Summary
Bacterial pathogens use secreted effector proteins to subvert host-cell defenses. VopL is an effector protein from Vibrio parahaemolyticus that nucleates actin filaments. VopL consists of a VopL C-terminal Domain (VCD) and a tandem array of three WASP homology 2 (WH2) motifs. Here we report the crystal structure of the VCD dimer bound to actin. The VCD binds three actin monomers in a spatial arrangement close to that in the canonical actin filament. In this configuration each actin can readily accommodate a WH2 motif. The data suggest a mechanism of nucleation wherein VopL creates filament-like structures, organized by the VCD and delivered by the WH2 array, that can template addition of new monomers. Similarities with Arp2/3 complex and formin proteins suggest that organization of monomers into filament-like structures is a general and central feature of actin nucleation.
Introduction
Many important cellular processes, including cell motility, vesicle trafficking, and cell division, depend upon precise spatial and temporal control of actin polymerization (Campellone and Welch, 2010; Dominguez, 2009; Pollard, 2007; Pollard and Cooper, 2009). Actin can polymerize on its own, but does so slowly, primarily due to kinetic barriers that hinder spontaneous nucleation (Pollard and Cooper, 1986; Sept and McCammon, 2001). Cellular actin nucleation factors accelerate filament formation by catalyzing nucleation in response to upstream regulatory signals. Their actions afford precise spatial and temporal control over actin filament dynamics in vivo (Padrick and Rosen, 2010; Pollard, 2007). The Arp2/3 complex, formin proteins, and WASP homology domain 2-based (WH2-based) nucleators are ubiquitous eukaryotic actin nucleation factors (Campellone and Welch, 2010). The structural mechanisms by which these systems mediate filament assembly are incompletely understood.
The Arp2/3 complex is a seven-protein assembly that contains two actin related proteins (Arp2 and Arp3), which are structurally similar to actin (Kelleher et al., 1995; Machesky et al., 1994). The VCA region of proteins in the Wiskott-Aldrich Syndrome Protein (WASP) family acts in concert with existing actin filaments to activate the Arp2/3 complex; the net result is nucleation of a new filament from the side of an existing one (Pollard, 2007). During Arp2/3 activation, the WH2 regions from two VCAs bind to and deliver actin monomers to Arp2 and Arp3 (Padrick et al., 2008; Padrick et al., 2011; Ti et al., 2011). Crystal structures of inactive Arp2/3 complex and EM analyses of the active form have shown that nucleation also involves substantial reorganization of the two Arp subunits to an arrangement that resembles successive “short pitch” monomers in an actin filament (Nolen and Pollard, 2007; Padrick et al., 2011; Robinson et al., 2001; Rodal et al., 2005; Rouiller et al., 2008; Xu et al., 2012). Nucleation thus appears to be based on an arrangement of the Arp subunits and recruited actins that mirrors the “barbed end” (or rapidly growing end) of the polarized actin filament, which readily incorporates additional monomers.
Formin proteins also act by recruiting and organizing actin monomers. These proteins nucleate filaments through a conserved formin homology 2 (FH2) domain, which tracks processively with the growing barbed end of the nascent polymer (Paul and Pollard, 2009). Crystal structures of formin-actin complexes indicate that the FH2 domain arranges monomers in a conformation that resembles a strained actin filament, leading to models of both nucleation and processive elongation (Otomo et al., 2005; Paul and Pollard, 2009; Thompson et al., 2013). In some formins, the FH2 domain acts in concert with sequence motifs proximal to, or overlapping with, an adjacent regulatory element (the DAD motif). These sequences, which appear to be related to the WH2 motif, can accelerate nucleation, and are thought to deliver actin to the FH2 domain (Chhabra et al., 2009; Gould et al., 2011; Heimsath and Higgs, 2012).
The WH2-based nucleation factors are defined by arrays of WH2 motifs. Well-studied examples include cordon-bleu (cobl), leiomodin (lmod), and SPIRE (Qualmann and Kessels, 2009). Members of this class vary in the number of WH2 motifs they possess, how these WH2 motifs are positioned relative to one another, and in nucleation potency. In some members (e.g. cobl), WH2 motifs are positioned in a manner that permits stabilization of a short-pitch actin-actin contact, which may be important for efficient nucleation (Carlier et al., 2011) (Qualmann and Kessels, 2009). In SPIRE, the arrangement of WH2 domains is more consistent with stabilization of longitudinal actin-actin contacts instead of the short-pitch actin dimer needed to produce a barbed end. This is consistent with EM analyses showing structures resembling a short, single actin strand in the presence of SPIRE as opposed to the pair of strands that compose an actin filament (Quinlan et al., 2005). In isolation, the SPIRE WH2 array exhibits relatively weak nucleation activity. But an interaction with the dimeric formin, Cappuccino, brings together two SPIRE WH2 arrays, greatly enhancing activity (Quinlan et al., 2007; Vizcarra et al., 2011). Thus, while different WH2-based nucleation mechanisms are possible, highest potency appears to involve stabilization of both strands of the nascent filament.
Vibrio parahaemolyticus is a gastrointestinal pathogen, and a cause of food-borne illness worldwide (Yeung and Boor, 2004). Transmission occurs primarily through the consumption of raw or undercooked shellfish harvested from contaminated marine waters, and results in diarrheal disease that is usually self-limiting (Yeung and Boor, 2004). Vibrio parahaemolyticus, like many bacterial pathogens, hijacks eukaryotic cytoskeletal processes through injection of effector proteins into host cells (Alto and Orth, 2012; Haglund and Welch, 2011). One of the Vibrio parahaemolyticus injected effector proteins is the actin nucleation factor VopL. VopL injection causes a substantial reorganization of the host cytoskeleton (Liverman et al., 2007) leading to the formation of characteristic actin stress fibers. VopL has two distinct domains: an N-terminal array of three WH2 motifs, and a unique VopL C-terminal domain (VCD). While the VCD is sufficient for nucleation activity, its potency is greatly enhanced by inclusion of the tandem WH2 arrays (Namgoong et al., 2011; Yu et al., 2011).
In order to understand the activity of VopL, and gain general insights into structural mechanisms of actin filament nucleation, we determined the crystal structure of the VopL VCD in complex with actin. In this structure, the VCD dimer binds to three actin monomers which show striking similarity to three consecutive monomers in an actin filament. Modeling shows that in this arrangement, each actin monomer is accessible to a WH2 domain. The structure and complementary biochemical data lead to a model in which the VCD functions as a low-affinity “organizer,” evolved to arrange actins in a filament-like configuration, and the WH2 arrays, while being poor organizers, bind actin monomers with high affinity and deliver them to the VCD. Together, the two elements enable full length VopL to potently template new actin filaments. The division of organization and delivery appears to be a general feature of actin nucleation factors.
Results
A new non-polymerizable actin mutant
To understand the mechanism of VopL-mediated nucleation, we determined the crystal structure of the VopL VCD in complex with actin. Crystallographic studies involving actin require some means of preventing polymerization. To achieve this, we introduced three mutations into the barbed-end of Drosophila melanogaster 5C actin (D287A, V288A, D289A). These mutations are located in the interface between actin monomers in a longitudinal contact (along the filament axis) (Figure 1A). Pelleting assays show that the D287A/V288A/D289A mutant actin does not form filaments under conditions that induce polymerization of wild type actin (Figure 1B). Although unable to polymerize, the mutant retains properties associated with wild type actin; it binds VopL WH2 motifs with nanomolar affinity (Figure S1), and is able to interact with the fast-growing barbed-end of existing actin filaments in a concentration dependent manner (Figures 1C and 1D), indicating that the mutant is stably folded and functionally intact.
Figure 1. Development of a barbed-end-blocked non-polymerizable actin. See also Figure S1.
