Abstract
Protein kinase C (PKC) signaling drives many important cellular processes and its dysregulation results in pathophysiologies such as cancer1. Because PKC is activated acutely and allosterically, it is difficult to monitor the cellular activity of endogenous PKC by conventional methodologies2. Rather, PKC signaling is best studied in situ using biosensors such as FRET-based reporters. We have generated several FRET-based reporters for studying PKC signaling in real time in live cells3. Using these reporters, we have demonstrated phase-locked oscillations in Ca2+ release and membrane-localized endogenous PKC activity in response to histamine4, as well as distinct signatures of endogenous PKC signaling at specific organelles in response to UTP5. Here we describe methods to image cells expressing the reporters and elaborate on data analyses, control experiments, and variations for imaging the activity of expressed PKC.
Keywords: protein kinase C (PKC), diacylglycerol (DAG), förester or fluorescence resonance energy transfer (FRET), FRET-based reporter, C kinase activity reporter (CKAR), diacylglycerol reporter (DAGR), targeted reporter, live-cell imaging, UTP
1. Introduction
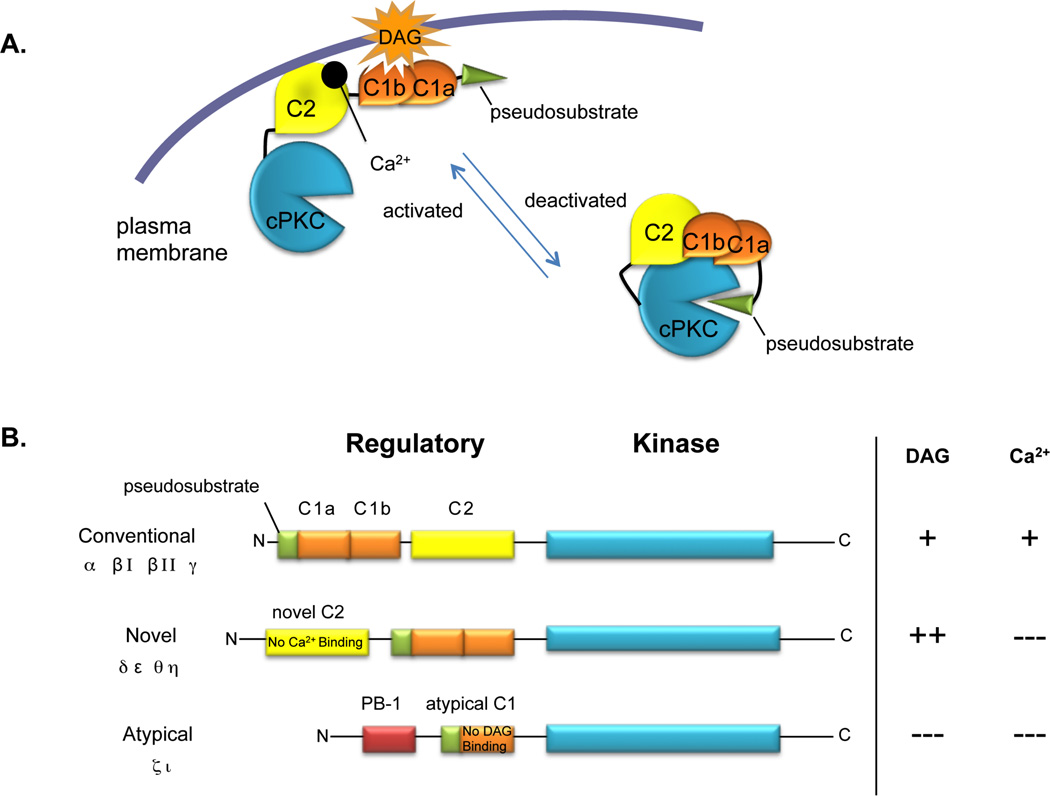
Signal transduction relies greatly upon the regulated enzymatic activity of protein kinases6. The protein kinase C (PKC) family is a group of ten mammalian isozymes sharing a highly-conserved kinase core that is ‘matured’ by a series of priming phosphorlation events promoted by the upstream kinases PDK-1 and mTORC2 (For review, see7). PKC isozymes contain membrane-targeting domains, C1 and C2, which sense levels of second messengers, diacylglycerol (DAG) and Ca2+, respectively, which are produced upon receptor activation (Figure 1). In response to changing levels of second messengers, PKC isozymes are allosterically and locally activated to phosphorylate downstream substrates (Figure 1A). Conventional PKC isoforms (cPKC: α, βI/II, γ) are activated by coincident elevation in intracellular Ca2+ and membrane-bound DAG; Novel PKC isoforms (nPKC: δ, ε, θ, η) respond robustly to DAG alone; Atypical PKC isoforms (aPKC: ζ, ɩ) respond to neither second messenger, although they are spatially localized by protein:protein interactions8 and appear to be regulated at the level of priming phosphorylation events (Figure 1B; for extensive review of PKC signaling, see9, 10). Phosphorylation of PKC substrates is tightly regulated by both spatio-temporally localized PKC activation and the opposing actions of cellular phosphatases4, 5.
Figure 1. Regulation of PKC isoforms.
