Abstract
There are a growing number of studies reporting the observation of purine-pyrimidine base-pairs that are seldom observed in unmodified nucleic acids because they entail the loss of energetically favorable interactions or require energetically costly base ionization or tautomerization. These high energy purine-pyrimidine base-pairs include G•C+ and A•T Hoogsteen base-pairs, which entail ~180° rotation of the purine base in a Watson-Crick base-pair, protonation of cytosine N3, and constriction of the C1′–C1′ distance by ~2.5 Å. Other high energy pure-pyrimidine base-pairs include G•T, G•U, and A•C mispairs that adopt Watson-Crick like geometry through either base ionization or tautomerization. Although difficult to detect and characterize using biophysical methods, high energy purine-pyrimidine base-pairs appear to be more common than once thought. They further expand the structural and functional diversity of canonical and noncanonical nucleic acid base-pairs.
Introduction
In naked unmodified nucleic acid duplexes, purines (guanine and adenine) pair up with pyrimidines (cytosine, thymine, and uracil) through complementary hydrogen bonds to form canonical G•C, A•T, and A•U Watson-Crick (WC) base-pairs (Figure 1A). Due to steric clashes involving imino and amino protons, and energetically unfavorable hydrogen bonding, the purine-pyrimidine (pur-pyr) mispairs G•T/U and A•C do not typically adopt a WC-like geometry when the bases are in their energetically dominant neutral tautomeric form. Rather they typically form G•T, G•U, and A+•C wobbles that deviate from the WC geometry (Figure 1B). This geometrical distinction between canonical WC base-pairs and non-canonical wobbles is exploited by polymerases, repair enzymes, and ribosomes to replicate, transcribe, and translate genetic information with high fidelity.
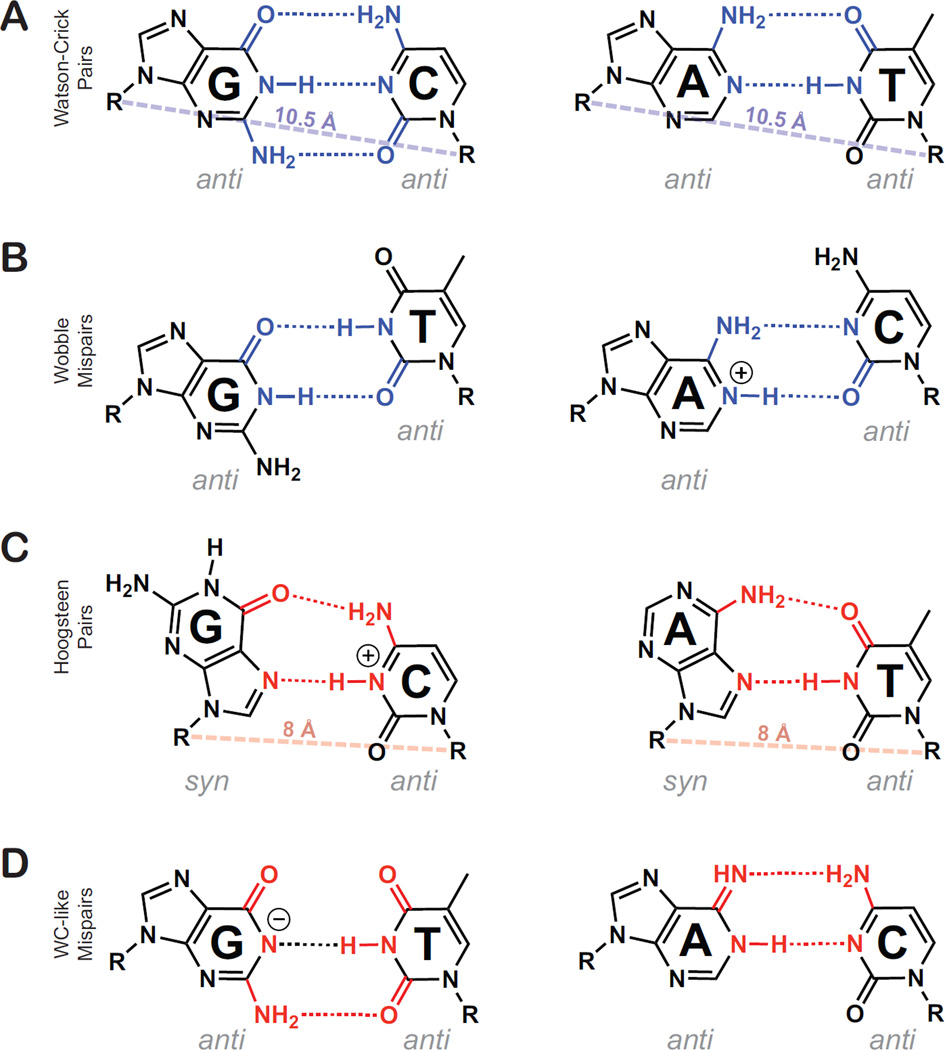
Figure 1.
Low and high energy pur-pyr pairing schemes. Hydrogen bonding partners for low energy WC and WB pairs are shaded blue, while high energy HG and WC-like mispairs’ hydrogen bonding partners are shaded red. (A) Canonical Watson-Crick G•C and A•T pairs. A rough average WC C1′–C1′ distance is shown as a dashed blue line for comparison with Hoogsteen pairs. (B) Neutral G•T wobble and ionized A+•C wobble. (C) G•C+ and A•T Hoogsteen pairing geometry, where purines are shown in the syn conformation. A rough average HG C1′–C1′ distance is shown as a dashed red line, reflecting the constriction of the C1′–C1′ distance by ~2.5 Å. (D) Deprotonated guanine in a WC-like G•T mispair is shown alongside an imino adenosine in a WC-like A•C mispair. While different tautomers and ionization states can explain the WC-like geometry, only two were shown for brevity. Possible ionized & tautomeric states that can explain the WC-like geometry for G•T mispairs include: G(enol), T(enol), G(N1-) and T(N3-). For A•C mispairs they include: A(Imino) and C(Imino).
