Abstract
Most head and neck squamous cell carcinomas (HNSCC) over-express ERBB1/EGFR, but EGFR- targeted therapies have yielded disappointing clinical results in treatment of this cancer. Here we describe a novel interaction between EGFR and the ligand EphrinB1 (EFNB1), and we show that EFNB1 phosphorylation and downstream signaling persists in the presence of cetuximab. Mechanistically, cetuximab drives a shift in EGFR dimerization partners within the signaling complex, suggesting that targeted drugs may trigger partner rearrangements that allow persistent pathway activation. EFNB1 attenuation slowed tumor growth and increased survival in a murine model of HNSCC, suggesting a substantial contribution of EFNB1 signaling to HNSCC development. Together, our findings suggest that EFNB1 is part of the EGFR signaling complex and may mediate drug resistance in HNSCC as well as other solid tumors.
Keywords: EphrinB1, cetuximab, MAP Kinase, head and neck cancer, ErbB
Introduction
A greater understanding of the molecular pathways driving tumor initiation and proliferation has heralded an age of targeted therapies for cancer. The therapeutic successes of these drugs are satisfying from both the scientific and medical perspectives. Their failures indicate an incomplete understanding of macromolecular complexes, the oncogenic pathways they drive, their crosstalk and drug-driven compensatory mechanisms. We have recently identified a novel component of one such macromolecular complex in breast cancer (1). Here, we further characterize its role in the context of head and neck squamous cell carcinoma (HNSCC).
In human papillomavirus (HPV) associated HNSCC, the high risk HPV type 16 (HPV16) E6 oncoprotein targets the cellular phosphatase, PTPN13, for degradation (2, 3). PTPN13 functions as a tumor suppressor (4–9) yet, the molecular mechanisms it modulates and how they become altered in cancer remain unclear. One important PTPN13 phosphatase substrate that may influence cancer initiation and/or progression is the signaling ligand EphrinB1 (EFNB1). EFNB1 belongs to a ligand family that binds and activates Eph receptor tyrosine kinases. Unlike most ligands, Ephrin ligands initiate their own downstream signaling following receptor engagement, a process called “reverse signaling” (10). PTPN13 transiently interacts with phosphorylated EFNB1, shutting “reverse signaling” off (11). Thus, in tumors with impaired PTPN13 expression or function, EFNB1 signaling may persist. In addition, Ephrin ligands are promiscuous in their associations (12, 13); for example, EFNB1 interacts with ERBB2 (14). Moreover, EFNB1 activation correlates with phosphorylation of ERK1/2 (14). Together, these studies suggest that the complex consisting of ERBB2/EFNB1 together with PTPN13 regulates intracellular signals critical for epithelial tumorigenesis.
ERBB2 belongs to the ERBB family of receptor tyrosine kinases which includes ERBB1/EGFR/HER1. Greater than 90% of HNSCC cases demonstrate up-regulation of ERBB1 (15, 16) which may impact tumor growth and patient outcomes. ERBB1 targeted therapies like cetuximab (Erbitux®), a chimeric anti-ERBB1 monoclonal antibody, and erlotinib (Tarceva®), an ERBB1 kinase inhibitor, are utilized as therapies in HNSCC patients although with modest clinical outcomes (17). These clinical findings emphasize the need to better define components of macromolecular complexes active in disease, understand the molecular pathways they drive, and characterize how they influence tumor initiation and progression. In the case of HNSCC, these types of studies may help define what role, if any, ERBB1 up-regulation plays in disease initiation and/or progression.
Our previous finding that EFNB1 associates with ERBB2 prompted us to ask whether it associates with other ERBB family members. Here we report that, like ERBB2, ERBB1 associates with EFNB1. Moreover, in the absence of PTPN13 function, EFNB1 phosphorylation is enhanced and ERK1/2 signaling is potentiated. These data suggest that EFNB1 exists in a complex together with ERBB1, ERBB2 or both. A combination of these associations likely exists which together regulate complex intracellular signals potentiated in the absence of PTPN13 function. Importantly, we show that while antibody therapies like cetuximab (Erbitux®, anti-ERBB1) and trastuzumab (Herceptin®, anti-ERBB2) potently block receptor activation, they do not attenuate EFNB1 activation or ERK1/2 phosphorylation. Moreover, we demonstrate that these ERBB targeted drugs promote the shifting of partners within ERBB/EFNB1 complexes and suggest that this mechanism supports persistent EFNB1 signaling despite potent ERBB receptor blockade. Thus, we propose that EFNB1 activation mediates signal transduction and drug resistance. In addition, we demonstrate that knock-down of EFNB1 significantly slows tumor growth and improves survival in a murine model of HNSCC. Together, our findings support a tumor suppressive role for PTPN13, suggest that EFNB1 may be a useful therapeutic target in HNSCC and other solid tumors and explain at least one mechanism by which HNSCCs fail cetuximab therapy.
Materials and Methods
Cell culture
HEK293, SCC1 and SCC47 cells were maintained with Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum and 1% penicillin/streptomycin. Primary mouse (MTE) and human tonsil epithelia (HTE) were maintained with KSFM medium while HEE and MEERL cells were maintained with E-medium (DMEM/Hams F12, 10% fetal calf serum, 1% penicillin/streptomycin, 0.5 µg/ml hydrocortisone, 8.4 ng/ml cholera toxin, 5 µg/ml transferrin, 5 µg/ml insulin, 1.36 ng/ml tri-iodo-thyonine, and 5 ng/ml EGF). HEE cells were generated from 1° HTE cells harvested from tonsillectomies performed for non-cancerous reasons and collected under institutional IRB approval with written consent at the University of Iowa (3). The stable HEE cells expressing HPV16 E6 and E7 were generated as previously described (3, 18). We have authenticated these cell lines by short tandem repeat (STR) DNA profiling and verified them with the reference STR profile. Authentication was performed in the summer of 2012 at Genetica DNA Laboratories (Cincinnati, OH, http://www.celllineauthentication.com/).
Plasmids
EFNB1 mutant constructs and PTPN13C/S has been previously described (6, 14). Wildtype human ERBB1 cDNA was obtained from Addgene (#11011, Cambridge, MA) and cloned by PCR into pCMV-HA (Clontech). ErbB1 TM mutants were generated from the HA-tagged wildtype ERBB1 construct using the Quick Change II XL Site-Directed Mutagenesis Kit (Agilent Technologies).
Transfection
HEK293 cells were transfected using Polyfect Transfection Reagent as per manufacturer’s instructions (Qiagen).
Immunoprecipitation and western blot
Cells were lysed in lysis buffer (50mM Tris HCl pH 7.5, 150mM NaCl, 5mM EDTA, 2mN Na3VO4, 100mM NaF, 10mM NaPPi, 10% glycerol, 1% Triton X-100) and soluble proteins assayed by BCA protein assay (Pierce). Equal total protein was used for IP and IB analysis by standard methodology. Antibodies for IP were as follows: Becton Dickinson anti-ERBB1, Dako anti-ERBB2, Santa Cruz anti-EFNB1; antibodies used for IB: Upstate anti-ERBB1, Invitrogen anti-ERBB2, Upstate anti-phospho-tyrosine, Ambion anti-GAPDH, Santa Cruz anti-EFNB1, Sigma anti-EFNB1, Cell Signaling anti-phospho-ERK1/2, Calbiochem anti-ERK1/2, Santa Cruz anti-phospho-Tyrosine317 EFNB1, Sigma anti-HA, Sigma anti-FLAG.
Immunofluorescence (IF) and Proximity Ligation Assay (PLA)
Antibodies for IF were: Becton Dickinson anti-ERBB1, Dako anti-ERBB2, Sigma anti-HA, Santa Cruz anti-EFNB1, Sigma anti-EFNB1. Surface EphrinB ligands were bound by EphB1-Fc (R&D Systems) on unfixed, unpermeabilized cells and detected with Millipore anti-human IgG-FITC. Antibodies used for PLA were: Becton Dickinson anti-ERBB1, Dako anti-ERBB2, Invitrogen anti-ERBB2, Santa Cruz anti-EFNB1. Standard IF and PLA protocols were followed.
