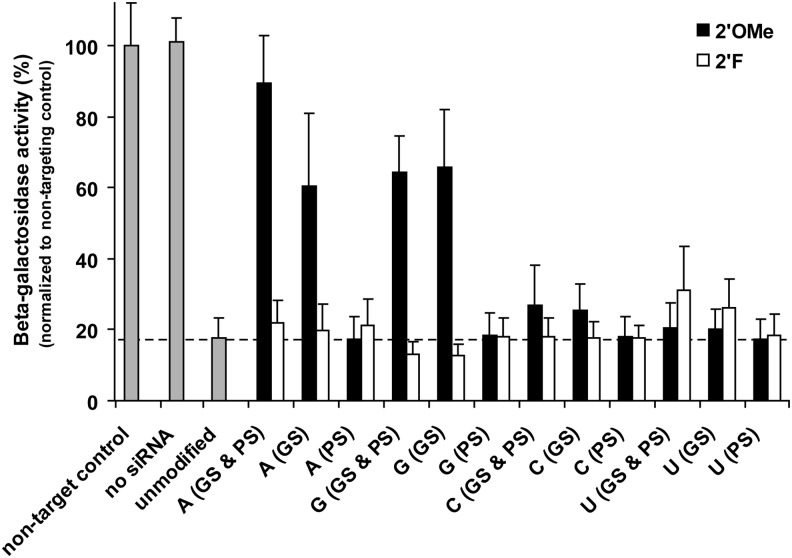
2′F modifications are more tolerated for siRNA knockdown
The knockdown activity of methoxy and fluoro modified versions of the 728 siRNA were compared by measuring β-galactosidase enzymatic activity in vitro (Fig. 3). Knockdown was also measured for the nontargeting β-gal control siRNA, and no significant inhibition of β-gal activity was observed (Supplementary Table S2). The unmodified β-gal 728 siRNA reduced enzyme activity levels to 18%. This baseline of β-gal knockdown was then compared with the series of 24 2′F or 2′OMe modified siRNAs. Relative to the unmodified siRNA, 2′OMe modifications of the guide strand—in particular adenosine and guanosine—had adverse affects on knockdown. In contrast, 2′F modifications were broadly tolerated and did not negatively impact the potential of knockdown for this siRNA.
FIG. 3.
Knockdown of β-galactosidase activity for βgal-728 siRNA. The modified positions (A for adenosine, G for guanosine, C for cytidine, and U for uridine) and location of modifications (GS for guide strand and PS for passenger strand) are noted. Unmodified β-gal 728, an unrelated nontargeting control siRNA, and no siRNA (β-gal plasmid alone) are included as controls. Gray bars indicate unmodified RNA, black bars represent 2′OMe modifications, and white bars represent 2′F modifications. Horizontal dotted line indicates the reduction in β-galactosidase enzymatic activity seen with the unmodified β-gal 728 siRNA. Knockdown values listed in Supplementary Table S3. β-gal, β-galactosidase.
Discussion
Using 63 different siRNAs, we conducted a systematic comparison of the impact of 2′OMe and 2′F modifications of guide and passenger strands on siRNA-mediated immune stimulation in human PBMC. We recapitulate earlier reports that cytidine modifications were ineffective in reducing RNA-mediated immune stimulation (Fig. 1A, B, Supplementary Figs. S1–S2). In contrast, when present in both strands, uridine and guanosine 2′OMe modifications were effective in abrogating immune stimulation—reducing TNFα levels by 42- to 77-fold for β-gal siRNAs (Supplementary Fig. S2) and by 70- to 72-fold for ApoB (Supplementary Fig. S1E). However, 2′OMe guanosine modifications were distinct from uridine in that the modification of only 2 guanosine residues was sufficient to reduce immune stimulation by 15- to 17-fold for β-gal siRNAs (Supplementary Fig. S2) and by 17-fold for ApoB (Supplementary Fig. S1E). In marked contrast, three 2′OMe uridine incorporations only reduced TNFα levels by 2-fold for β-gal siRNAs. Fluoro modification of guanosine or uridine in both strands reduced TNFα levels by 2- to 6-fold but did not completely silence the stimulatory response (Supplementary Fig. S2). 2′F uridine modifications have been reported as abrogating cytokine induction, but this was in the context of a single-stranded RNA of a different sequence, which may explain the differing results (Sioud et al., 2007).