There are other types of pur-pyr base-pairs that expand on the simple picture depicted above. These base-pairs that are seldom observed in unmodified nucleic acids because they entail the loss of energetically favorable interactions or require energetically costly base ionization or tautomerization. We will therefore refer to these base-pairs as ‘high energy’ pur-pyr base-pairs. High energy pur-pyr base-pairs include A•T and G•C+ Hoogsteen (HG) base-pairs [1–3], in which the purine base in a WC base-pair rotates roughly 180°about the N-glycosidic bond to adopt a syn rather than anti conformation (Figure 1C). While A•T HG base-pairs preserve two hydrogen-bonds, as in WC basepairs, formation of HG G•C+ base-pairs is accompanied by a net loss of one hydrogen bond and require protonation of C(N3) (Figure 1C). The HG base-pairs also require the translation of the complementary bases into closer proximity, causing constriction of the C1′–C1′ distance by ~2.5 Å (Figure 1C).
Other high energy pur-pyr base-pairs include G•T, G•U, and A•C mispairs that adopt a WC-like geometry through movement of otherwise sterically clashing imino (G•T and G•U) and amino (A•C) protons. For G•T and G•U mispairs, this can be accomplished by having either purine or pyrimidine adopt minor O6 or O4 enol tautomers, respectively, or through deprotonation of G(H1) or T/U(H3) (Figure 1D). On the other hand, WC-like A•C base-pairs can only arise by having either the A or C nucleobase adopt its rare imino tautomeric form (Figure 1D) [4••,5••].
While the existence and functional importance of such high energy pur-pyr base-pairs was hypothesized soon after the discovery of the double helix [1,6,7], there have been very few reports documenting their experimental observation. Recent studies suggest that such base-pairs can exist ubiquitously but transiently and in low abundance, and that they can be stabilized through intermolecular interactions or functionally important chemical modifications. Thus, there is good reason to believe that high energy pur-pyr base-pairs and base-mispairs exist in greater abundance than previously thought, and that they expand the structural and functional diversity of both canonical and non-canonical basepairs.
Methods for characterizing transient base-pairs and base ionization and tautomerization
There are several challenges in characterizing high energy pur-pyr base-pairs in nucleic acids. First, their energetic instability relative to other competing base-pairs in naked duplexes means that if they exist, they do so only transiently for short periods of time and in low abundance. Second, even when stabilized appreciably, characterizing such pur-pyr base-pairs can prove challenging by conventional spectroscopic techniques. For example, it can be difficult to distinguish HG from WC base-pairs by X-ray crystallography because the syn purine in a HG base-pair often entails small geometrical differences as compared to an anti purine in a WC base-pair, particularly for A•T base-pairs (Figure 2A) [8,9]. On the other hand, base ionization and tautomerization involves movement or loss of protons, which are not directly observed by X-ray crystallography. As a result, such base-pairs have to be inferred based on heavy atom positions, taking into account potential hydrogen bonding schemes and the local steric and electrostatic environment. While solution NMR can in principle be used to characterize such long-lived base-pairs, which have unique chemical shifts and/or Nuclear Overhauser Effects (NOEs) [10–12•], the appreciable stabilization of these base-pairs in unmodified nucleic acids typically occurs in large molecular systems that exceed the size limit of NMR application. Finally, while techniques such as UV and IR have successfully been used to visualize the ground-state tautomerization of aromatic heterocycles [13] and to characterize tautomeric states in individual nucleobases in isolation or when interacting with other small molecules [14,15], they cannot yet be applied to characterize tautomeric states of specific bases in nucleic acid polymers with atomic resolution.
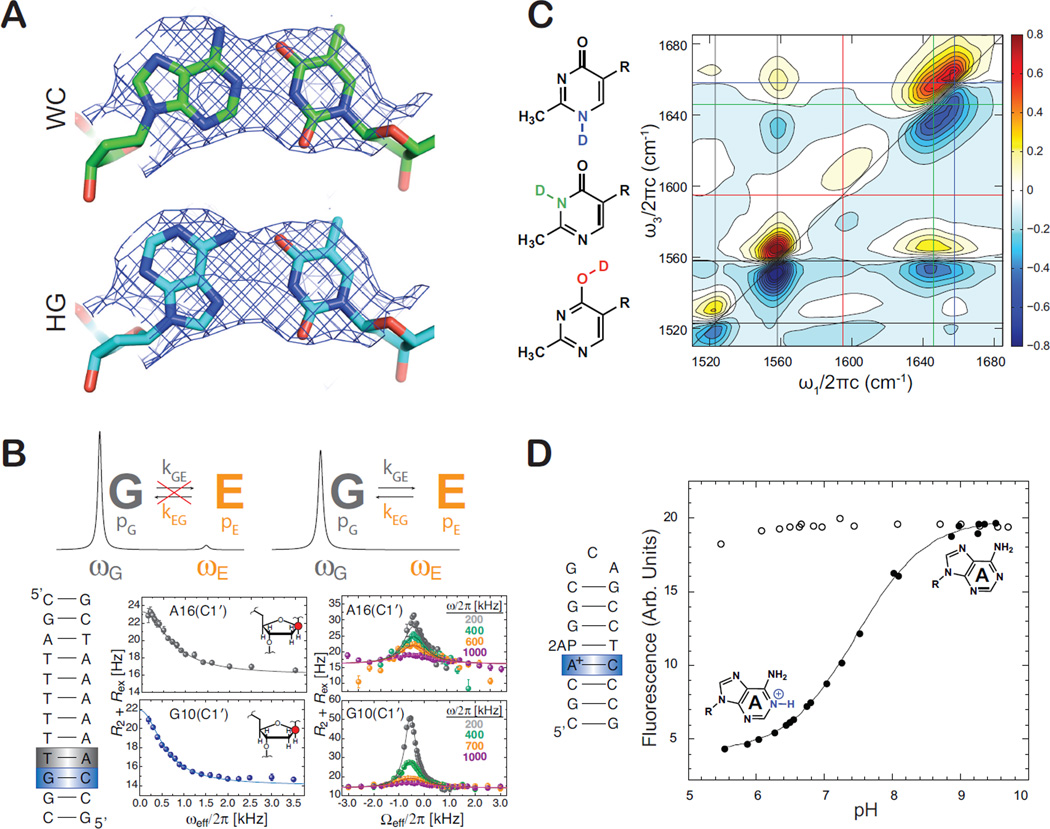
Figure 2.