Quantification of PLA
PLA positive signals (visualized as fluorescent red dots) were analyzed by confocal microscopy (Olympus FluoView1000, 60× oil objective, 2.5× magnified, Alexa 568 detector). At least 3 stacked Z series for each condition were analyzed, confocal IOB files converted to tiff format then analyzed by DuoLink Imagetool software (Olink Biosciences). The number of PLA positive signals from at least three different Z-stacked images per condition was averaged. Experiments were repeated at least 3 times with similar results. Students t-test was used to analyze the data and test for significance.
Targeted therapy treatment
Cells were grown on 60 mm tissue culture dishes to 80% confluence and treated with recombinant EGF (SAFC Biosciences) in the presence or absence of cetuximab (20 µg/ml, Imclone), trastuzumab (10 µg/ml, DAKO), or their combination for 4 hours at 37°C. Control cells received either no treatment or EGF alone. Cells were harvested as described above and analyzed by IP and western blot. For analysis by PLA, cells were seeded on 16 well chamber slides and treated as described followed by processing for PLA (as described above).
Generating stable cell lines
Cells were seeded onto 6 well tissue culture dishes and transfected using Polyfect transfection reagent as described above. Twenty-four hours post-transfection, cells were placed under antibiotic selection with Zeocin (250, 500 or 1000 µg/ml, Invitrogen), clones were picked and transferred to 48 well dishes, maintained under selection and passaged up to 100 mm dishes at which time one dish was lysed for biochemical analysis and another frozen for later use.
In vivo studies
All animal studies were reviewed and approved by the IACUC. Cells were trypsinized and harvested from tissue culture plates and resuspended. An 18-gauge needle with 100,000 cells was injected subcutaneously into the right hindlimb of each C57Bl/6 mouse (N=5 per group). C57Bl/6 male mice were 4– 6 weeks of age, weighed at least 20–25 gm and were purchased from Jackson Laboratories. Injected cells included: parental cells, non-silencing, wt mEfnB1, or sh EfnB1. Weekly caliper measurements were taken and tumor volume calculated as follows: (width)2(depth). Mice were humanely euthanized when tumors reached 2cm in the greatest dimension or the animal became emaciated or had functional impairment of the leg.
Results
EFNB1 associates with ERBB1 and ERBB2
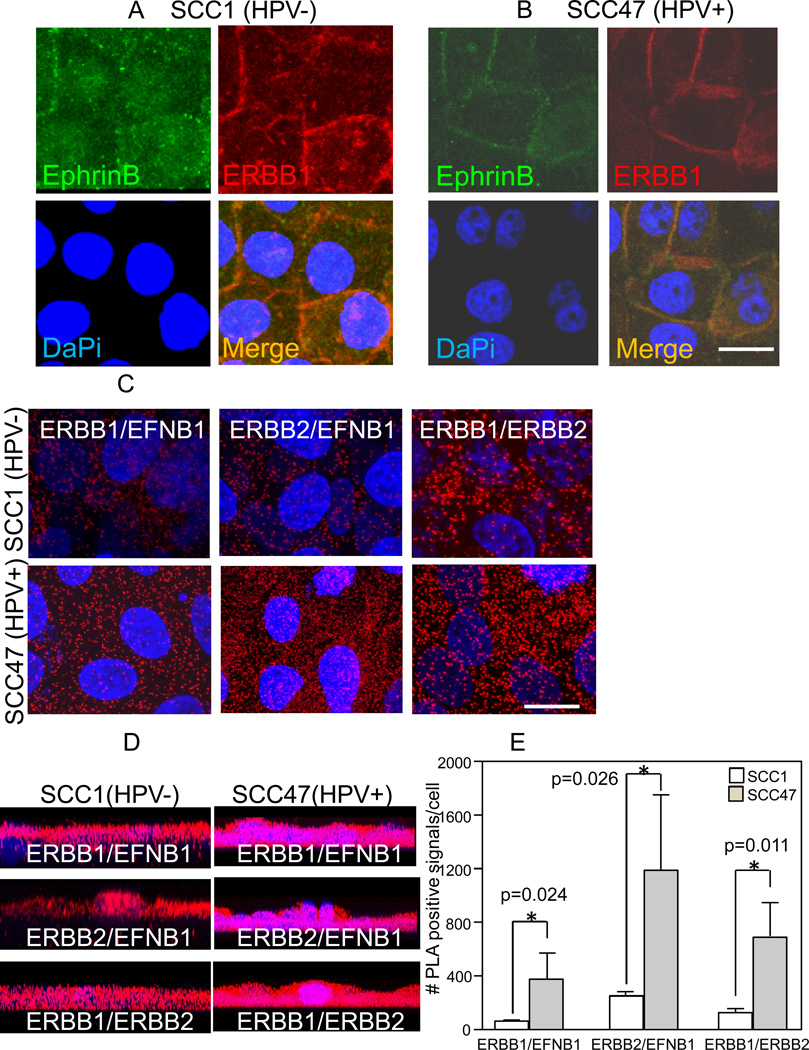
To test whether ERBB1 and EFNB1 interact, two HNSCC cell lines were processed for immunofluorescence (IF); the SCC1 cell line is HPV negative (HPV−), while the SCC47 cell line is HPV positive (HPV+). Surface, rather than total, EphrinB was localized because cells robustly express intracellular EphrinB ligands making the distinction of surface expression difficult. EphB1-Fc binding followed by IgG-FITC localizes surface EphrinB ligands. Both cell lines express ERBB1 and EphrinB which co-localize at cell-cell borders (Figs. 1A,B).
Figure 1.
ERBB1 and EFNB1 interact. A and B, En face confocal images of SCC1 (A) and SCC47. B, cells stained for surface EphrinB ligands (green) and total ERBB1 (red). Merged images (yellow). DaPi (blue) nuclear counterstain. Scale bar 10µm. C, En face stacked confocal images of SCC1 and SCC47 cells processed for proximity ligation assay (PLA) for ERBB1/EFNB1, ERBB2/EFNB1 and ERBB1/ERBB2. Positive PLA signals (red) and DaPi (blue) nuclear counterstain. Scale bar 10µm. D, Stacked confocal images in the XZ plane taken from Z-series collected in panel C. E, Quantification of PLA positive signals in panel C.
To verify that the co-localization by IF involved ERBB1 and EFNB1 specifically, proximity ligation assay (PLA) and co-immune precipitation studies were performed. Figure 1C demonstrates positive PLA signals for EFNB1 and ERBB1 in SCC1 (HPV−) and SCC47 (HPV+) cells. The previously discovered ERBB2/EFNB1 association (14) was confirmed and the presence of ERBB1/ERBB2 heterodimers was determined (Fig. 1C). In addition, stained Z-stacked images in the XZ plane indicate that these interactions are not restricted to the cell surface but also occur within the cytoplasm (Fig. 1D). These intracellular PLA signals likely represent internalized complexes. Importantly, PLA quantification demonstrates that infection with HPV16 correlates with enhanced ERBB/EFNB1 interactions (Fig. 1E). EFNB1, ERBB1 and ERBB2 protein levels in SCC1 (HPV−) and SCC47 (HPV+) cells are very similar (Fig. 3), thus increased protein expression cannot account for the enhanced interactions evident in SCC47 (HPV+) cells.