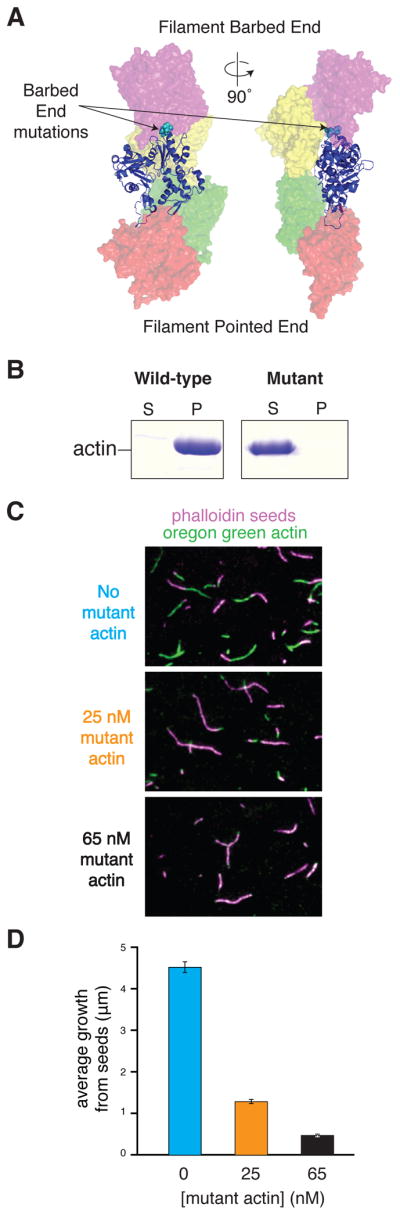
(A) A single actin monomer in an ideal filament (Oda et al., 2009) is shown in ribbon representation, with the four flanking actins shown as transparent surfaces. The locations of the sidechains mutated to prevent polymerization (cyan spheres) are shown.
(B) Actin pelleting assays comparing mutant Drosophila 5C actin to wild type rabbit muscle actin. 9 μM samples of rabbit muscle and barbed-end-blocked actin were allowed to polymerize in 50 mM KCl for 16 hours. Samples were separated by centrifugation into a soluble pool and pellet pool (containing filaments) and analyzed by SDS-PAGE.
(C) Mutant actin exhibits capping activity at filament barbed ends. Filament seeds (stained with Alexa-546 phalloidin) were mixed in G-buffer with 0.5 μM unlabeled rabbit actin, 0.15 μM rabbit actin labeled with Oregon green maleimide, and either 0 μM, 0.025 μM, or 0.065 μM mutant actin. Polymerization was initiated by addition of 10x KMEI buffer and allowed to proceed for 10 minutes, at which time individual filaments were imaged by TIRF microscopy.
(D) Average length of growth from the barbed ends of phalloidin seeds in the presence of increasing concentrations of mutant actin. Values shown are average filament lengths growing from the barbed end of phalloidin stabilized seeds, with 423, 275 and 70 seeds analyzed at 0, 25 and 65 nM mutant actin, respectively. Error bars represent the 1σ standard error in filament length. Average value for the 65 nM mutant actin sample is likely an overestimate as many seeds did not grow visibly at either end, and hence were rejected as barbed and pointed ends could not be distinguished from one another.
Structure of the VCD-actin complex
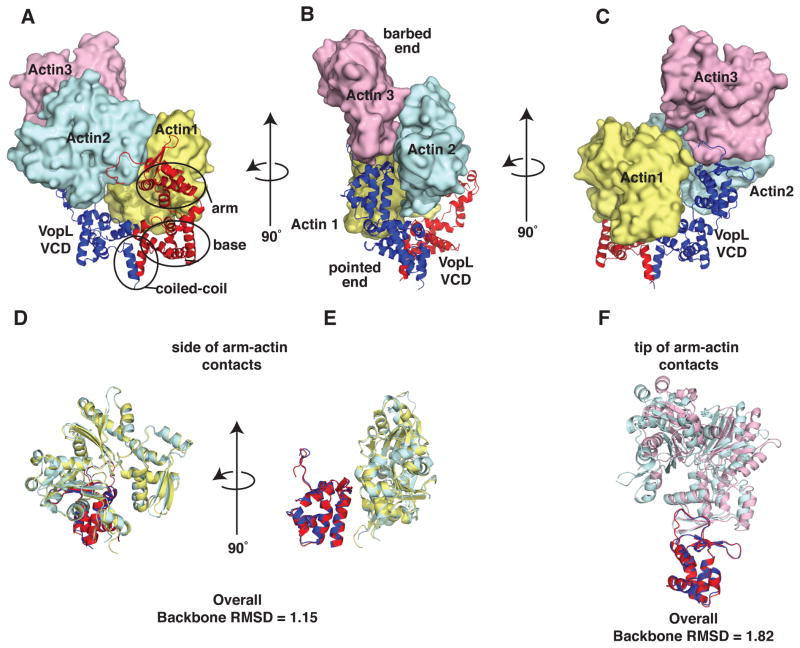
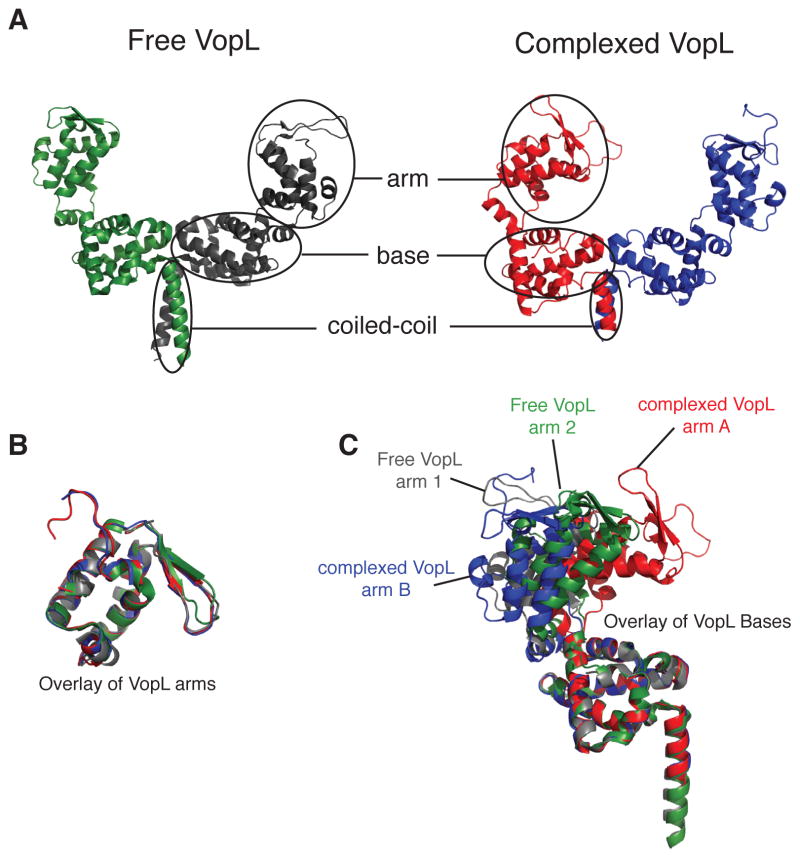
Co-crystallization of the mutant actin and VCD yielded crystals that contained both components and diffracted to 2.75 Å, enabling structure determination by molecular replacement (Figure 2, Table S1 and Figure S2B; RCSB ID code rcsb081512). In this structure, each asymmetric unit contains a VCD dimer and three actin monomers. The actin monomers adopt a spatial arrangement that resembles that of three successive subunits in an actin filament (Figure 2 and Figure S2C). Given the many possible arrangements actin might adopt in an arbitrary crystal, it is likely that this resemblance is functionally significant. As in free VopL, each VCD monomer consists of three structural units: arm, base, and carboxy-terminal helix (Figures 3 and S3) (Namgoong et al., 2011; Yu et al., 2011). Contacts between the base units form an elongated platform, stabilized by the C-terminal helices, which form a coiled-coil. The arms emerge from the ends of this platform and are directed opposite to the coiled-coil. In both the free protein and in complex with actin, the VCD dimer is asymmetric; all four arms (from the two structures) exhibit distinct orientations relative to the base platform, owing to rigid body rotations about the flexible arm-base linkers (Figure 3B and C).