New techniques to characterize high energy pur-pyr base-pairs. (A) Imperfect resolution of crystallographic electron density maps can lead to difficulties in identifying precise pair geometry. Highlighted in green is an A•T pair modeled to the electron density in the expected WC geometry. Highlighted in cyan is the same pair re-modeled as a Hoogsteen pair, showing it conforms to the same electron density map (PDB ID: 2EFW). (B) R1ρ NMR relaxation dispersion relies on measuring broadening of signals for dominant states due to exchange with a transient species. The line broadening contribution (Rex) to transverse relaxation (R2) is measured as a function of the spin lock power (ωeff) and both power and frequency (Ωeff) of an applied radio frequency. Shown are example relaxation dispersion profiles for sugar C1′ nuclei in guanine and adenine residues arising due to exchange with Hoogsteen base-pairs. (C) 2D IR methods aid characterization of three possible tautomeric states (color-coded) of the ligand oxythiamine. Shown are color-coded horizontal/vertical lines that connect the diagonal FTIR peaks to cross-peaks of coupled vibrations that can be correlated to each of the three tautomeric states. (D) Characterizing base ionization and measurements of pKa using a simple 2-aminopurine (2-AP) fluorescence assay. Ionization of a given base as a function of pH results in a change in 2-AP fluorescence in an adjacent 2-AP – T base-pair.
Recent advances in biophysical techniques are helping address these long-standing limitations. NMR techniques that involve the measurement of carbon and nitrogen rotating frame (R1ρ) relaxation dispersion [16,17] have made it possible to structurally, kinetically, and thermodynamically characterize transient A•T and G•C+ HG base-pairs in naked canonical duplex DNA that exist for short periods of time (~0.3 to ~1.5 ms) and in low abundance (<1%) [18••,19]. Here, transient HG base-pairs, or any other transient species with unique NMR chemical shifts, populations >0.1%, and exchange rates in the microsecond to millisecond timescale, can be detected and characterized as exchange broadening contributions to NMR signals that can be modulated by changing the power level and frequency of a continuous radiofrequency spin lock field that is applied during a relaxation period (Figure 2B) [16,17].
There has also been growing success in capturing high energy mispairs using increasingly sophisticated X-ray crystallographic methods and techniques. Many of these advances are made possible by innovations in sample engineering that make it possible to precisely trap high energy functional states and obtain high quality diffraction data to infer tautomerization or ionization [4••,5••,20••,21•–23], or even observe DNA polymerase mispair incorporation in ‘real time’ [24•].
There are other advances in techniques that may aid the characterization of base ionization and tautomerization in nucleic acids in the future. The rapidly growing field of 2D IR is allowing new insights into ultra-fast dynamics of isolated base pairs and nucleic acid oligomers [25,26]. For example, 2D IR techniques were recently successfully used in conjunction with variable temperature NMR and computational density functional theory (DFT) calculations to characterize and quantify three possible tautomeric states of oxythiamine (Figure 2C) [27•]. Using binding isotope effects, the authors were able to provide evidence that an RNA riboswitch preferentially binds one tautomeric form of the related thiamine pyrophosphate ligand, highlighting the importance of ligand tautomerization during RNA-ligand recognition.
A simple technique based on 2-aminopurine (2-AP) fluorescence has recently been introduced for characterizing base protonation in DNA and RNA [28•]. Here, a protonation event at a given base-pair is detected as a change in fluorescence by an adjacent 2-AP (Figure 2D). The authors demonstrated this approach in the measurement of pKa values for A+•C base pairs in DNA and a protonated RNA C+ base involved in pairing with the Hoogsteen face of a G in a RNA base-quartet. This approach can enable simple and fast characterization of ionized base-pairs in systems that are too large to study by NMR and possibly other techniques.
Hoogsteen base-pairs
Hoogsteen base-pairs were first proposed to explain how poly(rU) strands associate with poly(rA)-poly(rU) duplexes to form triplexes [29] and were subsequently visualized by Karst Hoogsteen through X-ray analysis of co-crystals containing 9-methyladenine and 1-methylthymine while searching for experimental evidence for WC base-pairs [1]. A•T and G•C+ HG base-pairs were subsequently observed in DNA bound to antibiotics [2] and proteins [30••,31], where they expand possibilities for sequence-specific DNA recognition by allowing unique contacts to form with the syn base [31,32], enhancing the negative electrostatic potential in the minor groove [30], and accommodating contacts that would otherwise result in steric clashes with a regular WC base-pair [32,33]. These X-ray structures suggest that HG base-pairs can be accommodated within the double helix with minor perturbations to nearby WC base-pairs [31]. There appears to be a good deal of promiscuity in the occurrence of such HG base-pairs. For example, while two neighboring A•T HG base-pairs are observed in X-ray structures of a palindromic CATG/CATG sequence bound to the DNA binding domain of p53 (Figure 3A) [30••], the same residues form WC base-pairs in X-ray structures with a longer spacer length [34] or a different intervening sequence between DNA half-sites [35]. There are also a few cases in which the HG base-pairs in X-ray structures of protein-DNA complexes may have been stabilized by crystal contacts [32] or by modifications introduced to aid crystallization [33]. This emphasizes the need to employ complementary techniques to characterize HG base-pairs in solution in the future [3].