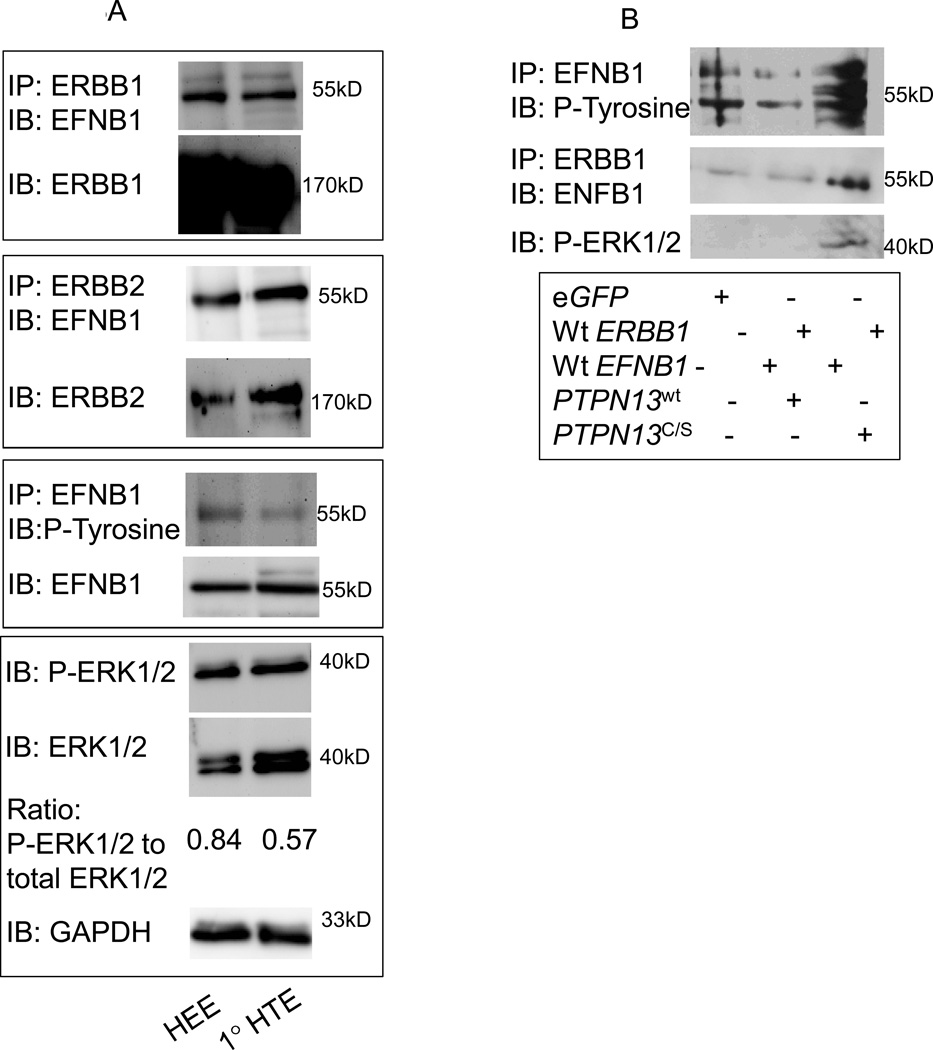
Figure 3.
EFNB1 associates with ERBB1 and ERBB2 and mediates ERK1/2 signaling in human tonsil epithelial cells.
A. Primary human tonsil epithelia (1°HTE), HTEs stably expressing HPV16 E6 and E7 (HEE), were analyzed by western blot as indicated. B, HEK293 cells were transfected as indicated and lysates analyzed by western blot as indicated.
EFNB1 and ERBB1 association is likely mediated via their transmembrane domains
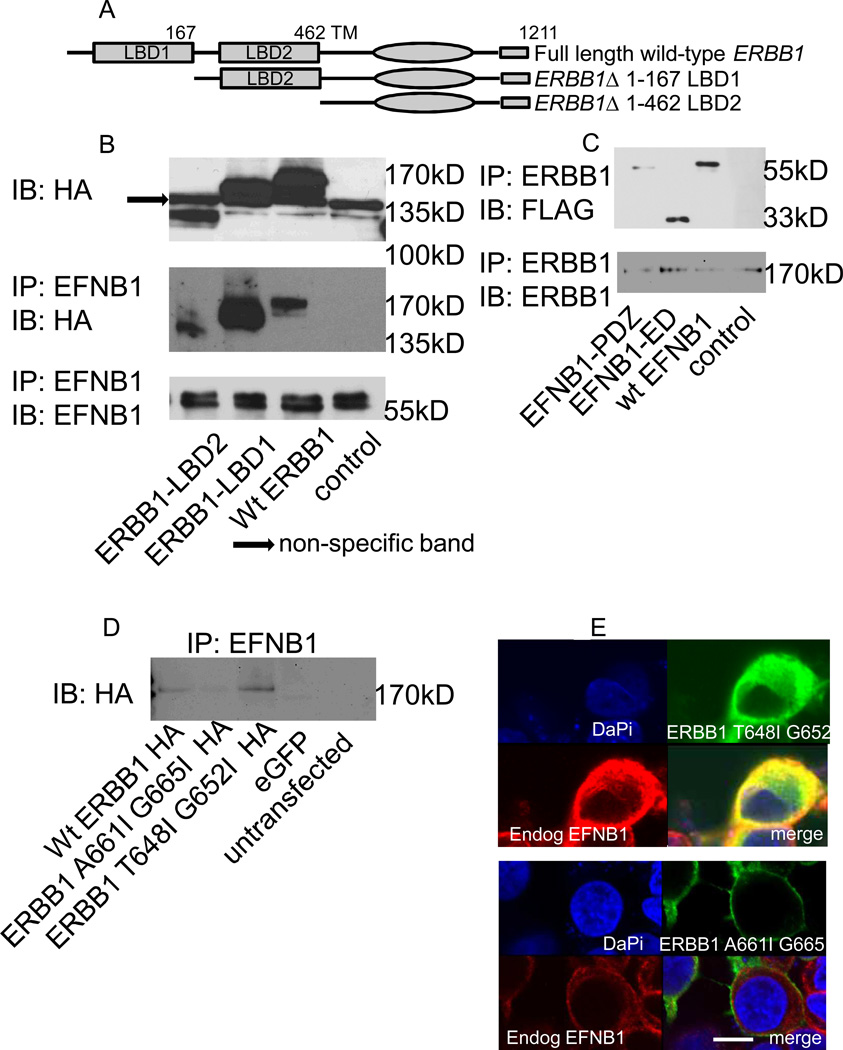
Due to the overwhelming expression of ERBB1 in HNSCCs, we focused further analysis on ERBB1 and EFNB1. To define the domains necessary and sufficient for ERBB1/EFNB1 association, we generated deletion mutants (Fig. 2A). All ERBB1 mutants, including the full-length wildtype, were HA-tagged; constructs express well, run at the predicted molecular weight and possess the HA tag (Fig. 2B). To test whether any of the ERBB1 deletions compromise its interaction with EFNB1, HEK293 cells were transfected, lysates immunoprecipitated for EFNB1 and immunoblotted for HA. Surprisingly, no loss of co-IP with any ERBB1 mutants occurred suggesting that the extracellular ligand binding domains of ERBB1 are not required for its association with EFNB1 (Fig. 2B). These data are consistent with our previous finding for the ERBB2/EFNB1 interaction (14). EFNB1 deletion mutants and wildtype constructs have been previously described and were all FLAG tagged (14). Briefly, either the entire extracellular domain (EFNB1-ED) or just the intracellular PDZ binding motif (EFNB1-PDZ) of EFNB1 was deleted. HEK293 cells were transfected with the EFNB1 constructs. Because HEK293 cells express nearly undetectable endogenous ERBB1, cells were also transfected with untagged wildtype ERBB1. Immunoprecipitation for ERBB1 followed by western blot for FLAG shows no loss of co-IP with any of the EFNB1 mutants suggesting that neither the extracellular domain nor the PDZ motif is required for its association with ERBB1.
Figure 2.