Figure 2. Overall structure of the VopL VCD in complex with an actin trimer. See also Figure S2 and Table S1.
(A–C) VopL VCD dimer is shown as a ribbon with one VopL monomer (chain A) red and the other (chain B) blue. Actin monomers 1, 2 and 3 are shown in surface representation and colored yellow, cyan, and pink, respectively. (B) Indicates barbed and pointed ends of the actin trimer.
(D and E) Side-of-arm contacts. Alignment of arm B + actin 2 (oriented as in (B)) with arm A + actin 1.
(F) Tip-of-arm contacts. Alignment of arm B and actin 3 (oriented as in (C)) with arm A + actin 2. The alignments were made using backbone atoms of the VCD arms only. ‘Overall Backbone RMSD’ is the result of aligning atoms from both actin and VCD arms.
Figure 3. VCD arms in different structure are related by rigid body rotations. See also Figure S3.
(A) Overview of free VopL (Yu et al., 2011) and VopL in the actin-bound structure. The arm (residues 283–382), base (residues 245–278, and 395–456), and coiled-coil (residues 462–474) domains are indicated.
(B) Alignments of the free and complexed VopL arms reveal a high degree of similarity.
(C) Backbone alignment of the four VopL monomers, restricting the alignment to the base, reveals that each of the four VopL arms adopts a distinct orientation relative to the base.
The actins are assembled onto this structure with their pointed ends directed toward the VCD (Figure 2B). Actin 1 binds to the face of the VCD dimer, making extensive contacts to both base units as well as both arms. Actin 2 does not contact the base. Rather, it sits with the pointed end of its subdomain 4 engaged with the tip of one arm. It also contacts the side of the opposite arm through the face of subdomain 2. Actin 3 sits analogously on the tip of the opposite arm, again through contacts of the pointed end of its subdomain 4. This arrangement is distinct from those in the two previously hypothesized models based on the free VCD structure (Namgoong et al., 2011; Yu et al., 2011).
In the VCD-actin complex, the VCD arms have undergone rigid body rotations relative to the arms in the free VCD structure (Figures 3A and 3C). This allows each arm to bind to two actins, and the resulting arm-actin assemblies are pseudo-symmetric: the arm A (red) contacts to actins 1 and 2 are analogous to those of arm B (blue) with actins 2 and 3 (Figure 2). The similarity of these arm-actin assemblies exists even at the level of detailed contacts. We classify the arm-actin contacts into two groups, actin-“tip-of-arm” contacts and actin-“side-of-arm” contacts (Figures 2D–F). Structural alignments of the tip of arm contacts show that the spatial arrangement of actin 2 (cyan) relative to the VCD arm A (red) mirrors that of actin 3 (pink) relative to the VCD arm B (blue), with an overall backbone RMSD of 1.82 Å (Figure 2F). The deviations derive from a small rigid body rotation of the actins, relative to the arms, apparent in an alignment based on the arms alone. Similarly, structural alignments of the side of arm contacts show that the spatial arrangement of actin 1 (light yellow) relative to VCD arm A (red) mirrors that of actin 2 (cyan) relative to VCD arm B (blue) (Figure 2D and 2E). Alignment of these elements yields a backbone RMSD of 1.15 Å. In this case, deviations are distributed throughout the structures, as evidenced by restriction of the alignments to the respective arms.
Actin adopts a filament-like arrangement in complex with the VCD
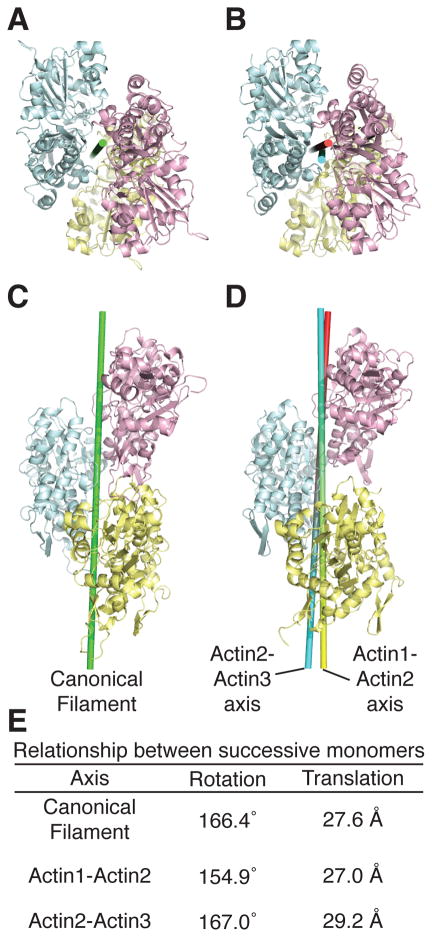
Strikingly, the two actins bound to each arm closely resemble the short pitch dimer present in an actin filament (Figure 4). In an idealized filament, successive monomers are related by a 166.4° rotation and 27.6 Å translation along the rotation axis (Fujii et al., 2010; Holmes et al., 1990; Oda et al., 2009) (Figure 4E). In the VCD complex, actins 1 and 2 are related by a 154.9° rotation and 27.0 Å translation (Figure 4E). Superposition of the actin1/2 dimer on a canonical actin filament model (Oda et al., 2009) gives a backbone RMSD of 3.33 Å. Actins 2 and 3 are similarly related by a rotation of 167.0° and translation of 29.2 Å, and superimpose on the canonical filament with backbone RMSD of 2.57 Å. These relationships suggest that the fundamental role of the VCD arm is to organize two actins into a short pitch dimer.
Figure 4. Actins in the VopL complex resemble the canonical actin filament. See also Figure S4.
(A and C) Canonical actin filament (Oda et al., 2009).
(B and D) Actins from the VopL complex, colored as in Fig. 1. Axes relating pairs of actin monomers are shown as cylinders.
(E) Rotations and translations associated with the depicted axes.
The VCD base separates the two arms, allowing them to share actin 2. This leads to an overall organization that bears close similarity to three successive monomers in a filament (Figure 4). The two respective short-pitch actin pairs are each associated with a rotation axis. Because the arrangement of the actins is close to that in a filament, these axes are nearly coincident (angle between axes = 5.1°, distance of closest approach between axes = 2.4 Å) (Figure 4). Further, this arrangement allows the “hydrophobic plug” of actin 2 (residues 264–270) to contact the interface between actins 1 and 3, as it does in the canonical filament model (Fujii et al., 2010; Oda et al., 2009) (Figure S4). Thus, the inherent symmetry of the VCD, coupled with the flexibility of the arm-base attachments, enforces a filament-like arrangement. This arrangement likely lies at the core of the VopL nucleation mechanism.
Mutagenic validation of VCD-actin contacts
In a previous study, mutation of charged VCD surface patches identified several residues important for nucleation activity (Table S2). With the VCD-actin structure in hand, it is now apparent that these mutations disrupt VCD-actin contacts, and thus validate the structure. The details of additional actin-VCD contacts that were not subject to mutagenesis appear in Figure S5.