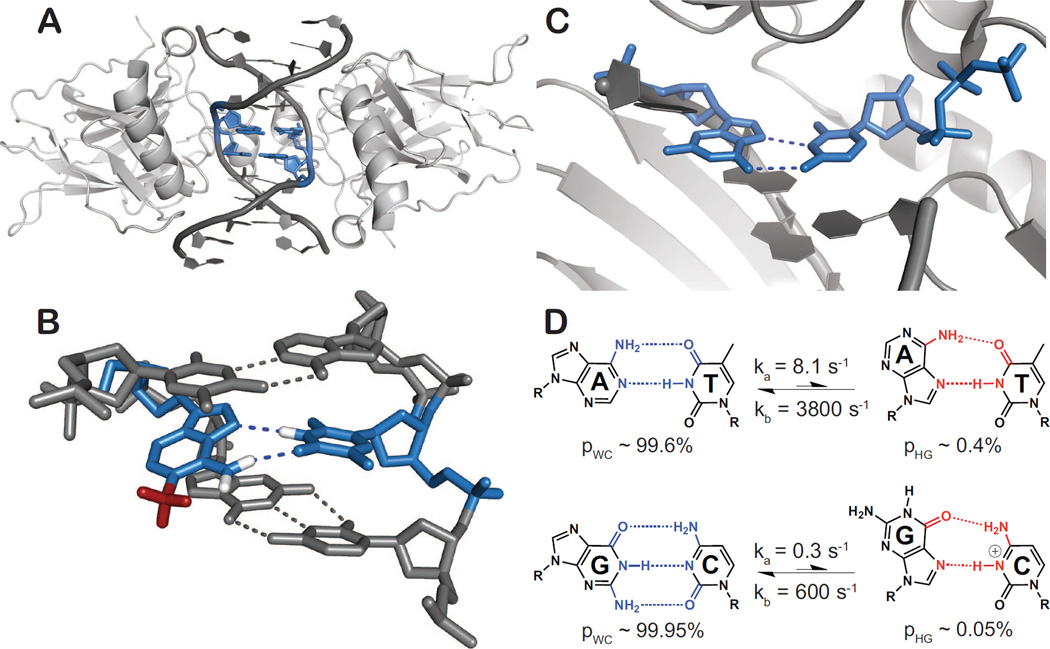
Figure 3.
Occurrence and functional roles for Hoogsteen base-pairs. A•T and G•C+ HG base-pairs are highlighted in blue. (A) Structure of DNA in complex with the p53 tumor suppressor protein provides a representative example of a DNA-protein complex containing HG base-pairs (PDB ID: 3IGL). (B) X-ray structure showing that the common DNA damaged purine, N1-methyladenine, prohibits the traditional A•T WC pairing by a steric clash of the N1-methyl group (red), and instead stabilizes Hoogsteen base-pairing with T. Protons were added to A(N1-methyl), A(N6) and T(N1) for illustrative purposes. (PDB ID: 3H8O). (C) Replication via human lesion bypass DNA polymerase ι (hPolι) proceeds through Hoogsteen (HG) base-pairing. hPolι is shown poised to insert an incoming dCTP against the template G via HG base-pair geometry (PDB ID: 2ALZ). (D) Two state conformational exchange between WC and transient HG A•T and G•C+ base-pairs in canonical duplex DNA showing the relative populations and exchange rate constants obtained from R1ρ; NMR relaxation dispersion experiments at pH 6.8 [18••,45•].
HG base-pairs have also been observed in damaged DNA, where they are believed to help maintain the integrity of the double helix and also provide recognition sites to recruit DNA repair enzymes (for review see [3]). For example, methylation of adenine N1 obstructs WC base-pairing but can readily be accommodated within the double helix through formation of HG base-pairs (Figure 3B) [36,37]. The enzyme AlkB, which repairs this mutagenic lesion, likely initially recognizes the unique HG base-pair formed between N1-methyladenine and thymine before flipping out the damaged purine for oxidative demethylation.
HG base-pairs are also frequently observed in purine-purine (pur-pur) mismatches, where the flipping of one purine base to a syn conformation leads to WC/HG type configuration that affords a shorter helical radius that can be more readily accommodated within BDNA as compared to the WC-WC configuration. Such HG-type mispairs are recognized by mismatch repair enzymes, such as MutS, which makes specific contacts with the syn adenine/guanine base in A•C, A•A, and G•G mismatches [38]. In a related, but vastly different system, WC/HG pur-pur mismatches which occur at the second and third positions of a codon/anti-codon mini-helix afford a WC-like C1′–C1′ distance that allows normally forbidden pur-pur base-pairs to be accommodated within the ribosomal decoding center, thus promoting read-through of stop codons containing pseudouridine at the first position [39•].
There is strong evidence that specialized polymerases, such as the human DNA Polι, can bypass certain forms of DNA damage by replicating DNA via HG rather than WC pairing (Figure 3C) [40,41]. Unlike replicative polymerases, Polι features a narrower active site, which is optimized for HG base-pairs that have characteristically shorter C1′–C1′ distances (Figure 1C). Several X-ray structures show that Polι can accommodate a wide range of purine alkylation and oxidation lesions in a syn conformation, whenever possible forming HG type base-pairs (Figure 3B) (reviewed in Makarova et al. [41]). These structural studies, together with biochemical studies showing the importance of Polι for cell survival in the presence of alkylating agents[42,43] and oxidative stress[44], provide the most compelling evidence to date for a biological function for HG base-pairs in duplex DNA.