A. Activation of PKC. In unstimulated cells, PKC (cPKC shown here) exists in a fully-phosphorylated, closed conformation (right species) with an N-terminal autoinhibitory pseudosubstrate peptide resting in the substrate binding cavity of the kinase core; this prevents phosphorylation of cellular substrates. Upon receptor stimulation and subsequent DAG and Ca2+ elevation, PKC translocates to cellular membranes (right species) using a Ca2+-sensitive C2 domain and DAG-sensitive C1 domains. Membrane binding relieves autoinhibition and permits phosphorylation of cellular substrates. B. Domain structure of PKC family. Conventional PKC isoforms contain a Ca2+-sensitive C2 domain and DAG-sensitive C1 domains. Novel PKC isoforms contain a Ca2+-insensitive C2 domain and DAG-sensitive C1 domains; however, these C1 domains bind membranes with two orders-of-magnitude higher affinity in the presence of DAG compared to those present in cPKC isozymes. Atypical PKC isoforms contain a DAG-insensitive C1 domain and a PB-1 domain; both of these serve as protein-protein interaction modules. Adapted from9.
Traditionally, the method of choice for demonstrating protein kinase activation has been monitoring the phosphorylation of residues that prime or activate kinases using Western blotting with phospho-specific antibodies that recognize these sites (as in11). However, conventional and novel PKC isoforms are most often constitutively phosphorylated as a maturation step, rendering analysis of phosphorylation sites an ineffective measure of prior cell activation. Rather, PKC is acutely activated by allosteric mechanisms resulting from binding membrane-embedded diacylglycerol (reviewed in2). As a result, the classic method for examining PKC activity has been to probe for the presence of PKC in membrane fractions of lysed cells, or by staining fixed cells with PKC antibodies as a measure of membrane translocation (as in12,13, 14). But, these methods can fall short of achieving precise spatial and temporal resolution of signaling events because they require fractionation or staining of cells that have been lysed or fixed at defined time points. The advent of green fluorescent protein (GFP; recently reviewed in15) presented PKC researchers with the ability to monitor membrane translocation of fluorescently-tagged PKC in live cells as a readout of activation, generating numerous elegant examples of PKC translocation over time in response to the addition of natural receptor agonists16, 17 or the addition of ligands which activate PKC by directly engaging the C1 domains of cPKC isozymes and nPKC isozymes, such as tumor-promoting phorbol esters18. But, these experiments rely on over-expressed, tagged PKC. Thus, these studies are blind to the activity of endogenous PKC and, importantly, the balance between PKC activity and the activity of cellular phosphatases. Therefore, live-cell imaging using biosensors or reporters to read out endogenous activity is ideal for examining PKC signaling.
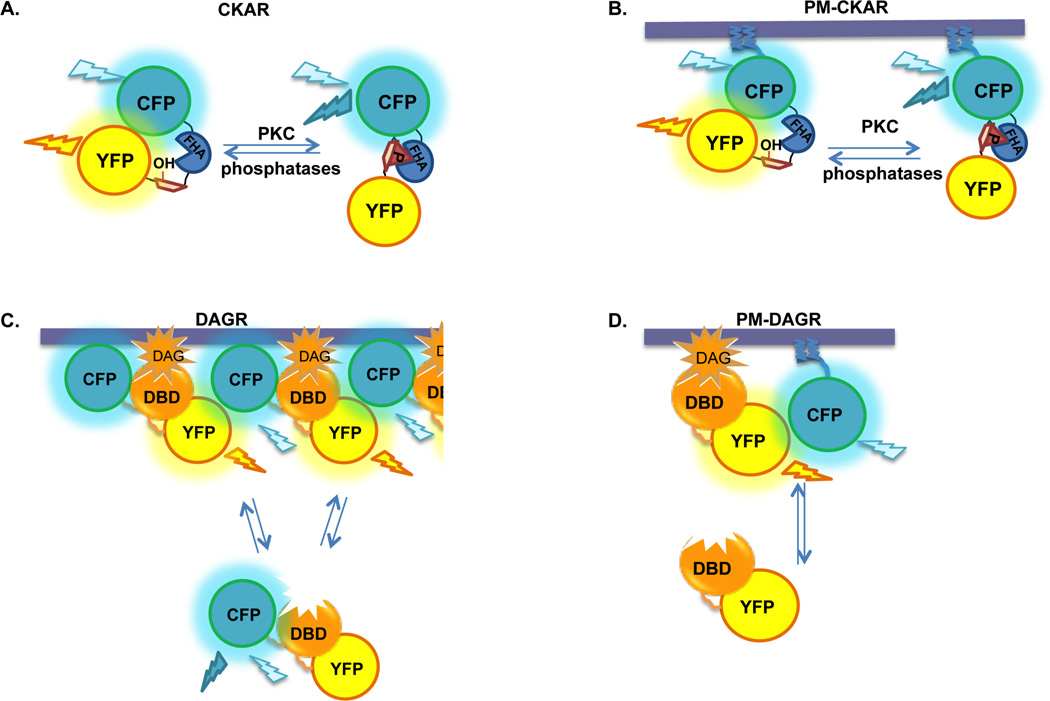
Our lab has developed and characterized förester resonance energy transfer (FRET)-based reporters for studying PKC signaling (Figure 2), which rely on changes in FRET from donor cyan fluorescent protein (CFP; ECFP variant) to acceptor yellow fluorescent protein (YFP; Citrine variant) to reflect changes in signaling activity. C Kinase Activity Reporter, CKAR, is a tool to measure directly the activity of PKC,4 and is similar in structure to the prototypical kinase reporter, A Kinase Activity Reporter, AKAR19. CKAR consists of CFP at the N-terminus, followed by an FHA2 phosphopeptide binding domain linked to a substrate sequence that is specific for PKC, followed by YFP at the C-terminus (Figure 2A). CKAR is phosphorylated by all conventional and novel protein kinase C isozymes tested, although with varying efficiency (Kajimoto, et al.; manuscript in preparation). Importantly, CKAR is not phosphorylated by other kinases predicted to have similar substrate specificity, such as PKA and CAMKII, in vitro4. Because CKAR is genetically encoded, we have been able to fuse short sequences to the N- or C-terminus of CKAR to target or enrich the reporter at specific regions of cells to monitor localized signaling4, 5 (plasma membrane-targeted reporter shown in Figure 2B). We also generated FRET-based tools to measure the production of the upstream second messenger, DAG. Diacylglycerol reporter, DAGR, consists of CFP and YFP flanking a diacylglycerol-binding domain (DBD), in this case, the C1 domain of PKCβ4 (Figure 2C). In addition to this original DAGR, we also generated targeted versions of DAGR. These consist of separate constructs encoding CFP targeted either to the plasma membrane or to the Golgi co-transfected with a YFP-tagged DBD, in this case, a mutated form of the C1b domain of PKCβ5 (Figure 2D). These tools have allowed us to monitor with great precision when and where PKC activity is elevated in live cells in response to agonist-mediated signaling, and to correlate this activity to regional differences in second messenger production and phosphatase activity.