ERBB1’s transmembrane domain mediates its association with EFNB1. A, schematic diagram of full length and mutant ERBB1 constructs. LBD, ligand binding domain; TM, transmembrane. B, HEK293 cells were transfected with ERBB1 wildtype and mutant constructs and lysates analyzed by western blot as indicated. Untransfected cells served as control. C, HEK293 cells were transfected with EFNB1 wildtype (wt EFNB1) and mutant constructs (EFNB1-ED, EFNB1-PDZ) and lysates analyzed by western blot as indicated. Untransfected celles served as control. ED, extracellular domain, PDZ, PDZ domain. D, HEK293 cells were transfected with ERBB1 transmembrane domain mutants and analyzed by western blot as indicated. Cells transfected with an enhanced green fluorescent protein (eGFP) construct as well as untransfected cells served as controls. E, En face confocal images of cells transfected as in panel D processed for immunofluorescence localizing ERBB1 constructs (HA, green) and endogenous EFNB1 (red). Merged panel (yellow); DaPi (blue) nuclear counterstain. Scale bars, 10 µm.
ERBB receptor transmembrane (TM) domains mediate homo- and hetero-dimerization (19, 20). In fact, ERBB TM domains contain two motifs, one mediating heterodimerization, the other homodimerization (21). To test whether ERBB1’s TM domain mediates its interaction with EFNB1, two additional ERBB1 mutants were generated: one within the first TM motif and another within the second TM motif. These constructs were HA tagged and transfected into HEK293 cells. Control cells were transfected with wildtype HA tagged ERBB1 or eGFP. Lysates were immune-precipitated for EFNB1 and analyzed by western blot for HA. While the first ERBB1 TM motif mutant, T648I G652I, retains co-IP with EFNB1, mutation within the second TM motif (A661I G665I) loses it (Fig. 2D). These data were verified by IF. HEK293 cells were transfected as described and cells processed for IF using an anti-HA antibody for ERBB1 detection and an anti-EFNB1 antibody for EFNB1 detection. Expression of the ERBB1 T648I G652I mutant (green) co-localizes with endogenous EFNB1 (red), evident in the merged panel (yellow). Expression of the ERBB1 A661I G665I (green) does not colocalize with EFNB1 (red) (Fig. 2E). Together, these data suggest that ERBB1’s second TM motif mediates the EFNB1 association.
Expression of HPV oncogenes correlates with EFNB1 and ERK1/2 phosphorylation in HNSCC. We and others have demonstrated that PTPN13 regulates EFNB1 phosphorylation. Moreover, the loss of PTPN13 expression, as seen in basal-like breast cancer, correlates with enhanced EFNB1 and ERK1/2 phosphorylation (14, 22). Importantly, breast tumors compromised for PTPN13 expression are associated with decreased overall survival (23). A similar dysregulation in signaling occurs in HNSCC, both in a human cell line (SCC84) and a murine model of head and neck cancer (14). Thus, we wondered whether EFNB1's phosphorylation state modulates its interactions with ERBB receptors. In addition, given that tonsil epithelial cells are the major site for HPV+ HNSCC, we tested whether ERBB/EFNB1 interactions exist in these cells. Thus, primary human tonsil epithelial cells (1°HTE) and HTEs stably expressing HPV16 E6 and E7 (HEE) were tested. Consistent with our previous studies, ERBB1 and ERBB2 associate with EFNB1 in both cell types (Fig. 3A). In addition, expression of E6 correlates with enhanced phosphorylation of EFNB1 and ERK1/2 consistent with previous reports (2, 3). These data show that ERBB1 and ERBB2 associate with EFNB1 in primary human tonsil cells as well as those expressing HPV oncogenes. Moreover EFNB1 associates with its ERBB partners regardless of its phosphorylation status. Importantly, expression of HPV16 oncogenes (HEE cells) enhances EFNB1 activation and subsequent ERK1/2 phosphorylation. These data are consistent with our previous findings that EFNB1 signals down the MAP Kinase pathway (14). Similar results were evident in primary murine tonsil epithelial cells (1°MTE) and a previously characterized mouse model of HPV related HNSCC (MTEs expressing HPV 16 E6, E7, Ras and luciferase or MEERL cells) (Supplemental Fig. 1) (18, 24, 25).
PTPN13 regulates EFNB1 phosphorylation and reverse signaling
HPV16 E6 mediates the degradation of PTPN13 (2, 3). The data suggest that enhanced EFNB1 activation and downstream signaling are a consequence of this degradation (Fig. 3A). To directly test this, we performed a series of transfections in HEK293 cells which express nearly undetectable ERBB receptors allowing us to control expression of the proteins involved. Briefly, HEK293 cells were transfected with wildtype ERBB1, wildtype EFNB1 and PTPN13 (either wildtype or a phosphatase null, PTPN13C/S) and lysates analyzed by western blot. In eGFP transfected controls, endogenous EFNB1 is phosphorylated (Fig. 3B). The multiple bands likely represent different phosphorylated forms of EFNB1 as suggested by Xu et al (26). In addition, a low level of EFNB1 co-IPs with ERBB1 yet ERK1/2 is not phosphorylated. Transfection with wildtype ERBB1, wildtype EFNB1 and wildtype PTPN13 decreases EFNB1 phosphorylation without altering its association with ERBB1; ERK1/2 remains unphosphorylated. Interestingly, transfection with wildtype ERBB1, wildtype EFNB1 and PTPN13C/S, a phosphatase null PTPN13 mutant, increases EFNB1 phosphorylation as well as its association with ERBB1 and initiates ERK1/2 phosphorylation. Together, these data are consistent with those of others demonstrating that PTPN13 regulates EFNB1 phosphorylation. In addition, the data suggest that phosphorylated EFNB1 associates more readily with ERBB1 and that the loss of PTPN13 function potentiates not only EFNB1 phosphorylation but also ERK1/2 signaling.