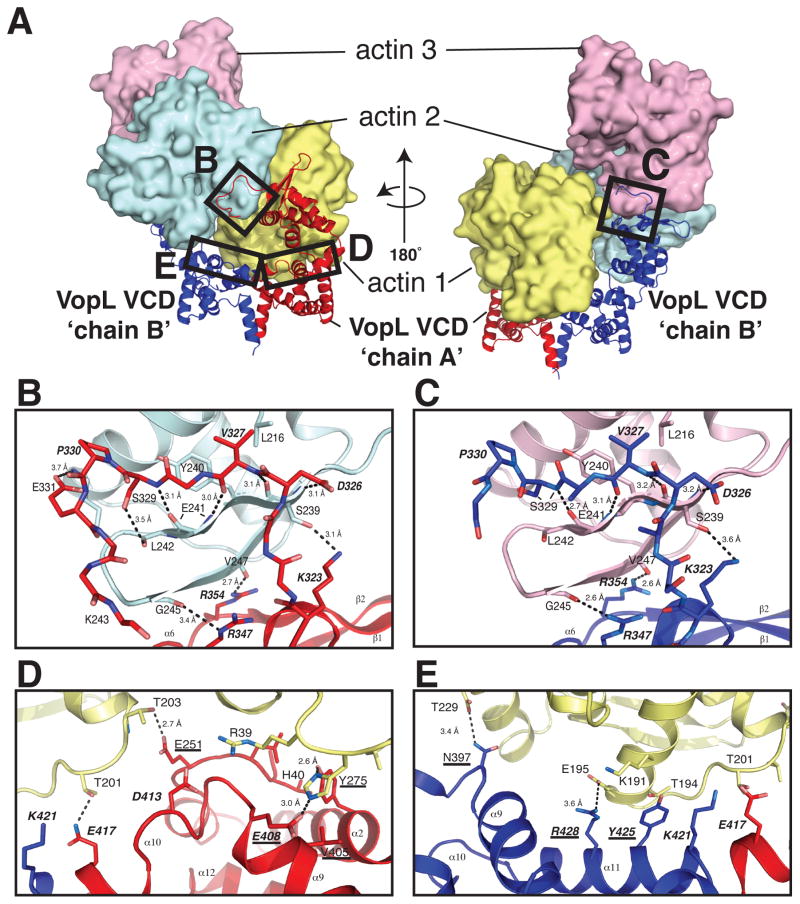
Two types of mutations were made, to the arm and to the base; the former will be discussed first. In our structure, the tip of the VCD chain A arm contacts subdomain 4 of actin 2, and, in a nearly identical fashion, the tip of the VCD chain B arm contacts subdomain 4 of actin 3. The VCD residues that compose the “tip of arm” contacts lie in a loop spanning residues Y322-A328, in the α6-helix (I341-T350), and in the first beta strand (R354-D360) (Figure 5A–C and Figure S2A). Two triple mutants affecting patches at the tip of the arm were K323E/R347E/R354E and D326G/V327G/P333G. Mutation of the first patch disrupts a hydrogen bond between VCD K323 and the sidechain of actin S239, and backbone hydrogen bonds between the side chains of VCD R347 and R354 and actin G245 and V247, respectively. Mutation of the second patch disrupts a hydrogen bond between VCD D326 and actin S239 and a hydrophobic interaction between VCD V327 and actin L216.
Figure 5. The details of the tip-of-arm actin contacts and the contacts of the VCD base to actin 1. See also Figure S5 and Table S2.
(A) Entire VCD-actin complex. The VopL chain identities and the actins are labeled. Boxes indicate the specific regions that appear in the close-up views in panels B–E.
(B) Interface between the tip of VCD chain A arm and actin 2.
(C) Interface between the tip of VCD chain B arm and actin 3.
(D) The interface between VopL chain A and actin 1.
(E) The interface between VopL chain B and actin 1.
In all panels the coloring scheme matches that in Figure 2. In stick presentations (panels B and C), carbon atoms match the ribbon color; nitrogen and oxygen atoms are colored blue and red, respectively. Residue numbers and secondary structural elements are indicated. Contacts within 4 Å are shown as dotted lines. Probable hydrogen bonds have distances indicated. Residues labeled with bold italics were mutated in the VCD homodimer to validate the structure (Table S2). Underlined labels indicate residues that were mutated in the VopL heterodimers to selectively impair either one or both of the symmetric binding sites for actin 1. Residues appearing in bold, underlined italics were mutated in both cases.
In contrast to the VopL arms, which lie in nearly identical binding sites, the base of VopL chain A and the base of VopL chain B interact differentially with actin 1, making contacts to its subdomains 2 and 4, respectively. Contacts between actin and the VopL chain A base involve the N-terminus of the VCD (L249-E251), the C-terminus of helix α2 (E262-A279), the C-terminus of helix α9 (K399-E408) and α10 (L409-K421) (Figures 5A and 5D). The contacts between chain B of the VCD base and actin are less extensive than those of chain A, and occur mainly through helix α11 (residues 421–433) and the α8-α9 loop (residues 395–399) (Figures 5A and 5E). The remaining mutations lie in the interface between the VCD base and actin 1, and target two surfaces patches. Mutation of the first patch (E408K/D413K/E417A) disrupts contacts between VCD chain A and actin subdomains 2 and 4: a hydrogen bond between VCD E408 and actin H40, a long-range electrostatic contact between VCD D413 and actin R39, and a hydrogen bond between VCD E417 and actin T201. Mutation of the second patch (Y425A/R428D and K421A/Y425A/R428D) disrupts contacts between subdomain 4 of actin and VCD chain B. VCD Y425 makes hydrophobic contacts with the hydrophobic portions of actin T194 and K191, and VCD R428 forms a hydrogen bond with actin T201. Mutation of K421 likely disrupts a salt bridge between the VCD dimer subunits, and is not directly involved with actin binding.
All of the mutations discussed above disrupt contacts between the VCD and actin. Accordingly, these mutations impair VopL nucleation activity, and thus establish that the VCD-actin contacts observed in the crystal are functionally important.
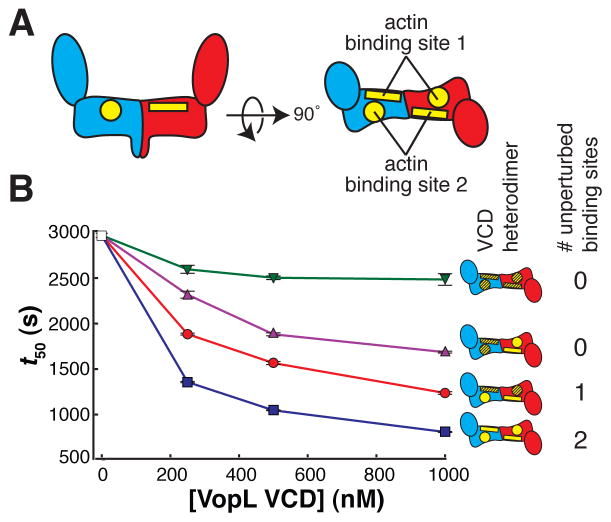
VCD heterodimers support the asymmetric engagement of actin
Although the mutagenesis studies described above confirm that the contacts in the crystal structure are important for VopL nucleation activity, we sought to develop a strategy that addresses more directly to the asymmetry of the structure and its role in nucleation. The VCD dimer, by virtue of its two-fold symmetry, can bind the actin trimer in two equivalent orientations. In any given nucleation event, VCD symmetry is broken through contacts of actin 1 to one face of the dimer. Thus, our structure predicts that mutagenic disruption of both faces of the dimer should strongly impair activity, while disruption of only one face should leave an equivalent binding site intact on the other face, and thereby confer less impairment. Mutagenic disruption of only one face requires the introduction of different sets of mutations into the respective VCD monomers within a dimer. To achieve this, we created heterodimeric VCD proteins by replacing the C-terminal coiled coil with an engineered coiled-coil heterodimer (O’Shea et al., 1993). Actin 1 contacts two patches of residues in the base platform, one on each VCD monomer (N397, Y425, and R428 on monomer A, and E251, Y275, V405, and D413 on monomer B) (Figure 5D–E). Mutating both patches in both monomers (E251A, Y275A, V405A, and E408A in one patch, N397E, Y425A, and R428E in the other patch) does not disrupt folding (Figure S6E), but does decrease activity relative to the wild-type heterodimer, as both faces of the VCD dimer are impaired (Figure 6A–B and Figure S6A–D). Similarly, mutating both patches on one monomer also decreases activity appreciably, again because both faces are affected. In contrast, and as predicted, when only one face of the dimer is disrupted, by mutating the first patch on one monomer and the second patch on the other monomer, activity is only modestly decreased. These data support the structure-based prediction that the VCD nucleates actin using one of two equivalent, mutually exclusive binding sites.