Recently, NMR carbon and nitrogen relaxation dispersion studies have shown that G•C+ and A•T WC base-pairs in canonical duplex DNA exist in dynamic equilibrium with HG base-pair forms (Figure 3D) [18••]. The transient HG base-pairs have lifetimes on the order of hundreds of microseconds to milliseconds, with populations of ~0.1 – 1%, where the G•C+ HG base-pairs are less abundant than A•T HG base-pairs at physiological pH by at least a factor of 20 (Figure 3D). pH dependent NMR studies of trapped HG base-pairs through N1-methylation indicate that the pKa of cytosine N3 in G•C+ HG base-pairs is ~7.2, and therefore the majority of transient G•C+ HG base-pairs exist in the protonated form at physiological pH [45•]. Studies suggest that the transient HG base-pairs occur across all DNA sequence contexts with small, albeit significant, sequence-specific differences in population and lifetimes (unpublished results).
Transient Hoogsteen base-pairs help explain the long and controversial observation of WC versus HG – small changes in conditions can favor one form over the other. Moreover, the HG base-pairs transiently expose the WC faces of purines to the major groove where they can make contacts with other molecules, and can potentially help explain the much greater abundance of N1 methylation in adenine versus guanine. A recent study employed computational mapping to show that HG base pair formation dramatically affects the accessibility of amino nitrogens in adjacent WC-paired cytosines for hydroxymethylation by formaldehyde [46•]. In this manner, transient HG base-pairs may help explain aspects of the DNA reaction with formaldehyde, which have remained a mystery over the past 25 years.
WC-like pur-pyr mispairs accommodated by base ionization or tautomerization
Soon after the discovery of the double helix, it was hypothesized that pur-pyr mispairs that adopt WC-like geometry through tautomerization or ionization (Figure 1D) may explain, in part, the occurrence of spontaneous base substitution mutations [7,47]. This infidelity arises because the shape of a potential base pair and its stereochemical complementarity (hydrogen bonding, π-π stacking, etc) with the polymerase active site is a primary determinant of nucleotide incorporation [4••,48]. While G•T and A+•C mispairs generally form non-WC like wobbles that are rejected from the polymerase active site owing to their incorrect shape (Figure 1B), by forming WC-like base-pairs, high energy pur-pyr mispairs can result in misincorporation and spontaneous mutations.
A WC-like G•T mispair was observed by X-ray crystallography in which an incoming dGTP is poised to be incorporated against a template T, with geometry nearly identical to that of a WC pair, in the active site of DNA pol λ (Figure 4A) [20••]. While the observed WC-like G•T mispair can arise from either ionization or tautomerization, the increased misincorporation rates with increasing pH suggests deprotonation of the imino nitrogen in either base. Such an ionized G•T WC-like base-pair would also be consistent with prior NMR and in vitro studies in which it was shown that G-5BrdU and G-5FdU pairs adopt WC-like geometry through ionization and increase the rate of misincorporation with increasing pH [49].
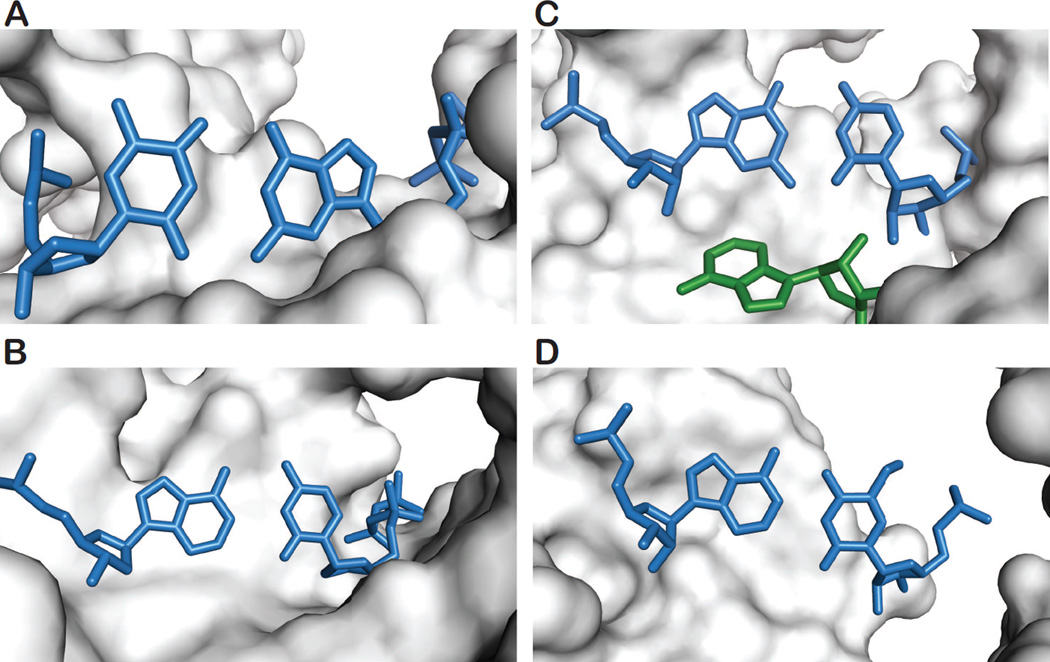
Figure 4.
An atomic view of high energy WC-like pur-pyr DNA and RNA mispairs in complex environments. Mismatched pur-pyr pairs are shown in blue. (A) A WC-like G•T mispair in the active site of DNA pol λ, showing an incoming dGTP poised for incorporation against the template T. Enzymatic pH studies suggest that this pair may be ionized, though a rare-tautomeric state could not be ruled out (PDB ID: 3PML). (B) Shown is a WC-like A•C mispair in the active site of DNA pol I with an incoming dCTP poised for incorporation against the template A. The WC-like geometry can be attributed to the rare-tautomeric state of either A or C (PDB ID: 3PX6). (C) A WC-like G-U mispair is shown at the first position of the anti-codon / codon mini-helix of a nearcognate tRNA. The WC-like geometry has been hypothesized to be due to a raretautomeric state of the pair. A ribosomal RNA is shown in green interacting with the minor groove of the mispaired guanosine (PDB ID: 3UYD). (D) A WC-like A-f5C mispair is shown at the wobble position of the human tRNAMet f5CAU where it can allow for the expanded decoding of an isoleucine codon (AUA) (PDB ID: 4GKK).