ERBB1 and ERBB2 antibody blockade do not attenuate downstream signaling
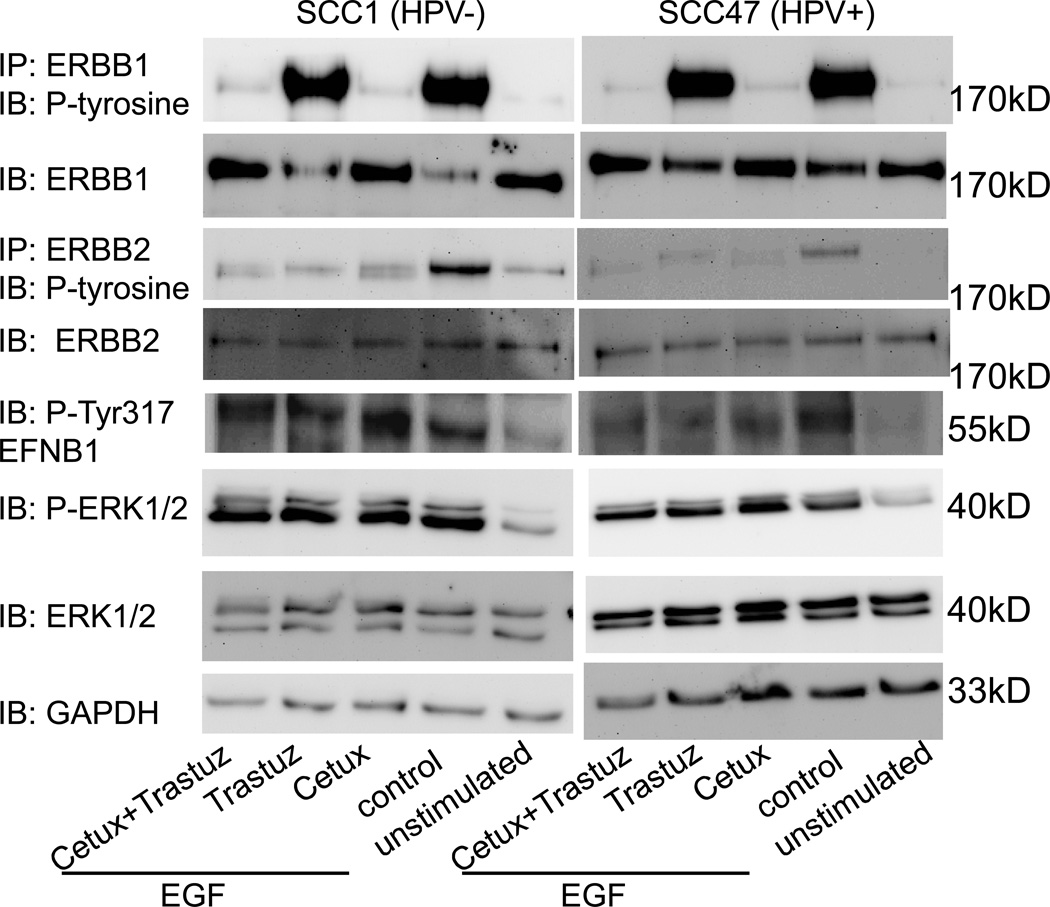
ERBB1 and ERBB2 targeted therapies are in clinical use for treatment of a variety of solid cancers including HNSCC. Our data suggest that PTPN13 modulates EFNB1-mediated signaling and that EFNB1 associates with ERBB1 and ERBB2. To test whether ERBB1 and/or ERBB2 targeted therapies alter EFNB1 activation and downstream signaling, we analyzed SCC1 (HPV−) and SCC47 (HPV+) cells. Cells were treated with EGF to stimulate ERBB receptor activation and then segregated into the following groups: 1) untreated (EGF alone, control), 2) treated with cetuximab, 3) treated with trastuzumab or 4) treated with both cetuximab and trastuzumab. Unstimulated (no EGF) and untreated cells (EGF alone) served as control. Figure 4 demonstrates western blot analysis of these groups. Both SCC1 (HPV−) and SCC47 (HPV+) cells show nearly undetectable endogenous ERBB1 or ERBB2 phosphorylation (unstimulated). In addition, and consistent with clinical data, cells abundantly express ERBB1 and modestly express ERBB2 (15, 16,27–31). EGF stimulation activates ERBB1 and ERBB2 suggesting that at least a fraction of ERBB1 heterodimerizes with ERBB2. Treatment with cetuximab alone potently attenuates EGF-mediated ERBB1 and ERBB2 phosphorylation suggesting that ERBB2 phosphorylation following EGF stimulation occurs via heterodimerization with ERBB1. As expected, trastuzumab has no effect on ERBB1 phosphorylation but attenuates ERBB2 activation. Treatment with a combination of cetuximab and trastuzumab is not additive. In other words, treatment with drugs in combination does not enhance the effect of each drug when given in isolation. Importantly, despite the ability of cetuximab and trastuzumab to attenuate receptor activation, neither demonstrated an effect on ERK phosphorylation; that is, ERK1/2 remains phosphorylated despite the presence of drug. These data suggest that downstream signaling persists despite ERBB1 and/or ERBB2 blockade. This finding is consistent with previous reports (32). Importantly, while EGF stimulation enhances phosphorylation of EFNB1 on tyrosine 317, neither cetuximab nor trastuzumab (alone or in combination) attenuate this. Based on our previous finding that EFNB1 signals via the MAP Kinase pathway (14), these data suggest that persistent signaling via EFNB1 drives ERK1/2 phosphorylation despite blockade of ERBB1 and ERBB2.
Figure 4.
EFNB1 and ERK1/2 phosphorylation persist in presence of cetuximab and trastuzumab therapies. SCC1 and SCC47 cells were treated with EGF alone (control) or in combination with cetuximab (cetux), trastuzumab (traztuz) or both (cetux+trastuz) and analyzed by western blot as indicated. Cells alone (unstimulated) served as an additional control.
Treatment with cetuximab or trastuzumab (together or in combination) drives the shifting of partners within ERBB/EFNB1 complexes
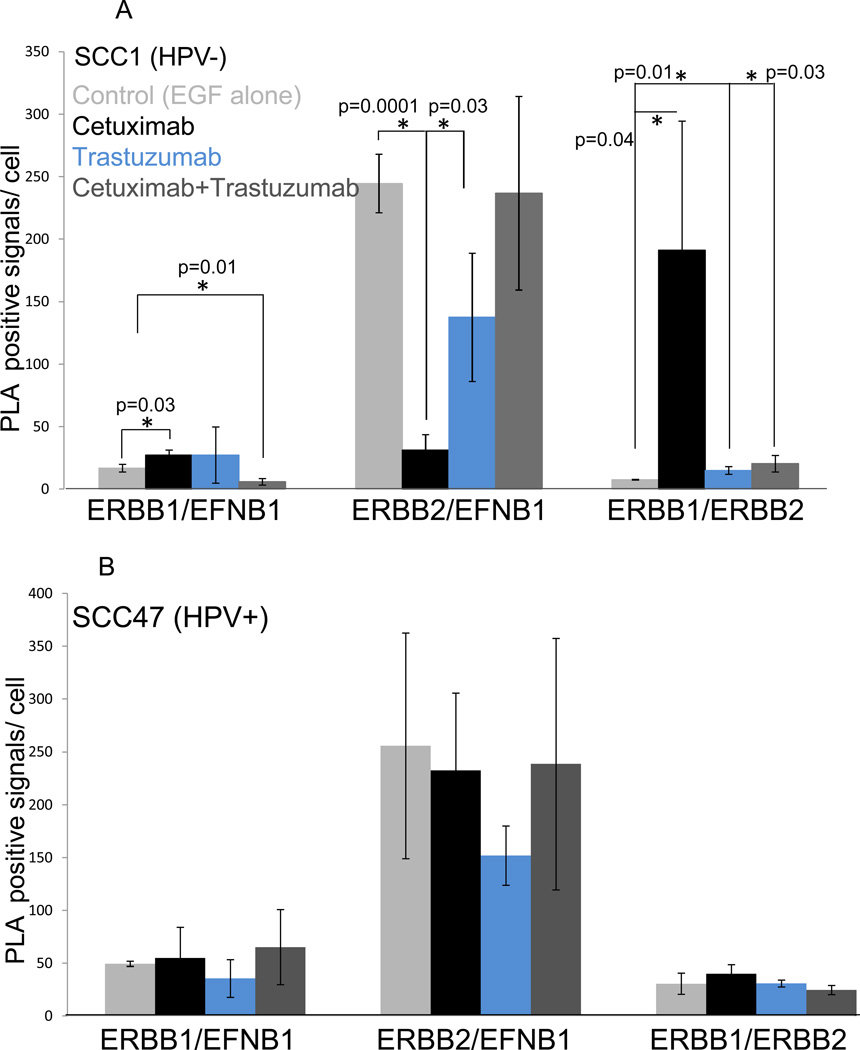
Given that EFNB1 associates with ERBB1 and ERBB2, we wondered whether treatment with targeted drugs mediates changes in partner associations within these complexes that might account for sustained signaling in the presence of drug. To test this, the experiments performed in Figure 4 were duplicated on 16 well chamber slides, cells were processed for PLA, and quantified. In SCC1 (HPV−) cells, stimulation with EGF alone (control, light gray) results in modest association of ERBB1 with EFNB1 (Fig. 5A). ERBB2 interactions with EFNB1 are more abundant in comparison suggesting that the preferred partner for EFNB1 is ERBB2. In addition, ERBB1 and ERBB2 heterodimers are in the minority. Treatment of cells with EGF+ cetuximab (black bars) leads to a significant increase in ERBB1/EFNB1 interactions (p=0.03), a significant decrease in ERBB2/EFNB1 interactions (p=0.0001) while ERBB1/ERBB2 heterodimerization is significantly enhanced (p=0.04). On the other hand, treatment with EGF+trastuzumab (blue bars) has no effect on the number of interactions formed between ERBB1 and EFNB1; however, EFNB1’s association with ERBB2 decreases (p=0.03) while ERBB1/ERBB2 heterodimers significantly increase (p=0.01). Lastly, treatment with a combination of EGF, cetuximab and trastuzumab (dark gray bars) also results in partner shifts. ERBB1/EFNB1 associations decrease (p=0.01) while ERBB1/ERBB2 heterodimerization increases (p=0.03). Interactions between ERBB2 and EFNB1 remain unchanged. In SCC47 (HPV+) cells (Fig. 5B), treatment with targeted drugs demonstrated similar trends as those evident with SCC1 (HPV−) cells though significance was not reached in any condition. Nonetheless, these PLA data together with the biochemical analysis of these experiments (Fig. 4) suggest that partner shifting occurs in the presence of targeted therapies and may explain the sustained activation of EFNB1 and ERK1/2 evident in the presence of drug.