Figure 6. The VopL VCD can bind to the actin trimer in two equivalent orientations. See also Figure S6.
(A) Schematic side (left) and top (right) views of the VCD. The VCD homodimer has two symmetry-related binding sites for actin 1. We subdivide each of these binding sites into two surfaces, one residing on each VCD chain. One surface is shown as a rectangle (including residues E251, Y275, V405, and E408), and the other surface is shown as a circle (including residues N397, Y425, and R428).
(B) Actin assembly was quantified by measuring the time to half-maximal polymerization (t50). The average and 1σ standard error (n = 4, error bars appear in front of the symbol) are plotted for four heterodimers possessing different numbers of unperturbed actin binding sites (wild type sites and mutant sites are yellow and hatched, respectively). Both patches on both monomers represented by green triangles, both patches on one monomer by magenta triangles, one patch on one monomer and one patch on the other monomer by red circles, and the unmutated heterodimer by blue squares. Experiments using 0 nM VopL (white square) were included in both data sets, but are the same data.
WH2 arrays deliver actin, which is organized by the VCD
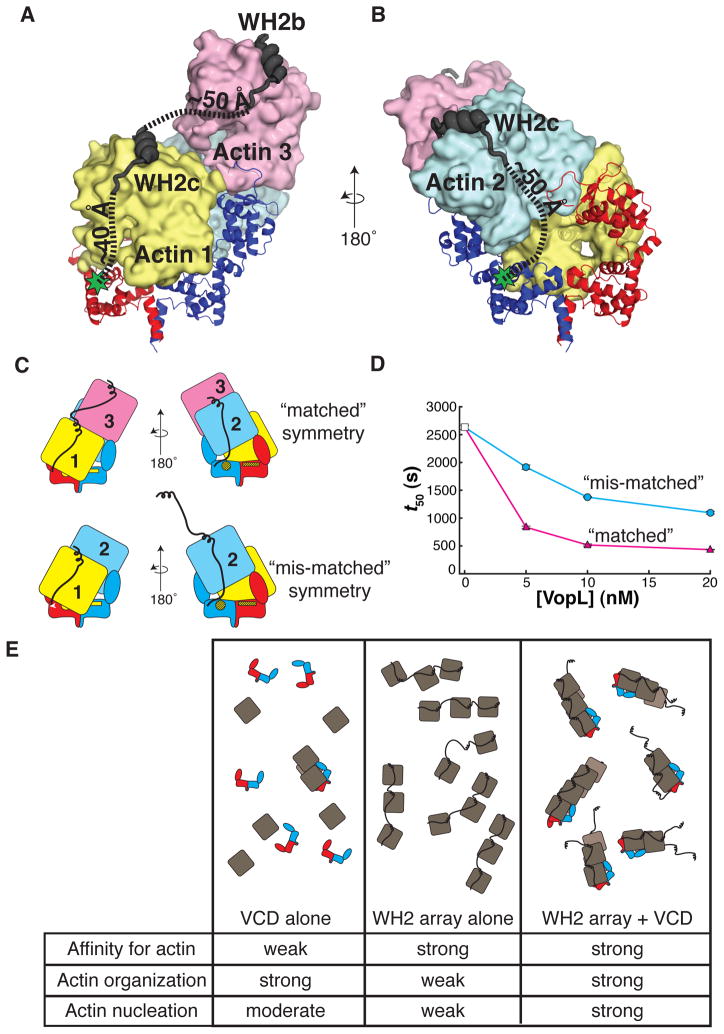
Adjacent to the VCD, each VopL monomer possesses three tandem WH2 motifs that bind actin with nanomolar affinity (Namgoong et al., 2011; Yu et al., 2011). This array exhibits little nucleation activity on its own, but when fused to the VCD, it increases the potency of VopL by over 100-fold (Namgoong et al., 2011; Yu et al., 2011). The WH2 motifs likely enhance activity by binding to and delivering actin monomers to the VCD, compensating for the modest affinity of the VCD for actin. To understand this process, we modeled WH2 motifs onto the actins in our structure, using a previously reported crystal structure of a VopL WH2-actin complex (Rebowski et al., 2010). WH2 motifs possess an amphipathic helix that binds in the cleft between subdomains 1 and 3 at the barbed end of the actin monomer, followed by an ‘LKKT motif’ which binds to the actin face (Carlier et al., 2011; Dominguez, 2009). Each of the three actins in the complex readily accommodates a WH2 motif without steric clashes. The 23 residue linker between the ‘LKKT’ sequence of the third WH2 motif and the N-terminus of the VCD can span the modeled distance between these points for both actin 1 and actin 2 (~40 and ~50 Å, respectively) (Figures 7A and B). We note that the structure contradicts our previous hypothesis regarding the aberrantly low activity of a fusion of the third WH2 motif with the VCD (Yu et al., 2011), an observation we are currently working to understand. The 20 residue linker between the second and third WH2 motifs (WH2b and WH2c, respectively) is sufficiently long to span the distance between the WH2 motifs modeled onto actins 1 and 3 (Figure 7A), but insufficient to span the distance between the WH2 motifs modeled onto actins 1 and 2. Thus, it is likely that one WH2 array delivers actins 1 and 3, while the opposite array delivers actin 2.
Figure 7. The VopL WH2 domains recruit and deliver actin monomers to the VCD.
(A) WH2b and WH2c modeled onto actin 3 and actin 1, respectively.
(B) WH2c modeled onto actin 2. (A and B) Green stars indicate the position of the VCD N-terminus. Dashed lines approximate the trajectories of the paths used to estimate the distances between structured elements in the model (VCD N-terminus to WH2c, and WH2c to WH2b).
(C) Illustration of VopL heterodimers that harbor mutations in the VCD that disrupt binding of actin 1 to one face, and additionally, are fused to a total of three WH2 motifs, an array of two WH2 motifs on one VCD monomer and a single WH2 motif on the other. The respective VCD monomers within a heterodimer are red or blue. Actins 1 (yellow), 2 (blue) and 3 (pink) are indicated. The unmutated face of the VCD is denoted by bright yellow patches. The mutated face of the VCD appears as yellow patches with black hash marks. Only the unmutated face could bind actin 1 in the mode observed in the crystal structure. The WH2 arrays are represented as black lines that emanate from the respective VCD monomers. In the construct with “matched” symmetry, the (WH2)2 array delivers actins 1 and 3 such that actin 1 contacts the unmutated face of the VCD; the single WH2 motif delivers actin 2, creating a stable trimer (Sept and McCammon, 2001). In the construct with “mis-matched” symmetry, if the system uses the wild type face of the VCD, there is no WH2 motif to deliver actin 3 longitudinally to actin 1; the (WH2)2 array could deliver an actin longitudinally to actin 2 (not shown in cartoon), but this would not create a stable actin trimer.