Figure 2. FRET-based reporters for PKC signaling pathways.
A. CKAR, C kinase activity reporter. CFP is linked to YFP by a substrate peptide specific for PKC and an FHA2 phosphopeptide-binding domain. Basal intramolecular FRET from CFP to YFP is reduced upon phosphorylation of the reporter by PKC, but can be restored upon dephosphorylation of the reporter by cellular phosphatases. B. PM-CKAR, plasma membrane-targeted CKAR. The N-terminal 7 residues of Lyn kinase encode sequences for myristoylation and palmitoylation; when these residues are fused to the N-terminus of CKAR, the reporter expressed in cells is enriched at the cytoplasmic side of the plasma membrane. C. DAGR, diacylglycerol reporter. CFP and YFP flank a diacylglycerol binding domain (DBD). Intermolecular FRET between reporters increases as they come in close proximity upon translocation to cellular membranes in response to DAG production. FRET decreases as DAG is metabolized and the reporters re-localize to the cytosol. D. PM-DAGR, plasma membrane-targeted DAGR. PM-DAGR consists of two separate constructs transfected together encoding YFP-tagged DBD and CFP targeted to the plasma membrane using the N-terminal 7 residues of Lyn kinase as described above. Intermolecular FRET from CFP to YFP increases as the reporter translocates to membranes in response to stimulated DAG production, and decreases upon DAG turnover and re-localization of YFP-DBD to the cytosol. Note that the DBD in B. and in C. are different (see text).
2. Materials
2.1 Cell culture
Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco/BRL, Bethesda, MD) supplemented with10% fetal bovine serum (FBS; HyClone, Ogden, UT).
Solution of trypsin (1%) and ethylenediamine tetraacetic acid (EDTA; 1 mM) from Gibco/BRL.
COS7 or other adherent cell line (ATCC, Manassas, VA).
Uncoated 35 mm glass-bottom imaging dishes (Matek, Ashland, MA).
Plasmid DNA encoding CKAR, CKAR T/A (Ala at phospho-acceptor site), DAGR, or targeted versions of these reporters (Addgene, Cambridge, MA).
2.2 Imaging
Hanks’ Balanced Salt Solution (HBSS; Sigma, St. Louis, MO) supplemented with 1 mM CaCl2 just prior to imaging.
Phorbol Myristate Acetate (PMA; EMD biosciences, Gibbstown, NJ) or Phorbol-12,13-dibutyrate (PdBu) from EMD biosciences dissolved in Dimethyl sulfoxide (DMSO) from Sigma to a stock concentration of 200 µM (dilute 1:1000 for working concentration).
Calyculin A from EMD biosciences at a stock concentration of 50 µM (dilute 1:1000 for working concentration) in DMSO.
Gö6976 in solution (500 µg/ml), from EMD biosciences (add 0.77 µL to 2 ml saline for working concentration of 500 nM).
Gö6983 from EMD biosciences at a stock concentration of 250 µM (dilute 1:1000 for working concentration) in DMSO.
Test agonist, such as UTP from EMD biosciences at a stock concentration of 100 mM in dH2O to be diluted 1:1000 for working concentration, or other receptor agonist.
**NOTE: Phorbol esters, Gö6976, Gö6983, and Calyculin A are toxic substances that should be handled with care and disposed of according to institutional guidelines. Stock solutions of these compounds are stored at −20 °C in small aliquots; repeated thawing and freezing is not recommended. UTP is stored at −20 °C in one large aliquot, and thawed on ice for use.