Figure 5.
Cetuximab and Trastuzumab treatment results in partner shifting. A, SCC1 cells were treated with EGF alone (control, light gray) or in combination with cetuximab (black), trastuzumab (blue) or both (cetuximab+trastuzumab, dark gray) and processed for PLA. Cells alone (unstimulated) served as an additional control. PLA positive signals were quantified. B, SCC47 cells were treated and analyzed as described in A.
Loss of EFNB1 decreases tumor growth in an in vivo model of HNSCC
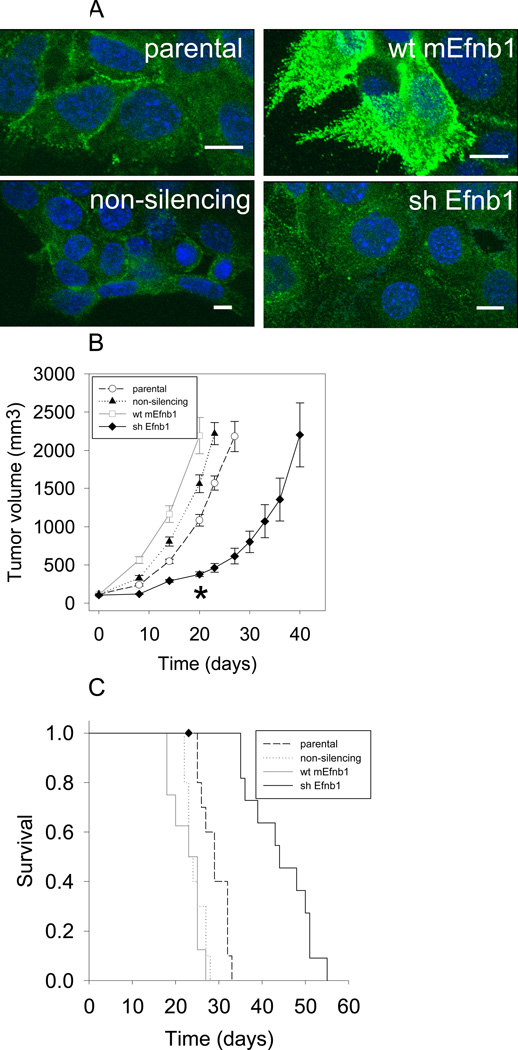
The data suggest that EFNB1 associates with ERBB1 and ERBB2 (alone or in combination) and that PTPN13 modulates these interactions by regulating EFNB1 phosphorylation and downstream signaling. While ERBB1 and ERBB2 roles in tumor growth have been appreciated for some times, we wanted to directly test EFNB1's function in tumor growth using a previously described murine model of HPV+ HNSCC (18, 24, 25). In these cells, HPV16E6-mediated degradation of PTPBL (the murine homolog of PTPN13) results in enhanced EFNB1 phosphorylation and downstream ERK1/2 signaling (14). Briefly, mouse tonsil epithelial cells stably expressing HPV16 E6, E7 together with RAS and luciferase (MEERL cells) were transfected with the pcDNA3.1 Zeocin expression vector containing either wildtype murine Efnb1 (wt mEnfb1) or an shRNA targeting Efnb1 (shEfnb1). Transfection with a non-silencing shRNA (non-silencing) as well as the parental cells served as controls. Cells were ring-cloned and clones tested biochemically. Over 50 clones were analyzed by western blot with only modest over-expression or knock-down of EFNB1 evident. However, IF localization of surface expression shows abundant increase in wt mEfnb1 clones and decrease in shEfnb1 clones (Fig. 6A). These findings are consistent with our previous observations that abundant intracellular EFNB1 expression precludes western blot detection of surface expression changes. Thus, selection of clones was based predominantly on IF surface EFNB1 localization. To test the role of EFNB1 in tumor growth in vivo, 100,000 cells of each clone were injected into the hindlimb of C57Bl/6 mice, N=5 mice per group. Tumor growth was measured weekly. Wt mEfnb1 tumors grew significantly faster than parental and shEfnb1 tumors (P<0.001). However, tumor growth of the wt mEfnb1 group was not significantly different than that of the non-silencing control group (p=0.42). Given that the wt mEfnb1 group was significantly different than the parental group, we speculate that the non-silencing construct mediates off-targets effects that account for this difference in tumor growth with the wt mEfnb1 group. Importantly, mice bearing shEfnb1 tumors had significantly slower tumor growth compared to all other groups (p<0.001) at all time points up to 20 days, at which time mice from the other groups reached sacrifice criteria (Fig. 6B). Consistent with tumor growth, mice bearing parental tumors survived significantly longer than those bearing wt mEfnb1 or non-silencing tumors (p<0.001) while shEfnb1 bearing mice survived significantly longer (p<0.001) than the other groups (Fig. 6C). These data demonstrate that EFNB1 significantly affects tumor growth and survival suggesting it is a good candidate for therapeutic targeting in HNSCC and other solid tumors.
Figure. 6.
Knock-down of Efnb1 slows tumor growth and prolongs survival in vivo. A, Mouse tonsil epithelial cells stably expressing HPV16 E6, E7, Ras and luciferase (MEERL, parental), over-expressing Efnb1 (wt mEfnb1), shRNA knocking down EphrinB1 (shEfnb1), or a non-silencing shRNA control (non-silencing) were analyzed by immunofluorescence for surface EFNB1 (green); DaPi (blue) nuclear counterstain. Scale bars, 10µm. B, Tumor volume as a function of time for C57Bl/6 mice injected with stable clones described in panels A. C, Kaplan-Meier survival plot of mice in panel B. ♦ indicates one mouse that was censored from the data. This mouse had an open sore which met the criterion for euthanasia prior to hitting maximal allowed tumor volume.
Discussion
We describe a novel association in HNSCC consisting of ERBB1 and EFNB1. ERBB2 is also a part of this complex, consistent with previous findings (14). Loss of PTPN13 function increases EFNB1 phosphorylation, enhances EFNB1’s interaction with ERBB1 and correlates with potentiated ERK1/2 activation. Our data do not test whether ERBB1directly phosphorylates EFNB1 or whether other components exist within this complex that mediate EFNB1 phosphorylation. However, previous studies suggest that SRC kinase phosphorylates EFNB1 following cognate receptor engagement (10) and our previous studies support this role for SRC (14). Whether SRC is recruited to EFNB1 when complexed to ERBB1 remains to be tested. Regardless of the mechanism, increased EFNB1 phosphorylation correlates with ERK1/2 phosphorylation.
Importantly, we show that ERBB1’s TM domain mediates the association with EFNB1. These data predict that ERBB1 targeted therapies (which bind extracellular, cetuximab, or intracellular, erlotinib, epitopes) alone will be inefficient in blocking intracellular signaling mediated by this complex as EFNB1 is likely impervious to them. Cetuximab’s binding site on ERBB1 has been solved (33). The extracellular domain of ERBB1 is subdivided into four subdomains (I-IV) (34). EGF binds epitopes on domains I and III while cetuximab binds only to domain III yet overlaps with the domain III epitope bound by EGF. Thus, cetuximab binding is sufficient to sterically inhibit EGF binding. ERBB1’s extracellular domains do not mediate its interaction with EFNB1 strongly suggesting that cetuximab binding will not inhibit it. Trastuzumab binds an epitope within the juxtamembrane region of extracellular domain IV of ERBB2 (35). Our previous findings demonstrate that ERBB2’s extracellular domains do not mediate its association with EFNB1 (14) and therefore, trastuzumab binding likely will not affect this interaction.