(D) Actin assembly was quantified by measuring the time to half-maximal polymerization (t50). The average and 1σ standard error (n = 4, error bars appear in front of the symbol) are plotted for two different VopL heterodimers. Both heterodimers harbor mutations that disrupt a single face of the VCD, and each possessing a single WH2 motif fused to one VCD monomer and two tandem WH2 motifs on the other. However, the two heterodimers differ from one another in how the WH2 arrays are positioned relative to the mutated surface on VCD, referred to as ‘matched’ (magenta triangles) or ‘mis-matched’ (cyan circles) as shown in panel C. Experiments using 0 nM VopL (white square) were included in both data sets, but are the same data.
(E) A summary of the roles of the VCD and the WH2 arrays in VopL-mediated actin nucleation. The cartoons represent the ensemble behavior, in the presence of actin, for the VCD alone, the WH2 array alone, and full length VopL. The rows in the table indicate the capacity to bind to actin, to organize actin into filament-like structures, and to nucleate actin filaments. Note that for visual clarity, in panels C and E, we represent the unbound WH2 motif as helical, although physical data suggest that in the free state WH2 motifs are disordered.
In order to test this model we returned to our heterodimer strategy. As described above (Figures 5 and 6), mutating both actin-binding patches on one face of a VCD heterodimer restricts assembly of the three actins to predominantly one orientation, dictated by binding of actin 1 to the wild type face. In a minimal system where a total of three WH2 motifs recruit the actin trimer, there exists a maximally active configuration where one VCD monomer within a dimer uses two WH2 motifs to deliver actins 1 and 3 to the wild type face, and the other VCD monomer uses a single WH2 motif to deliver actin 2. If the WH2 arrays were swapped, the actin geometry in the initial trimer would not match that of the recruiting elements, since there would be no WH2 motif to recruit actin 3 (Figure 7C). Thus, the “single-face” heterodimer, by forcing use of only one face of the VCD, provides a means of examining the relationship between actin geometry controlled by the VCD and delivery of actin by the WH2 arrays of the individual subunits.
As previously (Figure 6), in order to impair one face of a VCD heterodimer we mutated N397, Y425, and R428 on one subunit (monomer A, red in Figure 7B) and E251, Y275, V405, and D413 on its partner (monomer B, blue in Figure 7B). As shown in Figure 7 panels A and B, our model predicts that WH2 motifs of monomer A (red) should deliver actins 1 and 3 to this heterodimer, while those of monomer B (blue) should deliver actin 2. As shown in Figure 7D, when monomer A is joined to two WH2 motifs and monomer B is joined to one motif, activity of the heterodimer is high. But when the WH2 arrays are swapped, so that monomer A has only one WH2 motif and monomer B has two motifs, activity is low (Figure 7D). Thus, when the geometry of actins organized by the VCD is matched to the geometry of recruitment by WH2 motifs (Figure 7C, top), the two elements can act synergistically to promote nucleation. But when the geometries are mismatched (Figure 7C, bottom), synergy is less and activity is lower. Together, these data provide strong support for our model of WH2-mediated delivery of actin to the VCD.
We note that in the full length protein the system likely proceeds through multiple complexes containing between three and six actins delivered by the WH2 arrays and organized by the VCD according to this same general plan (Figure 7E, right). The distribution of these complexes will depend on the actin concentration and the relative rates of actin binding and nucleation, but there is not necessarily a single defined pathway involving all six WH2 motifs.
Deviations from the canonical filament model
The organization of actins in the VCD complex qualitatively resembles that in an actin filament. However, some details of the structure are quantitatively distinct from the filament. First, the rotational and translational relationships between monomers do not exactly match those in an ideal filament (although we note that monomers within individual actin filaments can exhibit substantial angular deviations from ideality (Schmid et al., 2004)). Second, the actin monomers adopt conformations that more closely resemble that of the unpolymerized actin monomer than the monomer in a filament, with backbone RMSD of 0.6–0.9 Å versus 2–3 Å, respectively. Third, the configuration in our structure positions the subdomain 2 DNAse I binding loop (D-loop) of actin 3 too far from the barbed end cleft of actin 1 for these elements to engage as they do in a filament (Figure S4F) (Fujii et al., 2010; Oda et al., 2009). Some of these deviations may result from our use of the actin mutant to obtain crystals. However, in order for WH2 domains to bind actin and participate in nucleation the D-loop cannot be bound in the barbed end cleft. Thus, accessibility of the cleft to WH2 domains and the absence of D-loop engagement likely reflect an authentic intermediate in the nucleation pathway.
Discussion
Here we have described the structure of the VopL VCD in complex with three actin monomers. The VCD organizes the actin monomers into a trimer that closely resembles an actin filament. This positioning of the actins is dictated by binding of each arm to an actin pair that closely resembles a short-pitch actin dimer, and sharing of one actin between the pairs. This positioning leaves all actin subdomain 1 - subdomain 3 clefts accessible to the array of WH2 motifs extending from the amino terminal end of each VCD subunit.
Together, these observations suggest a mechanism of nucleation that relies on synergistic actions of the VCD and WH2 domains. In this mechanism, the VCD organizes actin monomers to closely resemble an actin filament. But since the domain has low affinity for actin, it has low nucleation activity in isolation. Conversely, the WH2 domains capture actin monomers with high affinity, but are poor organizers due to their inherent flexibility. When together in the same molecule, the two elements can effectively bind and organize actins in a filament-like configuration (Figure 4). At some stage during nucleation, either before or after additional monomers bind, this structure must reorganize into a bona fide filament, where actin monomers are in the filamentous conformation and the D-loops are engaged in the barbed end clefts. This rearrangement could weaken VCD-actin interactions and would block interactions between the WH2 amphipathic helix and actin, contributing to the observed rapid dissociation of VopL from nascent filaments (Namgoong et al., 2011).
We posit that the structure of the VopL VCD bound to actin provides a snapshot of the nucleation process, suggesting three general mechanistic themes that describe the actions of nucleation factors. First, this structure provides strong evidence that nucleation factors act by organizing monomers into a filament-like configuration. Second, comparison to other nucleation factors suggests that the separation of actin organizing and recruiting functionalities that we observe for VopL may be general. Third, the dynamic association of a given nucleation factor with a filament end may depend on the degree to which the nucleation factor organizes actins to resemble the canonical filament structure.
A long-standing hypothesis is that actin filaments arise from filament-like nuclei, and that the fundamental purpose of nucleation factors is to facilitate the formation of these structures (Dominguez, 2009). The VCD-actin structure provides the strongest evidence to date that this idea is in fact correct; VopL positions three actins in a manner very similar to those in the canonical filament. Previous crystal structures of actin-nucleation factor complexes have been less compelling in this regard; actin-actin contacts have arisen from crystallographic symmetry and do not match those in the canonical filament to the degree that they do in this structure. The crystal structure of the formin Bni1p engaged with actin (Otomo et al., 2005) revealed that actin lies in a filament-like configuration, but with an 180° rotation dictated by a crystallographic symmetry axis. The actin monomers in the structure of FMNL3 bound to actin also made contacts along on a crystallographic axis, but did not adopt a filament-like configuration (Thompson et al., 2013). Electron microscopy reconstruction of Arp2/3 complex engaged with a nucleated filament (at 26 Å) and in a soluble activated state (at ~20 Å), provide additional views of nucleation (Rouiller et al., 2008; Xu et al., 2012) that are consistent with the idea that nucleation factors function by organizing filament-like nuclei. That several of these nucleation factors appear to induce short-pitch-like contacts strongly supports the idea that actin nucleation occurs through filament-like nuclei, and that actin nucleation factors function by generating such structures.