3. Methods
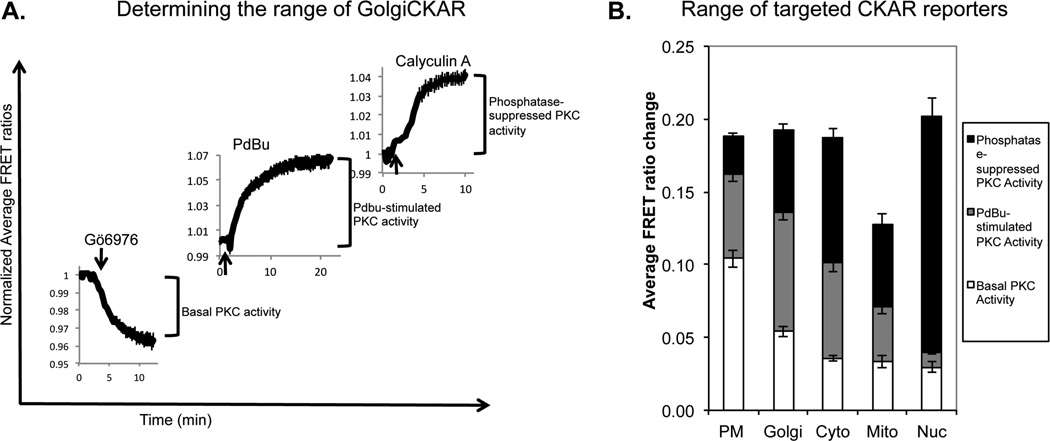
It is important to note that the maximal FRET ratio change possible with CKAR is 20%, and typical agonist-stimulated responses reach 5%. Because of this relatively low signal-to-noise ratio, it is critical to average data from multiple cells across 3 or more dishes and calculate error before attempting to compare PKC responses. For initial characterization, the full range of the CKAR response should be determined(as in5, Figure 3). The response of test agonists will fall within this range, and the detection limit will be apparent. Other experiments that must be done to ensure confidence in positive CKAR responses are to reverse or prevent the PKC response using PKC inhibitors, such as Gö6983 and Gö6976. One should also image the response of a mutant form of the reporter containing an Ala residue at the phospho-acceptor site (CKAR T/A), which should not change upon addition of agonist.
Figure 3. Experimentally verifying the range of targeted CKARs.
A. The range of a kinase activity reporter is determined in three experiments. ‘Basal PKC activity’ is the magnitude of the decrease in FRET ratio upon adding PKC inhibitor after acquiring baseline FRET ratios. ‘PdBu-stimulated activity’ is the FRET ratio increase from baseline upon addition of PdBu. ‘Phosphatase-suppressed PKC activity’ is the FRET ratio increase from the plateau of the maximal response to PdBu upon addition of Calyculin A. FRET ratio changes from GolgiCKAR depicted here are normalized to 1, and represent the average responses of multiple COS7 cells across three or more dishes; maximal FRET ratio changes are determined by fitting the data to monoexponential curves. Experimentally-determined ranges for targeted CKAR responses in COS7 cells. Note that Mito-CKAR has a decreased range compared to all other reporters, which all exhibit an approximately 20% maximal FRET ratio change; this likely results from slight proteolysis of Mito-CKAR in COS7 cells. Adapted from5.
Expression of these reporters in cells has the inherent possibility of altering signaling by competing with endogenous signaling molecules for access to PKC (CKAR) or DAG (DAGR). Therefore, when carrying out these experiments, it is important to select cells that express low levels of CKAR and DAGR. In fact, we have calibrated the YFP fluorescence intensity with reporter concentration using purified protein4, and consistently image cells expressing no more than approximately 1 µM CKAR. This is well below concentrations of an abundant cellular substrate, MARCKS, which can reach concentrations of 20 µM20. In addition, CKAR expression levels ranging from approximately 0.5–5 µM report consistent FRET ratio changes in response to agonist, supporting that this level of expression does not appreciably interfere with signaling4. Expression of DAGR also has the potential to alter signaling by acting as a “sponge” to soak up agonist-stimulated DAG, preventing endogenous downstream effectors from responding. Indeed, we have experimentally verified this effect by expressing high levels of C1 domain and monitoring effects on PKC activity5. Thus, for both CKAR and DAGR, it is critical to evaluate signaling only in cells expressing low reporter levels.
While the strength of this imaging approach is the ability to measure signaling carried out by endogenous signaling molecules in real time, it is also possible to measure the activity of expressed PKC isoforms. For example, one might want to compare the activity of a mutant PKC to wild-type PKC. The variation described in section 3.5 describes how to explore these differences in activity using CKAR. The challenges with this method are 1] dealing with unequal expression of PKC amongst individual cells and 2] separating the activity contributed by overexpressed PKC from that contributed by the endogenous PKC. The first challenge is easily overcome by tagging PKC with a spectrally-compatible fluorophore (i.e. mCherry21) whose fluorescence intensity can be monitored as a measure of PKC expression level without interfering with the FRET reporter readout. The second challenge can be overcome by generating a “dose-response” curve for the endogenous PKC vs. the expressed PKC using the agonist of choice. This will allow the determination of an agonist concentration that is sufficient to activate the overexpressed and more abundant PKC from the endogenous PKC.
3.1 Cell culturing
COS7 and other adherent cell lines are passaged just prior to confluence using trypsin/EDTA to dissociate cells. These cells are diluted in fresh media in 10 cm dishes for maintenance of the culture and split into 35 mm glass bottom dishes for experimental setup. Cells are plated sparsely for imaging; for example, a 1:40 split of a 70–80% confluent 10 cm dish of COS7 cells provides conditions that are optimal for efficient transfection and imaging of individual cells.
Once cells have adhered to the glass (either later on the day of plating or on the following day), cells are transfected according to manufacturers’ protocols with plasmid DNA encoding the reporter of interest. For example, COS7 cells are transfected with 0.5–1 ug of reporter DNA using 3 uL FUGENE6 (Roche pharmaceuticals, Palo Alto, CA) per 35 mm dish. Overnight expression is typically sufficient to obtain cells expressing appropriate levels of the reporters.