The low energy A+•C wobble mispair is readily rejected from the polymerase active site not only because of its non-WC geometry, but likely also because protonated bases within the hydrophobic active site have been shown to destabilize the polymerase closed ternary complex [50]. However, an X-ray structure has shown an A•C mispair adopting a clear WC-like geometry within the active site of DNA pol I, with the incoming dCTP poised for incorporation against a template A (Figure 4B) [4••]. Unlike G•T mispairs, only tautomerization of A or C can explain the observed WC-like A•C mispair, which is necessary to avoid steric clashes with exocyclic amino protons. While the rare imino adenine and cytosine tautomers are energetically unstable in isolation [51], the unique the local electrostatic environment within the polymerase active site may alter the tautomeric equilibrium. This is entirely plausible given that the tautomeric equilibrium is dependent upon the pKa of the nitrogens involved in the proton movement, and it has been shown that such pKas can be significantly perturbed by their environment [12•,52]. Together, the observation of WC-like G•T and A•C mispairs in polymerase active sites provide compelling evidence in support of the mutagenesis hypothesis put forward by Watson and Crick half-a-century ago.
The fidelity of translation also relies on proper WC pairing between the mRNA codon and tRNA anti-codon, particularly at the first and second position. This allows key interactions to take place between ribosomal RNA (rRNA) residues and the minor-groove of the tRNA/mRNA mini-helix. However, near-cognate tRNAs, including ones with G(mRNA) – U(tRNA) mismatches at the first or second position, have been shown to be prone to amino acid misincorporation [53]. Recent X-ray structures of near-cognate tRNAs paired with mRNA show WC-like G•U mispairs at the first and second position of the codon/anti-codon mini-helix in the 70S ribosome that may involve tautomerization (Figure 4C) [21•]. Although the role of tautomerization in tRNA proofreading requires further investigation [21•,54], it may help explain how incorrect aminoacyl-tRNAs containing G•U mispairs at the first and second position of the codon–anticodon minihelix can adopt a WC-like conformation and thereby facilitate amino acid misincorporation [55].
In contrast to the first and second positions of the codon/anti-codon mini-helix requiring WC geometry for translational fidelity, it has long been known that the wobble position is not as geometrically restrained. As there are typically fewer tRNAs than the 43 possible codons of the genetic code, many tRNAs must be able to recognize multiple codons [5••]. The ability of the wobble position to form pairs outside of G•C and A•U pairs plays a crucial role in facilitating tRNA recognition of degenerate codons. Furthermore, it has been shown that post-transcriptional modifications of the wobble base can broaden, or constrain, tRNA decoding. In one such instance, a post-transcriptional 5-formylcytidine (f5C) modification at the wobble position of human mitochondrial tRNAMet (hmtRNAMet f5CAU) allows for expanded decoding of methionine (AUG) as well as isoleucine (AUA) codons (Figure 4D) [5••]. Here, the f5C modification enables pairing of hmtRNAMet f5CAU to AUA codons through a WC-like A•f5C mispair at the wobble position. In addition to f5C, two other natural pyrimidine modifications (cmo5U[22] and mcm5s2U[23]) have been shown to enable increased decoding capabilities through, in part, the formation of WC-like pur-pyr mispairs wherein the pyrimidines adopt minor enol (G•modU) or imino tautomers (A•modC). The energetic penalty for adopting the corresponding rare-tautomeric forms is believed to be attenuated by intermolecular interactions and/or the electronic and steric modulation of the base by the modifications. Such modifications result in the deviation of the genetic code and allow for expanded decoding capabilities of translational RNAs. They provide a compelling example of how high energy pur-pyr mispairs can also be harnessed to expand the functionality of nucleic acids.
Future perspective
While HG pairs and WC-like mispairs are not new, recent years have seen a veritable boom in their structural and functional occurrence. Rapidly advancing biophysical techniques and creative sample engineering coupled with functional assays are making it possible to visualize such high energy pur-pyr base-pairs under a wide variety of biologically important structural and functional contexts. The observation of transient HG base-pairs in duplex DNA, with comparable energetics to canonical WC pairs, when combined with the current difficulties in resolving WC from HG based on X-ray diffraction data, raises the possibility that HG base-pairs exist in much greater abundance in vivo. In particular they are likely to be found in A•T rich regions of the genome where they may facilitate sequence-specific DNA recognition and other sequence-specific transactions. The heavy reliance on WC geometry in ensuring replicative, transcriptional and translational fidelity means that high energy pur-pyr mispairs that can adopt WC-like geometry not only give rise to deleterious effects, but are also a source of genetic ‘flexibility’ by being the rare exception to the rule. As with HG base-pairs, it is likely that relatively close energetic differences between these WC-like mispairs to their canonical counterparts undoubtedly means they will be observed with increasing frequency in even more complex and significant environments.
Highlights.
High energy purine-pyrimidine (pur-pyr) pairs are more common than once thought.
New spectroscopic methods provide novel insights into pur-pyr pairs.
Hoogsteen G•C+ & A•T pairs can exist transiently in canonical duplex DNA. Hoogsteen G•C+ & A•T pairs are important for protein-DNA recognition.
WC-like pur-pyr mispairs are important in replicative and translational fidelity.