In cancers with decreased or lost PTPN13 function, ERBB/EFNB1 associations and signaling may be potentiated. We tested these possibilities and found that indeed, while treatment with cetuximab potently blocked ERBB1 activation, EFNB1 phosphorylation persisted as did that of ERK1/2. Trastuzumab treatment yielded similar results and the combination of these targeted therapies demonstrated no ill effect on EFNB1 phosphorylation or downstream signaling. The findings were strikingly similar in both HPV+ and – cells suggesting that HPV status does not alter drug efficacy. However, the PLA data demonstrate that HPV infection promotes ERBB/EFNB1 interactions (Fig. 1E) while analysis of these interactions in the context of drug treatment, demonstrated no significant differences in the HPV+ cells (Fig. 5B). We speculate this is a reflection of the increased number of associations due to HPV infection in these cells and suggest that a maximal threshold of total ERBB/EFNB1 interactions per cell may exist such that once this threshold is reached, conditions that would otherwise drive significant changes become difficult to discern. In addition, we noted a higher variability in the number of interactions for some conditions which also affected attainment of statistical significance. Taken together, however, these data explain, at least in part, the modest clinical benefits evident with cetuximab in the HNSCC setting. In addition, the data suggest that dual targeting of ERBB1 and ERBB2 likely will not yield better results than either drug alone. This has in fact been demonstrated for HNSCC cell lines (36). Moreover, a randomized phase II study using lapatinib, a dual ERBB1 and ERBB2 inhibitor, demonstrated no significant difference in objective response rate between lapatinib and placebo (37). These findings are not unique to HNSCC but appear to be the rule rather than the exception in a majority of solid tumors. For example, in a subset of molecularly classified breast tumors that over-expressed ERBB2, trastuzumab therapy proved beneficial for only a minority of patients while most demonstrated de novo or acquired trastuzumab resistance (38).
The finding that targeted drugs promote shifting of partners within ERBB/EFNB1 complexes emphasizes the complexity of macromolecular complexes, their roles in disease and suggests that targeted drugs may drive the progression of tumors. As with other solid tumors, it is increasingly evident that targeting a single element lacks long term clinical efficacy. These findings strongly demonstrate that silencing or eliminating a single oncogenic driver merely imposes evolutionary forces that promote alternative mechanisms to maintain, or even potentiate, the cancerous phenotype. This may be best exemplified by non-small cell lung cancer in which patients with classic ERBB1 mutations (L858R and exon 19 deletions) initially respond to ERBB1 targeted therapies yet eventually acquire a secondary mutation, T790M, which confers resistance (39). Multi-pronged approaches for effective treatment have been proposed for several cancers (40–43). Our current findings that targeted therapies drive shifting of partners within a multi-kinase/ligand/phosphatase complex strongly support these proposals. Along these lines, the use of pertuzumab in combination with trastuzumab demonstrates promise for HER2 breast cancer patients (44, 45). Pertuzumab binds an extracellular epitope within domain II of ERBB2 and blocks dimerization with other ERBB family members (46). Our data demonstrate that in SCC1 cells, ERBB1/2 heterodimers are in the minority and only become a major dimer group following cetuximab treatment (Fig. 5A). The current study did not evaluate ERBB3 or ERBB4, therefore it is unclear what role they may play in the multi-kinase/ligand/phosphatase complex described. However, given our finding that the TM domain of ERBB1 mediates the association with EFNB1, it seems unlikely that pertuzumab will abrogate ERBB/EFNB1 interactions. The role of ERBB TM domains in mediating dimer formation is well established (47–49). Our finding that the TM also is critical for EFNB1 associations strongly suggests that small molecules able to abolish TM interactions may effectively block dimer associations of multiple signaling molecules not limited to the ERBB family. Such drugs may have potent clinical efficacy. Our data also suggest that EFNB1 targeted therapy may be of clinical value. In all, the data suggest that blocking formation of the complex as opposed to blocking individual components within the complex may provide better tumor growth control and clinical outcomes.
In summary, the present findings are consistent with published data and support a tumor suppressive function for PTPN13 (2, 14). These studies confirm our previous findings that EFNB1 associates with members of the ERBB receptor tyrosine kinases. For those cancers with up-regulation of ERBB1 (like HNSCC), its association with EFNB1 may significantly affect tumorigenesis. Moreover, acquisition of PTPN13 loss-of-function mutations or its decreased expression (due to HPV infection or epigenetic silencing) may further enhance ERBB1/EFNB1-mediated signals. Our study suggests that EFNB1 is an important target for therapeutic intervention and that combinatorial targeted therapy approaches must include ERBB1, ERBB2, as well as EFNB1 and possibly SRC for quenching complex mediated oncogenic signals. While these studies focus on HNSCC, EFNB1 complexes with ERBB2 in breast cancer where loss of PTPN13 correlates with decreased overall survival (14) and thus, we speculate that a similar complex exists in breast (and possibly other solid tumors) suggesting that a multi-targeted drug approach may be beneficial in this setting as well.
Supplementary Material
Acknowledgements
We thank Catherine Christopherson for excellent administrative assistance and Nichole Haag for technical assistance.
Grant Support
This work was supported by R01 DE018386 NIH/NIDCR (J.H. Lee, B.G. Wieking, and D.W. Vermeer) and 8P20GM103548 Center of Biomedical Research Excellence NIH/Research Centers in Minority Institutions and Institutional Development Award (P.D. Vermeer, P.L. Colbert, and B.G. Wieking).
References
- 1.Sorani MD. Informatics technology mimics ecology: dense, mutualistic collaboration networks are associated with higher publication rates. PLoS One. 2012;7:e30463. doi: 10.1371/journal.pone.0030463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spanos WC, Hoover A, Harris GF, Wu S, Strand GL, Anderson ME, et al. The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with ras for invasive growth. J Virol. 2008;82:2493–2500. doi: 10.1128/JVI.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spanos WC, Geiger J, Anderson ME, Harris GF, Bossler AD, Smith RB, et al. Deletion of the PDZ motif of HPV16 E6 preventing immortalization and anchorage-independent growth in human tonsil epithelial cells. Head Neck. 2008;30:139–147. doi: 10.1002/hed.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 5.Yeh SH, Wu DC, Tsai CY, Kuo TJ, Yu WC, Chang YS, et al. Genetic characterization of fas-associated phosphatase-1 as a putative tumor suppressor gene on chromosome 4q21.3 in hepatocellular carcinoma. Clin Cancer Res. 2006;12:1097–1108. doi: 10.1158/1078-0432.CCR-05-1383. [DOI] [PubMed] [Google Scholar]