When preparing to image targeted DAGR constructs, more YFP-DBD than targeted CFP is transfected (typically a 3:1 ratio is sufficient) to maximize the range of responses.
3.2 Imaging CKAR/DAGR
Cells expressing the reporter of interest are rinsed once with HBSS + 1 mM CaCl2 and imaged in 2 ml of this solution. Once cells expressing an optimal level of the reporter are selected, a series of CFP, FRET, and YFP images are acquired.
Background signal is subtracted from areas of the image lacking cells or from areas with untransfected cells.
If using Metafluor software, one region per cell is selected for monitoring FRET ratios in real time.
Acquisition of CFP, FRET, and YFP images over fixed time intervals, typically 10–15 s, is carried out through the entire experiment. If using Metafluor software, the average FRET ratios (CFP/FRET for CKAR or FRET/CFP for DAGR) for the selected cellular regions are plotted as a readout of the signaling response. The YFP intensity is also graphed to monitor for photobleaching.
Baseline FRET ratio readings are acquired for 5–15 min. Agonist or inhibitor is not added until the baseline is either flat or has a consistent linear slope.
To add agonist or inhibitor, approximately 0.5 ml–1 ml of HBSS is withdrawn from the dish and used to pre-dilute the drug taken from the stock. This pre-diluted drug is added carefully in between image acquisitions, and the time of addition is noted.
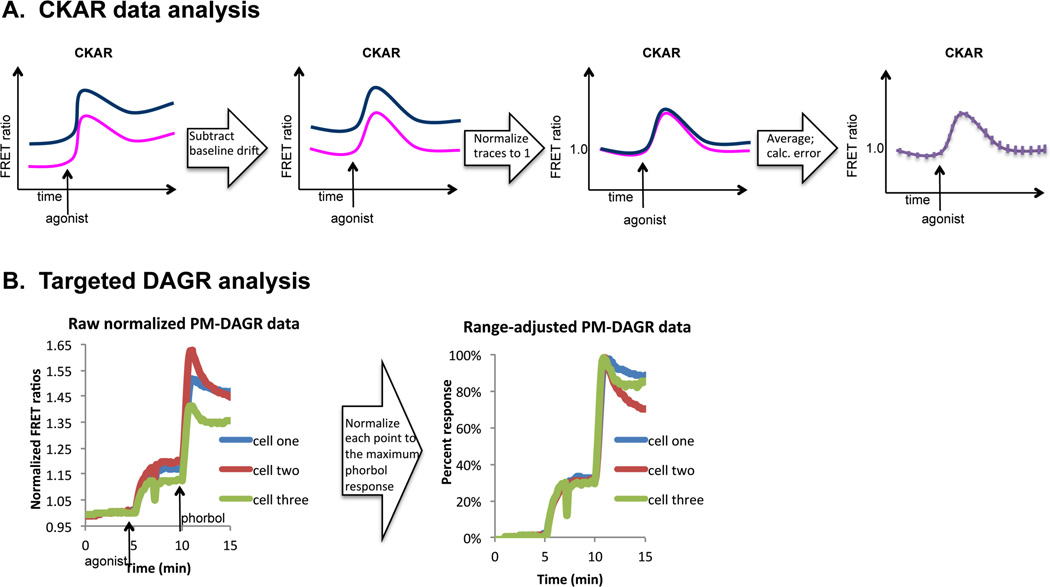
3.3 CKAR data analysis (Figure 4A)
Figure 4. Data analysis.
A. Flow chart depicting typical analysis steps that yield average FRET ratios suitable for comparison across different scenarios. The far left panel is an illustration of raw FRET ratio values that drift over time. The middle left panel depicts drift-corrected FRET ratios. The middle right panel shows normalized FRET ratios, and the far right panel shows the normalized average FRET ratios with calculated error. This analysis step is described in detail in section 3.3. B. Normalization step for targeted DAGR analysis. The left plot contains raw FRET ratios from two cells transfected with PM-DAGR and stimulated with UTP (100 µM) followed by PdBu (200 nM). The right plot contains these same data normalized for the ranges of the individual cells. This analysis step is described in detail in section 3.4.
The average FRET ratio for each region selected is plotted over time using a graphing program such as Microsoft Excel.
For each average FRET ratio, the slope of the baseline approximately 5 min before the addition timepoint is determined. Ideally, this slope will be zero, but occasionally the baseline FRET ratio exhibits drift. The drift often has a steeper slope initially and then levels off to a fairly linear slope that is consistent throughout the course of the experiment. One can mathematically determine the slope by graphing the linear portion of the baseline FRET ratio drift. Then, the linear drift (i.e. slope × time) is simply subtracted systematically from each data point in the response curve.
Responses (corrected for drift, if necessary) are all normalized to 1 by dividing by the initial FRET ratio.
The normalized FRET ratio responses are averaged across all cells imaged, referencing data from different dishes to the agonist or inhibitor addition timepoint. If agonist or inhibitor was added at different time points for different dishes, baseline FRET ratios can be deleted from the beginning of the experiment. The normalized average FRET ratio from all cells imaged is plotted against time in a new graph.
The standard error of the mean is calculated and plotted for each data point in the average FRET ratio.
3.4 DAGR data analysis
Original DAGR is imaged and analyzed in a similar manner as described above for CKAR.