Based on the structure and biochemistry, we propose that VopL functions via a division of labor, whereby the WH2 domains bind actin monomers with high affinity and deliver them to the VCD, the latter serving to enforce a filament-like arrangement. These two distinct functionalities, present within the same molecule, give rise to potent nucleation activity (Figure 7E). Such division of labor also appears to be important to both the Arp2/3 complex and formins. In Arp2/3-mediated nucleation, Arp2 and Arp3 serve a function analogous to that of the VCD, in that they act as an organizing entity of low inherent affinity for actin monomers (Kaiser et al., 1999; Mullins et al., 1997; Padrick et al., 2011). When Arp2/3 complex is activated by WASP family protein VCA peptides, the WH2 motifs in VCA bind actin monomers with high affinity and deliver them to the Arp2/Arp3 organizing template. In formin proteins, the FH2 domain serves a function analogous to that of the VCD, in that it too induces a filament-like actin nucleus (Otomo et al., 2005). Despite high affinity for the filament barbed end, many FH2 domains exhibit only modest affinity for actin monomers, or only bind a single monomer ((Thompson et al., 2013) and AAY, TO, unpublished observations). In some formins, such as Inf2 (Chhabra et al., 2009), FMNL3 (Heimsath and Higgs, 2012), and mDia1 (Gould et al., 2011), there is a WH2-like sequence C-terminal to the FH2 domain. In these proteins, the FH2 domains have weak activity on their own, but have high activity in concert with the additional sequences. These data are consistent with the FH2 domains acting as an organizing template, and the WH2-like sequences delivering actin monomers. Furthermore, and consistent with the overall idea, the formin mDia1 may use the nucleation factor APC as an actin recruiting factor (Okada et al., 2010). Taken together, this “division of labor” appears to be a mechanistic theme that applies to diverse nucleation factors.
Different nucleation factors exhibit distinct dynamic properties at the end of the nascent filament. Structural data on the Arp2/3 complex (Rouiller et al., 2008), the formin Bni1p (Otomo et al., 2005) and now VopL suggest that deviations from an ideal filament, or lack thereof, may underlie this variability. The Arp2/3 complex remains persistently associated with the pointed end of the filament it nucleates. Arp2/3 complex in a filament branch organizes the actin homologs Arp2 and Arp3 in a configuration indistinguishable from that of actins in the filament (Rouiller et al., 2008; Xu et al., 2012). The formin FH2 dimer moves with the end of the nascent filament, through cyclic release of individual formin subunits from the terminal two actins, and subsequent binding of this subunit to an incoming actin monomer (Goode and Eck, 2007; Otomo et al., 2005; Xu et al., 2004). In complex with the Bni1p FH2 domain, actins adopt a filament-like, but strained configuration; relief of this strain has been invoked to explain the binding and release dynamics of the FH2 subunits at the filament end (Otomo et al., 2005; Thompson et al., 2013). We have now found that VopL, which dissociates from filaments shortly after nucleation (Namgoong et al., 2011), also arranges actin monomers in a configuration that deviates from the canonical filament. Together, these behaviors suggest that strain in the nucleus and/or nascent filament may play an important role in dictating the dynamic behaviors of nucleation factors. Those that are structured to bind an ideal filament may remain persistently attached, while those that bind strained configurations may be dynamic. Such features could be selected for during evolution to produce the specific behaviors needed for distinct biological activities.
Experimental Procedures
Protein purification
VopL VCD (residues 247–484) was expressed in Escherichia coli and purified as previously described (Yu et al., 2011) Briefly, VCD was expressed as a GST fusion in BL21(DE3)-T1R E. coli, at 18° overnight. VCD was purified over glutathione sepharose (GE) and cleaved off the beads with TEV protease. Further purification was accomplished using SOURCE15Q ion exchange followed by Superdex 200 gel filtration (GE) chromatographies. VCD mutants were made using site directed mutagenesis or gene synthesis (Genscript). VCD heterodimers and VopL heterodimers bearing the minimal set of three WH2 domains were produced by replacing the VopL coiled coil with one of two compatible ‘peptide velcro’ coiled coil pairs, and co-expressing these proteins in BL21(DE3)-T1R bacteria from two origin-compatible plasmids, pMAL-c2 (NEB) and pCDF1 (Novagen). TEV protease cleavable maltose binding protein- fusions with an engineered acidic helix (O’Shea et al., 1993) were cloned into a pMAL-C2 derived vector (NEB). TEV protease cleavable His6- fusions with an engineered basic helix (O’Shea et al., 1993) were cloned into a pCDF1 (Novagen) derived vector. VCD heterodimers, and heterodimers bearing the minimal set of three WH2 domains were purified using amylose (NEB) affinity chromatography, followed by Ni-NTA affinity chromatography (Qiagen), and affinity tags were removed using TEV protease. Purification to homogeneity was accomplished using SOURCE15Q and SOURCE15S ion exchange chromatography (GE), for the VCD constructs, and the WH2-VCD constructs, respectively. Subsequently, heterodimers were subjected to gel filtration chromatography using Superdex 200 pg resin (GE) in KMEI buffer (50 mM KCl, 10 mM Imidazole pH 7.0, 1 mM EGTA, 1 mM MgCl2). Barbed end blocked His6-actin (Drosophila melanogaster 5C actin with D287A, V288A, D289A mutations) was expressed using a recombinant baculovirus produced in Sf9 cells, using the Bac-to-Bac system and a modified pFastBacHT expression vector (Invitrogen) that included an L21 enhancer sequence (Sano et al., 2002) driving expression of the mutant 5C actin. Mutant actin was purified by nickel affinity chromatography. Then, TEV protease was used to remove the His6-tag. The purification was completed using anion exchange followed by gel filtration chromatography. For crystallization, a 1:1 molar mixture of VCD and actin were dialyzed for 16 hours into 10 mM Tris pH 8.0, 50 mM KCl, 0.2 mM ATP, 2 mM TCEP, 0.1 mM CaCl2, and concentrated to 20 mg/mL.
A VopL WH2c peptide (199–226) bearing an introduced C-terminal tyrosine residue to aid in quantitation by absorbance was expressed in E. coli as a GST fusion. Following cell lysis, the fusion was affinity-purified using 5 mL glutathione agarose following manufacturer recommendations. Following elution from glutathione agarose, the fusion was concentrated to 1 mL using a centrifugal concentrator (Amicon Ultra 15, 30000 MWCO UFC903024, Millipore) and the peptide was cleaved from the GST fusion with TEV protease at 4°C overnight. The peptide was subsequently purified by two cycles of gel filtration chromatography (Superdex 75 10/300 GL, GE Healthcare) in KMEI buffer supplemented with 0.2 mM ATP.
Structure determination
The VCD-actin mixture was crystallized from hanging drops containing 1 μL protein and 1 μL of well solution (0.1 M MMT buffer, pH 8.0, and 24% PEG1500) yielding crystals with dimensions up to 500 × 500 × 300 μm3. Crystals were flash frozen directly from the drop in liquid nitrogen. Diffraction data were collected at beamline 19-ID at the Advanced Proton Source (APS), Argonne National Laboratory. The data were processed and scaled using HKL3000 (Minor, 1997). The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) as implemented in Phenix (Adams et al., 2010) with actin (Nair et al., 2008) and VCD arm fragments and VCD base/coiled-coil dimers (Yu et al., 2011) as search models. The model was improved through iterative cycles of model building in Coot (Emsley et al., 2010) and subsequent positional and TLS atomic displacement parameter refinement in Phenix.