Unlike the 3 other nucleotides, both 2′OMe and 2′F modification of adenosine resulted in a striking reduction of TNFα levels (Fig. 2C, D and Supplementary Tables S2–S3). This pronounced effect was present even with the modification of individual strands. For the 2 β-gal siRNAs, 2′OMe adenosines reduced TNFα induction by 47- to 133-fold whether present in both strands or limited to individual strands. The incorporation of 2′OMe adenosines in the ApoB siRNA reduced TNFα levels by 70- to 74-fold (Supplementary Fig. S1D and Table S4). Uridine or guanosine 2′OMe modifications significantly reduced TNFα levels, but their immunosuppression was less consistent across individual strands than the effect observed for 2′OMe adenosine (Supplementary Figs. S1E and S2). 2′F adenosine was not as effective in reducing TNFα levels as was 2′OMe. This was most evident for β-gal 728; however, even then, the 2′F adenosine suppression of the RNA-mediated immune response was 4- to 13-fold. Furthermore, the limited 2′F or 2′OMe modification of just 3 adenosines effectively reduced immune stimulation (Fig. 2C).
Particular sequence motifs within siRNAs have been reported as being a specific source of immune stimulation—most pronounced, being the “UGUGU” motif present in the β-gal siRNAs used in our study (Judge et al., 2005). A putative “GUGUG” motif has been highlighted in the ApoB siRNA as well (Judge et al., 2006). A more extensive “GUCCUUCA” immunostimulatory motif potently induced IFNα levels in mouse plasmacytoid dendritic cells (Hornung et al., 2005); however, this longer motif is not present within the siRNAs used in our study. Despite the current literature supporting these stimulatory motifs, the UGUGU motif is frequently unmodified within the β-gal siRNAs and yet TNFα are significantly reduced (Figs. 1–2). Most dramatically, the 2′OMe modification of only 2 guanosines on the passenger strand greatly reduces immune stimulation, while the UGUGU motif on the guide strand remains unmodified (Fig. 2B). Similar results are seen with 3 adenosine modifications of β-gal control siRNA (Fig. 2C) and two 2′OMe guanosines within ApoB siRNA (Supplementary Fig. S1C). These findings are also consistent with our previously published analysis of the impact on the modifications of a micro RNA miR-122, which contains putative “UGU” motifs (Peacock et al., 2011).
Fluoro modifications (in particular, purines) are more broadly tolerated in the siRNA guide strand and have a less deleterious effect on siRNA knockdown activity than comparable 2′OMe modifications (Fig. 3). Modified siRNAs containing 2′-fluoro have previously been reported and maintain excellent in vitro knockdown activity and potency (Chiu and Rana, 2003; Allerson et al., 2005; Prakash et al., 2005; Muhonen et al., 2007). Overall, this suggests that 2′F modifications (especially adenosines) are ideally suited for the abrogation of immune stimulation while retaining the activity of siRNA knockdown. Thus, the immune stealth conferred by 2′F adenosines coupled with their broad tolerance in maintaining RNAi knockdown activity highlights the potential value of incorporating 2′F adenosine modifications into future siRNA designs, including possible RNA therapeutics.
Supplementary Material
Acknowledgments
The authors thank Enrique Vazquez, Mark Levorse, and Becky Arvary for their synthesis of oligonucleotides; Dipali Ruhela, Silvia Chang, Zhi Yu Hu, Radu Mihalia, and Alan Wei for their role in providing material and expertise for LNP formulation; Jeremy Caldwell and Peter Haeberli for their valuable discussion and feedback; and Stanford Blood Bank for providing buffy coats. The authors were employees of Merck & Co., Inc., within the RNA Therapeutics Department, which provided research funding for this work.