For targeted versions of DAGR, variability will exist in the range of responses for each cell (Figure 4B, left panel). This results from the variations in relative expression levels of CFP and YFP. It is possible to work around these variations by selecting cells with similar expression levels (as was done for the data in Figure 5A). However, because phorbol esters such as PMA and PdBu cause an overriding maximal translocation of DAG-binding C1 domains to cellular membranes, it is also possible to correct for these differences. Figure 4B shows two different cells on the same dish responding to the addition of a natural agonist (UTP) and the subsequent stimulus of PdBu. Note that in the left panel, the cell with the better response to UTP also responds better to PdBu (Cell one). Because both cells are responding to the same concentration of PdBu, this indicates that ‘Cell one’ has a greater capacity to respond because of a more favorable ratio of CFP to YFP. Normalizing for the range of each cell by taking the baseline to be “0% response” and the maximal PdBu-stimulated FRET ratio change to be “100% response”, causes the responses of individual cells to UTP to become nearly super-imposable (Figure 4B, right panel). This correction controls for variations in the range of responses resulting from cell-to-cell variability in CFP relative to YFP expression levels22, providing a meaningful way to compare differences in the magnitudes of the responses to different ligands using membrane-targeted DAGR.
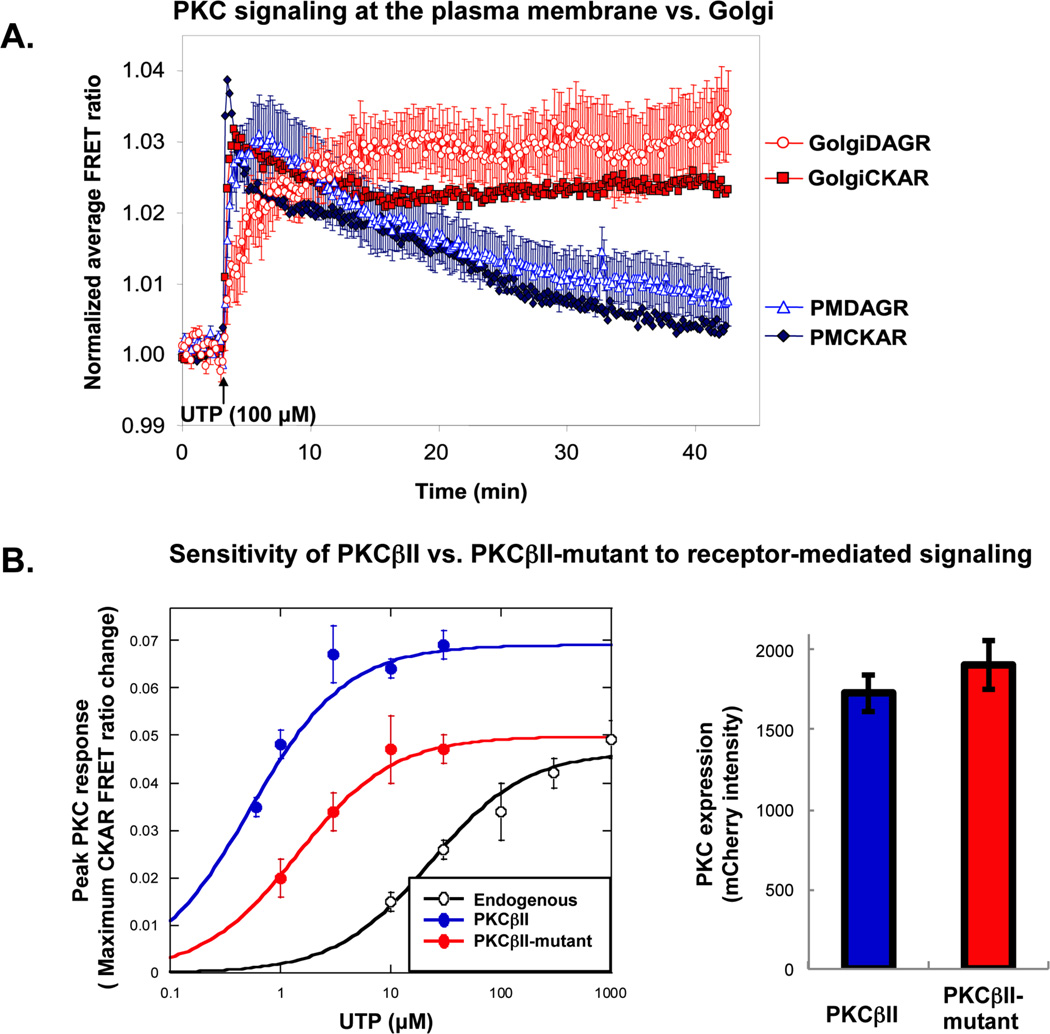
Figure 5. Examples of data generated using FRET-based reporters for PKC signaling.
A. Localized PKC signaling in response to UTP in COS7 cells. COS7 cells were transfected with PMCKAR (closed black diamonds), PMDAGR (open black triangles), Golgi-CKAR (gray squares) or Golgi-DAGR (open gray circles), and stimulated with UTP (100 µM); the average FRET ratio change was plotted over time. Here, targeted DAGR responses were not normalized for range, but cells expressing similar levels of CFP and YFP were imaged. Error bars represent SEM of data obtained from over 10 cells across three dishes per response. Note that, in this experiment, PKC activity and DAG production persist longer at the Golgi compared to the plasma membrane. Adapted from5. B. Using CKAR to test the effects of mutation on exogenous PKC. COS7 cells were transfected with CKAR alone (to monitor endogenous PKC activity, open circles), CKAR and mCherry-WT PKCII (closed black circles), or CKAR and m-Cherry PKCβII-mutant (closed gray circles). The left panel shows the maximal FRET ratio change in response to increasing concentrations of UTP. Each data point represents the maximum response determined by curve-fitting the average FRET ratio change within the first two minutes as described in section 3.5. Note that, in this case, the mutant PKC is desensitized to receptor-mediated signaling compared to the wild-type PKC. The right panel is a graph of mCherry intensity values, demonstrating equivalent expression of mCherry-tagged PKC constructs.