Biochemistry
Actin assembly assays contained 2 μM rabbit muscle actin (10% pyrene labeled) in KMEI supplemented with 100 μM ATP, following a previously described method adapted to a multi-well plate format (Leung et al., 2006). Actin spin-down assays were performed using 9 μM actin in KMEI. Polymerization was allowed to proceed for 16 hours at room temperature, and samples were centrifuged at 100,000 g for 2 hour at 20° C. Supernatant and pellet fractions were collected and analyzed using Coomassie stained SDS-PAGE gels.
Isothermal titration calorimetry
ITC experiments were conducted using a VP-ITC microcalorimeter (Microcal) at 20 °C. Prior to each experiment, WH2 peptides and mutant actin were subjected to gel filtration chromatography to exchange them into KMEI buffer supplemented with 0.2 mM ATP. In each experiment, 100 μM of WH2 peptide was titrated into 10 μM actin in KMEI supplemented with 0.2 mM ATP. Data were analyzed and fit to a single-site model using Origin 7 for ITC (Microcal).
Individual filament growth assay
Rabbit muscle actin was purified as previously described (Leung et al., 2006), rabbit actin (10 mL of 40 μM in G-buffer without sodium azide and without DTT) was polymerized by dialysis overnight in KMEI supplemented with 0.2 mM ATP. With gentle stirring, a 5-fold molar excess of Oregon green C5-maleimide in DMSO was added, and labeling was allowed to proceed overnight in the dark at 4°C. This reaction was then dialyzed against 2 L of G-buffer for three days, clarified by centrifugation and purified by gel filtration column (Superdex 200 pg 26/600, GE Healthcare). Fluorescently labeled actin seeds were prepared by adding ~0.6 μM AlexaFluor 546-phalloidin (#A22236, Life Technologies) to a 4 μM solution of rabbit actin in KMEI in a final volume of 200 μL. Polymerization was allowed to proceed for 2 hours. Reactions to be imaged by TIRF microscopy were prepared with 0.5 μM unlabeled rabbit actin, 0.15 μM Oregon green-labeled actin, 10 μL of phalloidin seed stock, and either 0 nM, 25 nM, or 65 nM purified mutant actin in G-Mg buffer (2 mM TrisHCl pH 8.0, 0.2 mM ATP, 1 mM MgCl2) in a final volume of 200 μL. Seed growth was initiated by addition of 20 μL 10X KMEI to 180 μL of the aforementioned mixture. Reactions were allowed to proceed for 10 minutes, at which time they were diluted 1:200 in TIRF buffer (KMEI supplemented with 15 mM glucose, 100 μg/ml glucose oxidase, 20 μg/ml catalase). Diluted filaments were imaged in flow cells, the assembly, NEM-myosin coating and loading of which is previously described (Hung et al., 2011). Filaments were imaged using an Olympus IX-71 fitted with an Olympus TIRF arm, a PlanApo 100x oil objective (n.a. 1.45), and a Photometrics Cascade II:512 EMCCD camera. The microscope was run using Micro-Manager (Vale lab). Dual-color imaging was done by switching between excitation lasers using shutters, and filtered using a dual-color filter cube (Semrock). Pixel size was calibrated with a micrometer. Exposure times were kept constant between conditions. All images were processed in NIH ImageJ using a Gaussian filter to reduce shot-noise, and background-subtracted. Filament lengths were quantified by total pixel number after simple-thresholding, conversion to binary, and using the skeletonize function. 75–200 individual filament-lengths were quantified for each condition.
Circular dichroism
VopL heterodimers (wild type and mutant) were purified as described above, but purified by gel filtration into 50 mM sodium phosphate, pH 8.0, 100 mM sodium chloride and 1 mM TCEP. 0.1 mg/mL samples were placed into 1 mm pathlength quartz cuvettes, and CD signal was measured at 220 nm as temperature was raised from 25° to 90° C in 2° increments. Temperature was allowed to stabilize for 10 seconds before measuring CD signal, and CD signal was averaged for 32 seconds. Data was fit to a six-parameter sigmoid, and Tm reported was the inflection point of the melting transition.
Molecular modeling
The VopL WH2c motif (PDB ID# 3M1F) (Rebowski et al., 2010) was modeled onto the VCD structure using Pymol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.) by superimposing actins in the WH2-actin and VCD-actin complexes. Actin-actin rotation axes in the VCD complex were determined by aligning successive actins to each other in QtMG30 (Potterton et al., 2002) using the ‘Superpose’ tool.
Supplementary Material
Highlights.
Crystal structure of an actin trimer bound to the VopL C-terminal domain (VCD)
Actins adopt a configuration that approximates that in an actin filament
During nucleation the WH2 motifs deliver actin into a structure organized by the VCD
Acknowledgments
We thank Dr. Luke Rice for discussion and assistance with crystallographic model building and refinement, Dr. Dominika Borek for assistance with crystallographic data processing, and Lynda Doolittle for assistance with actin assembly assays. Work was supported by the Howard Hughes Medical Institute and grants from the NIH (R01-GM56322) and Welch Foundation (I–1544). Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Orth K. Subversion of cell signaling by pathogens. Cold Spring Harbor perspectives in biology. 2012;4:a006114. doi: 10.1101/cshperspect.a006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Husson C, Renault L, Didry D. Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. International review of cell and molecular biology. 2011;290:55–85. doi: 10.1016/B978-0-12-386037-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. Journal of cell science. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annual review of biochemistry. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Current biology: CB. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. The Journal of cell biology. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsath EG, Jr, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. The Journal of biological chemistry. 2012;287:3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser DA, Vinson VK, Murphy DB, Pollard TD. Profilin is predominantly associated with monomeric actin in Acanthamoeba. Journal of cell science. 1999;112(Pt 21):3779–3790. doi: 10.1242/jcs.112.21.3779. [DOI] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. The Journal of cell biology. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Morgan DM, Rosen MK. Biochemical properties and inhibitors of (N-)WASP. Methods Enzymol. 2006;406:281–296. doi: 10.1016/S0076-6879(06)06021-6. [DOI] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. The Journal of cell biology. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Otwinowski Z. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. The Journal of cell biology. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair UB, Joel PB, Wan Q, Lowey S, Rould MA, Trybus KM. Crystal structures of monomeric actin bound to cytochalasin D. J Mol Biol. 2008;384:848–864. doi: 10.1016/j.jmb.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, Dominguez R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nature structural & molecular biology. 2011;18:1060–1067. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: design of a heterodimeric coiled coil. Current biology: CB. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. The Journal of cell biology. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annual review of biochemistry. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell motility and the cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annual review of biochemistry. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends in cell biology. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. The Journal of cell biology. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G, Namgoong S, Boczkowska M, Leavis PC, Navaza J, Dominguez R. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J Mol Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nature structural & molecular biology. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. The Journal of cell biology. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano K, Maeda K, Oki M, Maeda Y. Enhancement of protein expression in insect cells by a lobster tropomyosin cDNA leader sequence. FEBS Lett. 2002;532:143–146. doi: 10.1016/s0014-5793(02)03659-1. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophysical journal. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME, Heimsath EG, Gauvin TJ, Higgs HN, Kull FJ. FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation. Nature structural & molecular biology. 2013;20:111–118. doi: 10.1038/nsmb.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E463–471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. Structure and function of the interacting domains of Spire and Fmn-family formins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11884–11889. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Rouiller I, Slaughter BD, Egile C, Kim E, Unruh JR, Fan X, Pollard TD, Li R, Hanein D, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yeung PS, Boor KJ. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog Dis. 2004;1:74–88. doi: 10.1089/153531404323143594. [DOI] [PubMed] [Google Scholar]
- Yu B, Cheng HC, Brautigam CA, Tomchick DR, Rosen MK. Mechanism of actin filament nucleation by the bacterial effector VopL. Nature structural & molecular biology. 2011;18:1068–1074. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.