Acknowledgements
We thank Anthony Mustoe for his input. We acknowledge support from NIH (R01065607) and an Agilent Thought Leader Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallographica. 1963;16:907–916. [Google Scholar]
- 2.Quigley GJ, Ughetto G, van der Marel GA, van Boom JH, Wang A, Rich A. Non-Watson-Crick GC and AT base pairs in a DNA-antibiotic complex. Science. 1986;232:1255–1258. doi: 10.1126/science.3704650. [DOI] [PubMed] [Google Scholar]
- 3.Nikolova EN, Zhou H, Gottardo FL, Alvey HS, Kimsey IJ, Al-Hashimi HM. A historical account of hoogsteen base-pairs in duplex DNA. Biopolymers. 2013;99:955–968. doi: 10.1002/bip.22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci U S A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. A crystallographic study provides direct evidence in support of the Watson and Crick hypothesis for spontaneous mutations. In addition to observing expected dATP•T and A•dTTP WC pairs, a WC-like A•dCTP mispair is clearly observed in the presence of maganese in the active site of DNA pol I. The imino tautomer of either base is invoked to explain the WC-like geometry of the mispair.
- 5. Cantara WA, Murphy FV, Demirci H, Agris PF. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222641110. A crystallographic study which shows that post-transcriptionally modified wobble position cytidines can allow for expanded decoding, specifically the recognition of the codon AUA by human mitochondrial tRNAMet f5CAU (hm tRNAMet f5CAU). The structural basis of this expanded decoding lies in the observed WC-like geometry of an A•f5C mispair at the wobble position of hmtRNAMet f5CAU.
- 6.Watson JD, Crick FH. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 7.Watson JD, Crick FH. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 8.Ethayathulla AS, Tse P-W, Monti P, Nguyen S, Inga A, Fronza G, Viadiu H. Structure of p73 DNA-binding domain tetramer modulates p73 transactivation. Proc Natl Acad Sci U S A. 2012;109:6066–6071. doi: 10.1073/pnas.1115463109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J. DNA polymerases: Hoogsteen base-pairing in DNA replication? Nature. 2005;437:E6–E7. doi: 10.1038/nature04199. [DOI] [PubMed] [Google Scholar]
- 10.Fazakerley GV, Gdaniec Z, Sowers LC. Base-pair induced shifts in the tautomeric equilibrium of a modified DNA base. J Mol Biol. 1993;230:6–10. doi: 10.1006/jmbi.1993.1119. [DOI] [PubMed] [Google Scholar]
- 11.Legault P, Pardi A. In situ probing of adenine protonation in RNA by 13C NMR. J Am Chem Soc. 1994;116:8390–8391. [Google Scholar]
- 12. Siegfried NA, O'Hare B, Bevilacqua PC. Driving Forces for Nucleic Acid pKa Shifting in an A+·C Wobble: Effects of Helix Position, Temperature, and Ionic Strength. Biochemistry. 2010;49:3225–3236. doi: 10.1021/bi901920g. A comprehensive study on the effect of local helical environment, temperature and solution conditions on the pKa perturbation of an adenosine in an A+•C mispair within a DNA helix. Results show that the pKa of A(N1) can be significantly perturbed (~4 units) depending on the aforementioned conditions.
- 13.Peng CS, Baiz CR, Tokmakoff A. Direct observation of ground-state lactam-lactim tautomerization using temperature-jump transient 2D IR spectroscopy. Proc Natl Acad Sci U S A. 2013;110:9243–9248. doi: 10.1073/pnas.1303235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mons M, Dimicoli I, Piuzzi F, Tardivel B, Elhanine M. Tautomerism of the DNA base guanine and its methylated derivatives as studied by gas-phase infrared and ultraviolet spectroscopy. J Phys Chem A. 2002;106:5088–5094. [Google Scholar]
- 15.Crews BO, Abo-Riziq A, Pluháčková K, Thompson P, Hill G, Hobza P, de Vries MS. Guanine–aspartic acid interactions probed with IR–UV resonance spectroscopy. Phys Chem Chem Phys. 2010;12:3597–3605. doi: 10.1039/b925340h. [DOI] [PubMed] [Google Scholar]
- 16.Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci U S A. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer AG, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chemical reviews. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 18. Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. An NMR study employing rotating-frame relaxation dispersion (R1ρ) shows that WC A•T and G•C base-pairs in canonical duplex DNA transiently morph into corresponding A•T and G•C+ HG base-pairs through the 180 degree rotation of the purine base from an ant to syn conformation.
- 19.Nikolova EN, Gottardo FL, Al-Hashimi HM. Probing Transient Hoogsteen Hydrogen Bonds in Canonical Duplex DNA Using NMR Relaxation Dispersion and Single-Atom Substitution. J Am Chem Soc. 2012;134:3667–3670. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bebenek K, Pedersen LC, Kunkel TA. Replication infidelity via a mismatch with Watson-Crick geometry. Proc Natl Acad Sci U S A. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. A crystallographic and in vitro study shows that a dGTP•T mispair can adopt WC-like geometry in the active site of DNA pol λ, likely through ionization, and can lead to replicative infidelity, especially at basic conditions.
- 21. Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. A crystallographic study of the 70S ribosome which shows that G-U mispairs at the first and second position of near-cognate tRNAs can form WC-like geometry, likely through tautomerization.
- 22.Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vendeix FA, Murphy FV, IV, Cantara WA, Leszczyńska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNALys3UUU is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism. J Mol Biol. 2012;416:467. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA Polymerase Choose Right from Wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. A very intriguing study employing timescale crystallography to track the progress and structural changes of DNA pol β during replication by visualizing multiple trapped catalytic intermediates with natural substrates at different times during catalysis. Using this method the authors are able to observe both nucleotide incorporation and misincorporation without the use of substrate analogues.