3.5 Variation: using CKAR to measure the activity of expressed PKC (Figure 5B)
The maximum response to a low concentration of agonist in cells transfected with CKAR alone is determined. In practice, the maximum response is calculated by curve-fitting the average response from 9 or more cells over 3 or more dishes. The maximum PKC response to GPCR agonists typically occurs within two minutes of agonist addition. Determination of the maximum response is carried out for increasing concentrations of agonist.
The maximum PKC responses are plotted on the y-axis and concentration of agonist on the x-axis using a semi-log scale. This generates a “dose-response” curve for endogenous PKC.
The “dose-response” curve for expressed PKC is similarly determined, selecting cells expressing similar levels of tagged PKC. Because the exogenous PKC is expressed more highly than the endogenous, or, in some cases, perhaps because particular PKC isoforms are better at phosphorylating CKAR than the endogenous isoforms present, the expressed PKC will respond to lower concentrations of agonist, and the response curve will shift to the left. The difference between the response curves for the expressed wild-type PKC and the endogenous isoforms provides a “window” for monitoring effects of mutations on PKC activity.
The “dose-response” curve for the tagged mutant PKC is determined next, selecting cells that express equivalent levels of mutant PKC compared to the wildtype. Mutations that decrease the ability of PKC to be activated will shift the curve to the right (as in Figure 5B), while mutations that sensitize PKC to agonist will shift the curve left-ward. These dose-response curves may be fitted with Michaelis-Menten kinetics, and can provide a quantitative measure of differences in PKC activity amongst mutants by revealing the concentration of agonist yielding half-maximal response.
Table 1.
Filters for imaging CKAR and DAGR
| Fluorochrome | Excitation (nm) | Dichronic mirror (nm) |
Emission (nm) |
|---|---|---|---|
| CFP | 420/20 | 450 | 475/40 |
| YFP (FRET-based) | 420/20 | 450 | 535/25 |
| YFP (Direct excitation) | 495/10 | 505 | 535/25 |
Acknowledgements
We thank Maya Kunkel for critical reading of this manuscript. This work was supported by NIH GM43154 and NIH PO1 DK54441.
Footnotes
The minimal setup for carrying out these experiments consists of a wide-field inverted epifluorescence microscope equipped with an automated filter wheel, appropriate filters, and a CCD camera controlled by image acquisition software. The filters must enable sequential collection of CFP and YFP fluorescent emission upon excitation of CFP; collecting YFP emission upon direct excitation of YFP is also beneficial as a way to monitor photobleaching. These experiments are carried out in our lab on a Zeiss Axiovert microscope (Carl Zeiss Microimaging, Inc.). Excitation and emission filters are switched in filter wheels (Lambda 10-2; Sutter), and images are acquired through a 10% neutral density filter. Optical filters (Chroma Technologies) are listed in Table 1. Images are captured using a MicroMax digital camera (Roper-Princeton Instruments) controlled by MetaFluor software (Universal Imaging, Corp.). Integration times are 200 ms for CFP and FRET, and 50 ms for YFP. Using this setup, cells having approximate YFP intensity of 1000 contain a cellular concentration of approximately 1 µM reporter; these levels of reporter provide consistent responses that are subject to regulation by endogenous receptors, PKC isoforms, and phosphatases.
If using a different setup than that described above, one needs to determine conditions that will provide sufficient fluorescence intensity for consistent FRET ratio changes over time without photobleaching. This is determined by varying neutral density filters (typically less than 30%) and exposure time (typically less than 1 s) until a good signal-to-noise ratio with minimal photobleaching is achieved over the appropriate period of time.
With these described acquisition settings and analyses, baseline FRET ratios are not quantitatively meaningful; only stimulated changes in FRET give a quantitative readout of changes in activity.
For each new application of CKAR (i.e. testing a new agonist), it is very important to test that the control CKAR T/A reporter does not yield a FRET ratio change and that the PKC inhibitor blocks and/or reverses the response.
For un-targeted reporters, selection of a dim cytoplasmic region (rather than the entire cell) provides the most robust and consistent responses, since CKAR in the nucleus does not often respond to stimulation (see Figure 3B). The ratiometric nature of the reporter controls for effects of minor movement of the population of reporters in and out of the selected region. However, it is important that the cell itself does not move in and out of the region selected.
For imaging targeted reporters in new cell lines, visual verification and/or extensive validation of targeting should be performed. Dyes that highlight the mitochondria and Golgi are available which are spectrally compatible with CKAR (Invitrogen, Carlsbad, CA), or one may co-localize the reporter with immunofluorescence staining for organelle marker proteins. Additionally, one should always keep in mind that, for CKAR, CFP and YFP are linked in the same molecule; therefore, CFP intensity should co-localize with YFP. Failure of these two images to co-localize indicates cleavage of the reporter resulting from targeting it near a cellular protease (as can happen on the surface of the mitochondria, for example5); this can also be tested by western blot.