Author Disclosure Statement
No competing financial interests exist.
References
- ABRAMS M.T. KOSER M.L. SEITZER J. WILLIAMS S.C. DIPIETRO M.A. WANG W. SHAW A.W. MAO X. JADHAV V. DAVIDE J.P., et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLERSON C.R. SIOUFI N. JARRES R. PRAKASH T.P. NAIK N. BERDEJA A. WANDERS L. GRIFFEY R.H. SWAYZE E.E. BHAT B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- BEHLKE M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- BRAMSEN J.B. KJEMS J. Chemical modification of small interfering RNA. Methods Mol. Biol. 2011;721:77–103. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- CEKAITE L. FURSET G. HOVIG E. SIOUD M. Gene expression analysis in blood cells in response to unmodified and 2′-modified siRNAs reveals TLR-dependent and independent effects. J. Mol. Biol. 2007;365:90–108. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- CHIU Y.L. RANA T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EBERLE F. GIESSLER K. DECK C. HEEG K. PETER M. RICHERT C. DALPKE A.H. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J. Immunol. 2008;180:3229–3237. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- HORNUNG V. GUENTHNER-BILLER M. BOURQUIN C. ABLASSER A. SCHLEE M. UEMATSU S. NORONHA A. MANOHARAN M. AKIRA S. DE FOUGEROLLES A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- JUDGE A. MACLACHLAN I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- JUDGE A.D. BOLA G. LEE A.C. MACLACHLAN I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- JUDGE A.D. SOOD V. SHAW J.R. FANG D. MCCLINTOCK K. MACLACHLAN I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- KENSKI D.M. COOPER A.J. LI J.J. WILLINGHAM A.T. HARINGSMA H.J. YOUNG T.A. KUKLIN N.A. JONES J.J. CANCILLA M.T. MCMASTERS D.R., et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5′-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010;38:660–671. doi: 10.1093/nar/gkp913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRISSEY D.V. LOCKRIDGE J.A. SHAW L. BLANCHARD K. JENSEN K. BREEN W. HARTSOUGH K. MACHEMER L. RADKA S. JADHAV V., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- MUHONEN P. TENNILA T. AZHAYEVA E. PARTHASARATHY R.N. JANCKILA A.J. VAANANEN H.K. AZHAYEV A. LAITALA-LEINONEN T. RNA interference tolerates 2′-fluoro modifications at the Argonaute2 cleavage site. Chem. Biodivers. 2007;4:858–873. doi: 10.1002/cbdv.200790073. [DOI] [PubMed] [Google Scholar]
- PEACOCK H. FUCINI R.V. JAYALATH P. IBARRA-SOZA J.M. HARINGSMA H.J. FLANAGAN W.M. WILLINGHAM A. BEAL P.A. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J. Am. Chem. Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRAKASH T.P. ALLERSON C.R. DANDE P. VICKERS T.A. SIOUFI N. JARRES R. BAKER B.F. SWAYZE E.E. GRIFFEY R.H. BHAT B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- ROBBINS M. JUDGE A. LIANG L. MCCLINTOCK K. YAWORSKI E. MACLACHLAN I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- SHIN D. KIM S.I. PARK M. KIM M. Immunostimulatory properties and antiviral activity of modified HBV-specific siRNAs. Biochem. Biophys. Res. Commun. 2007;364:436–442. doi: 10.1016/j.bbrc.2007.10.012. [DOI] [PubMed] [Google Scholar]
- SIOUD M. FURSET G. CEKAITE L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 2007;361:122–126. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- WATTS J.K. DELEAVEY G.F. DAMHA M.J. Chemically modified siRNA: tools and applications. Drug. Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- WINCOTT F. DIRENZO A. SHAFFER C. GRIMM S. TRACZ D. WORKMAN C. SWEEDLER D. GONZALEZ C. SCARINGE S. USMAN N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.