- 25.Yang M, Szyc Ł, Elsaesser T. Femtosecond Two-Dimensional Infrared Spectroscopy of Adenine-Thymine Base Pairs in DNA Oligomers. J Phys Chem B. 2011;115:1262–1267. doi: 10.1021/jp1090697. [DOI] [PubMed] [Google Scholar]
- 26.Greve C, Preketes NK, Fidder H, Costard R, Koeppe B, Heisler IA, Mukamel S, Temps F, Nibbering ET, Elsaesser T. N-H Stretching Excitations in Adenosine-Thymidine Base Pairs in Solution: Pair Geometries, Infrared Line Shapes, and Ultrafast Vibrational Dynamics. J Phys Chem A. 2013;117:594–606. doi: 10.1021/jp310177e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh V, Peng CS, Li D, Mitra K, Silvestre KJ, Tokmakoff A, Essigmann JM. Direct Observation of Multiple Tautomers of Oxythiamine and their Recognition by the Thiamine Pyrophosphate Riboswitch. ACS Chem Biol. 2013 doi: 10.1021/cb400581f. Study employing 2D IR, FT IR, NMR, BIEs and DFT to characterize the tautomeric states of an oxythiamine pyrophosphate ligand in isolation and when bound to the thiamine pyrophosphate riboswitch.
- 28. Wilcox JL, Bevilacqua PC. A Simple Fluorescence Method for pKa Determination in RNA and DNA Reveals Highly Shifted pKas. J Am Chem Soc. 2013;135:7390–7393. doi: 10.1021/ja3125299. Useful and simple method for characterizing the pKa and/or protonated DNA/RNA bases by monitoring a change in fluorescence of an adjacent 2-aminopurine reporter.
- 29.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J Am Chem Soc. 1957;79:2023–2024. [Google Scholar]
- 30. Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. A computational and crystallographic study of the tumor suppressor protein, p53, tetramers bound to target DNA which elaborates on the role of HG base pairs in protein-DNA and protein-protein interactions by changing the local helical shape and electrostatic environment.
- 31.Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30:5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patikoglou GA, Kim JL, Sun L, Yang S-H, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice PA, Yang S-w, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 34.Malecka KA, Ho WC, Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2008;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure. 2010;18:246–256. doi: 10.1016/j.str.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Zhan Y, Fenn D, Chi LM, Lam SL. Effect of 1-methyladenine on double-helical DNA structures. FEBS Lett. 2008;582:1629–1633. doi: 10.1016/j.febslet.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Lu L, Yi C, Jian X, Zheng G, He C. Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 2010;38:4415–4425. doi: 10.1093/nar/gkq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Natrajan G, Lamers MH, Enzlin JH, Winterwerp HH, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez IS, Ng CL, Kelley AC, Wu G, Yu Y-T, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. Crystallographic and biochemical studies characterize the role of a uridine to pseudouridine modification in the initial position of a stop codon. The modification allows for the efficient read-through of the stop codon by allowing certain bases to form previously restricted geometry, including a pur-pur HG pair.
- 40.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G. C+ Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Makarova A, Kulbachinskiy A. Structure of human DNA polymerase iota and the mechanism of DNA synthesis. Biochemistry (Mosc) 2012;77:547–561. doi: 10.1134/S0006297912060016. [DOI] [PubMed] [Google Scholar]
- 42.Johnson RE, Yu S-L, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2'-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Res. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P. Human DNA polymerase iota protects cells against oxidative stress. EMBO J. 2008;27:2883–2895. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nikolova EN, Goh GB, Brooks CL, III, Al-Hashimi HM. Characterizing the Protonation State of Cytosine in Transient G·C Hoogsteen Base Pairs in Duplex DNA. J Am Chem Soc. 2013;135:6766–6769. doi: 10.1021/ja400994e. NMR and computational study reveals that the pKa of cytosine N3 is >7.0. Therefore, the majority of transient G•C+ HG base-pairs in canonical duplex DNA are protontaed.
- 46. Bohnuud T, Beglov D, Ngan CH, Zerbe B, Hall DR, Brenke R, Vajda S, Frank-Kamenetskii MD, Kozakov D. Computational mapping reveals dramatic effect of Hoogsteen breathing on duplex DNA reactivity with formaldehyde. Nucleic Acids Res. 2012;40:7644–7652. doi: 10.1093/nar/gks519. Using computational mapping, this study shows that G•C+ HG formation leaves the amino nitrogen of an adjacent WC paired cytosine prone to hydroxymethylation by formaldehyde.
- 47.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 48.Kirmizialtin S, Nguyen V, Johnson KA, Elber R. How conformational dynamics of DNA polymerase select correct substrates: experiments and simulations. Structure. 2012;20:618–627. doi: 10.1016/j.str.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Eritja R, Bloom LB, Goodman MF. Ionization of bromouracil and fluorouracil stimulates base mispairing frequencies with guanine. J Biol Chem. 1993;268:15935–15943. [PubMed] [Google Scholar]
- 50.Xia S, Wang J, Konigsberg WH. DNA Mismatch Synthesis Complexes Provide Insights into Base Selectivity of a B Family DNA Polymerase. J Am Chem Soc. 2012;135:193–202. doi: 10.1021/ja3079048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fonseca Guerra C, Bickelhaupt F, Saha S, Wang F. Adenine tautomers: relative stabilities, ionization energies, and mismatch with Cytosine. J Phys Chem A. 2006;110:4012–4020. doi: 10.1021/jp057275r. [DOI] [PubMed] [Google Scholar]
- 52.Wilcox JL, Bevilacqua PC. pKa Shifting in Double-Stranded RNA Is Highly Dependent upon Nearest Neighbors and Bulge Positioning. Biochemistry. 2013;52:7470–7476. doi: 10.1021/bi400768q. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Shah B, Bondarenko PV. G/U and Certain Wobble Position Mismatches as Possible Main Causes of Amino Acid Misincorporations. Biochemistry. 2013;52:8165–8176. doi: 10.1021/bi401002c. [DOI] [PubMed] [Google Scholar]
- 54.Voorhees RM, Ramakrishnan V. Structural Basis of the Translational Elongation Cycle*. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 55.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. New structural insights into the decoding mechanism: Translation infidelity via a G-U pair with Watson-Crick geometry. FEBS Lett. 2013;587:1848–1857. doi: 10.1016/j.febslet.2013.05.009. [DOI] [PubMed] [Google Scholar]