If the agonist or inhibitor has intrinsic fluorescence, it may interfere with the CKAR readout. The ability of a fluorescent drug to interfere with CKAR readout can be tested by either selecting a region of the image containing no cells and monitoring CFP and FRET, or by monitoring the FRET ratio of cells transfected with CKAR T/A. If these values consistently change upon addition of the agonist or inhibitor, then the agonist or inhibitor cannot be used for FRET imaging experiments. The exception is that if the fluorescence of an inhibitor is fairly low and very minimally affects the FRET ratio of CKAR T/A, it is appropriate to pretreat samples with this inhibitor to verify that the inhibitor blocks the response to an agonist.
For targeted DAGR constructs, it is difficult to compare the magnitude of the FRET ratio change between the plasma membrane and Golgi. This is because the C1 domain may already be pre-localized to a particular subset of membranes and therefore the stimulated response will appear to be small in magnitude. We have only used these reporters to test the kinetics of the response at a particular region of the cell, correlating the duration of the DAG response with duration of the PKC response to the same agonist5 (Figure 5A).
It is difficult to image cells that adhere loosely to glass or that move during the duration of the experiment (usually less than 30 min). In particular, treatment of loosely adherent cells with phosphatase inhibitors such as Calyculin A often results in release of the cells from the dish. One can attempt to improve conditions for imaging by plating cells on glass coated with a matrix such as Matrigel or poly-lysine. In our experience, coating glass coverslips with matrix has not interfered with the FRET ratio readout of CKAR.
During data analysis, it is important to not over-subtract drift. Adding inhibitor to reverse the agonist-induced response back to baseline not only controls for specificity but also provides a good readout of whether the baseline has been over-subtracted. The FRET ratio over time after PKC inhibition should level off to a line with a flat slope if the subtraction has been carried out properly.
The experiments described here were carried out at room temperature and in HBSS supplemented with Ca2+, but the use of other media and temperature controls may improve the quality of the data for certain experiments. In addition, other accessories such as controlled flow perfusion systems (available from Bioscience Tools, San Diego, CA) may enable additional types of experiments to be carried out, such as pulse-addition of agonist.
References
- 1.Gokmen-Polar Y, Murray NR, Velasco MA, Gatalica Z, Fields AP. Elevated protein kinase C betaII is an early promotive event in colon carcinogenesis. Cancer Res. 2001;61:1375–1381. [PubMed] [Google Scholar]
- 2.Newton AC. Analyzing protein kinase C activation. Methods Enzymol. 2002;345:499–506. doi: 10.1016/s0076-6879(02)45041-0. [DOI] [PubMed] [Google Scholar]
- 3.Violin JD, Newton AC. Pathway illuminated: visualizing protein kinase C signaling. IUBMB Life. 2003;55:653–660. doi: 10.1080/152165401310001642216. [DOI] [PubMed] [Google Scholar]
- 4.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- 6.Manning BD. Challenges and opportunities in defining the essential cancer kinome. Sci Signal. 2009;2:pe15. doi: 10.1126/scisignal.263pe15. [DOI] [PubMed] [Google Scholar]
- 7.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 9.Gallegos LL, Newton AC. Spatiotemporal dynamics of lipid signaling: protein kinase C as a paradigm. IUBMB Life. 2008;60:782–789. doi: 10.1002/iub.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton AC. Protein kinase C. IUBMB Life. 2008;60:765–768. doi: 10.1002/iub.118. [DOI] [PubMed] [Google Scholar]
- 11.Mattingly RR. Mitogen-activated protein kinase signaling in drug-resistant neuroblastoma cells. Methods Mol Biol. 2003;218:71–83. doi: 10.1385/1-59259-356-9:71. [DOI] [PubMed] [Google Scholar]
- 12.Pan TT, Neo KL, Hu LF, Yong QC, Bian JS. H2S preconditioning-induced PKC activation regulates intracellular calcium handling in rat cardiomyocytes. Am J Physiol Cell Physiol. 2008;294:C169–C177. doi: 10.1152/ajpcell.00282.2007. [DOI] [PubMed] [Google Scholar]
- 13.Hosoda K, Saito N, Kose A, et al. Immunocytochemical localization of the beta I subspecies of protein kinase C in rat brain. Proc Natl Acad Sci U S A. 1989;86:1393–1397. doi: 10.1073/pnas.86.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito N, Kose A, Ito A, et al. Immunocytochemical localization of beta II subspecies of protein kinase C in rat brain. Proc Natl Acad Sci U S A. 1989;86:3409–3413. doi: 10.1073/pnas.86.9.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyawaki A. Green fluorescent protein glows gold. Cell. 2008;135:987–990. doi: 10.1016/j.cell.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Sakai N, Sasaki K, Ikegaki N, Shirai Y, Ono Y, Saito N. Direct visualization of the translocation of the gamma-subspecies of protein kinase C in living cells using fusion proteins with green fluorescent protein. J Cell Biol. 1997;139:1465–1476. doi: 10.1083/jcb.139.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang QJ, Bhattacharyya D, Garfield S, Nacro K, Marquez VE, Blumberg PM. Differential localization of protein kinase C delta by phorbol esters and related compounds using a fusion protein with green fluorescent protein. J Biol Chem. 1999;274:37233–37239. doi: 10.1074/jbc.274.52.37233. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Gambhir A, Hangyas-Mihalyne G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J Biol Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 21.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 22.Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem. 2007;282:826–830. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]