- 6.Hoover AC, Strand GL, Nowicki PN, Anderson ME, Vermeer PD, Klingelhutz AJ, et al. Impaired PTPN13 phosphatase activity in spontaneous or HPV-induced squamous cell carcinomas potentiates oncogene signaling through the MAP kinase pathway. Oncogene. 2009;28:3960–3970. doi: 10.1038/onc.2009.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu J, Huang YJ, Wang LE, Sturgis EM, Wei Q. Genetic polymorphisms in the PTPN13 gene and risk of squamous cell carcinoma of head and neck. Carcinogenesis. 2009;30:2053–2058. doi: 10.1093/carcin/bgp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glondu-Lassis M, Dromard M, Lacroix-Triki M, Nirde P, Puech C, Knani D, et al. PTPL1/PTPN13 regulates breast cancer cell aggressiveness through direct inactivation of Src kinase. Cancer Res. 2010;70:5116–5126. doi: 10.1158/0008-5472.CAN-09-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mita Y, Yasuda Y, Sakai A, Yamamoto H, Toyooka S, Gunduz M, et al. Missense polymorphisms of PTPRJ and PTPN13 genes affect susceptibility to a variety of human cancers. J Cancer Res Clin Oncol. 2010;136:249–259. doi: 10.1007/s00432-009-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdmann KS. The protein tyrosine phosphatase PTP-Basophil/Basophil-like. Interacting proteins and molecular functions. Eur J Biochem. 2003;270:4789–4798. doi: 10.1046/j.1432-1033.2003.03895.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee HS, Mood K, Battu G, Ji YJ, Singh A, Daar IO. Fibroblast growth factor receptor-induced phosphorylation of ephrinB1 modulates its interaction with Dishevelled. Mol Biol Cell. 2009;20:124–133. doi: 10.1091/mbc.E08-06-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka M, Kamata R, Sakai R. Phosphorylation of ephrin-B1 via the interaction with claudin following cell-cell contact formation. Embo J. 2005;24:3700–3711. doi: 10.1038/sj.emboj.7600831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeer PD, Bell M, Lee K, Vermeer DW, Wieking BG, Bilal E, et al. ErbB2, EphrinB1, Src kinase and PTPN13 signaling complex regulates MAP kinase signaling in human cancers. PLoS One. 2012;7:e30447. doi: 10.1371/journal.pone.0030447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 16.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–3584. [PubMed] [Google Scholar]
- 17.Lee J, Moon C. Current status of experimental therapeutics for head and neck cancer. Exp Biol Med (Maywood) 2011;236:375–389. doi: 10.1258/ebm.2010.010354. [DOI] [PubMed] [Google Scholar]
- 18.Hoover AC, Spanos WC, Harris GF, Anderson ME, Klingelhutz AJ, Lee JH. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch Otolaryngol Head Neck Surg. 2007;133:495–502. doi: 10.1001/archotol.133.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 20.Escher C, Cymer F, Schneider D. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J Mol Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J Biol Chem. 2004;279:21177–21182. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 22.Palmer A, Zimmer M, Erdmann KS, Eulenburg V, Porthin A, Heumann R, et al. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Mol Cell. 2002;9:725–737. doi: 10.1016/s1097-2765(02)00488-4. [DOI] [PubMed] [Google Scholar]
- 23.Revillion F, Puech C, Rabenoelina F, Chalbos D, Peyrat JP, Freiss G. Expression of the putative tumor suppressor gene PTPN13/PTPL1 is an independent prognostic marker for overall survival in breast cancer. Int J Cancer. 2009;124:638–643. doi: 10.1002/ijc.23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams R, Lee DW, Elzey BD, Anderson ME, Hostager BS, Lee JH. Preclinical models of HPV+ and HPV− HNSCC in mice: an immune clearance of HPV+ HNSCC. Head Neck. 2009;31:911–918. doi: 10.1002/hed.21040. [DOI] [PubMed] [Google Scholar]
- 25.Spanos WC, Nowicki P, Lee DW, Hoover A, Hostager B, Gupta A, et al. Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Arch Otolaryngol Head Neck Surg. 2009;135:1137–1146. doi: 10.1001/archoto.2009.159. [DOI] [PubMed] [Google Scholar]
- 26.Xu Z, Lai KO, Zhou HM, Lin SC, Ip NY. Ephrin-B1 reverse signaling activates JNK through a novel mechanism that is independent of tyrosine phosphorylation. J Biol Chem. 2003;278:24767–24775. doi: 10.1074/jbc.M302454200. [DOI] [PubMed] [Google Scholar]
- 27.Silva SD, Cunha IW, Younes RN, Soares FA, Kowalski LP, Graner E. ErbB receptors and fatty acid synthase expression in aggressive head and neck squamous cell carcinomas. Oral Dis. 2010;16:774–780. doi: 10.1111/j.1601-0825.2010.01687.x. [DOI] [PubMed] [Google Scholar]
- 28.Sheikh Ali MA, Gunduz M, Nagatsuka H, Gunduz E, Cengiz B, Fukushima K, et al. Expression and mutation analysis of epidermal growth factor receptor in head and neck squamous cell carcinoma. Cancer Sci. 2008;99:1589–1594. doi: 10.1111/j.1349-7006.2008.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CS, Redshaw A, Boag G. Epidermal growth factor receptor immunoreactivity in human laryngeal squamous cell carcinoma. Pathology. 1997;29:251–254. doi: 10.1080/00313029700169005. [DOI] [PubMed] [Google Scholar]
- 30.Xia W, Lau YK, Zhang HZ, Liu AR, Li L, Kiyokawa N, et al. Strong correlation between c-erbB-2 overexpression and overall survival of patients with oral squamous cell carcinoma. Clin Cancer Res. 1997;3:3–9. [PubMed] [Google Scholar]
- 31.Cavalot A, Martone T, Roggero N, Brondino G, Pagano M, Cortesina G. Prognostic impact of HER-2/neu expression on squamous head and neck carcinomas. Head Neck. 2007;29:655–664. doi: 10.1002/hed.20574. [DOI] [PubMed] [Google Scholar]
- 32.Mandic R, Rodgarkia-Dara CJ, Zhu L, Folz BJ, Bette M, Weihe E, et al. Treatment of HNSCC cell lines with the EGFR-specific inhibitor cetuximab (Erbitux) results in paradox phosphorylation of tyrosine 1173 in the receptor. FEBS Lett. 2006;580:4793–4800. doi: 10.1016/j.febslet.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 35.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 36.Kondo N, Tsukuda M, Sakakibara A, Takahashi H, Hyakusoku H, Komatsu M, et al. Combined molecular targeted drug therapy for EGFR and HER-2 in head and neck squamous cell carcinoma cell lines. Int J Oncol. 2012;40:1805–1812. doi: 10.3892/ijo.2012.1376. [DOI] [PubMed] [Google Scholar]
- 37.Del Campo JM, Hitt R, Sebastian P, Carracedo C, Lokanatha D, Bourhis J, et al. Effects of lapatinib monotherapy: results of a randomised phase II study in therapy-naive patients with locally advanced squamous cell carcinoma of the head and neck. Br J Cancer. 2011;105:618–627. doi: 10.1038/bjc.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu T, Claret FX. Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol. 2012;2:62. doi: 10.3389/fonc.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oxnard GR. Strategies for overcoming acquired resistance to epidermal growth factor receptor: targeted therapies in lung cancer. Arch Pathol Lab Med. 2012;136:1205–1209. doi: 10.5858/arpa.2012-0254-RA. [DOI] [PubMed] [Google Scholar]
- 41.Hurvitz SA, Hu Y, O'Brien N, Finn RS. Current approaches and future directions in the treatment of HER2-positive breast cancer. Cancer Treat Rev. 2013;39:219–229. doi: 10.1016/j.ctrv.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabinowits G, Haddad RI. Overcoming resistance to EGFR inhibitor in head and neck cancer: a review of the literature. Oral Oncol. 2012;48:1085–1089. doi: 10.1016/j.oraloncology.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Pegram M. Can we circumvent resistance to ErbB2-targeted agents by targeting novel pathways? Clin Breast Cancer. 2008;8(Suppl 3):S121–S130. doi: 10.3816/cbc.2008.s.008. [DOI] [PubMed] [Google Scholar]
- 44.Baselga J, Cortes J, Kim SB, Im SA, Hegg R, Im YH, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keating GM. Pertuzumab: in the first-line treatment of HER2-positive metastatic breast cancer. Drugs. 2012;72:353–360. doi: 10.2165/11209000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 46.Fuentes G, Scaltriti M, Baselga J, Verma CS. Synergy between trastuzumab and pertuzumab for human epidermal growth factor 2 (Her2) from colocalization: an in silico based mechanism. Breast Cancer Res. 2011;13:R54. doi: 10.1186/bcr2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakash A, Janosi L, Doxastakis M. GxxxG motifs, phenylalanine, and cholesterol guide the self-association of transmembrane domains of ErbB2 receptors. Biophys J. 2011;101:1949–1958. doi: 10.1016/j.bpj.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prakash A, Janosi L, Doxastakis M. Self-association of models of transmembrane domains of ErbB receptors in a lipid bilayer. Biophys J. 2010;99:3657–3665. doi: 10.1016/j.bpj.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cymer F, Schneider D. Transmembrane helix-helix interactions involved in ErbB receptor signaling. Cell Adh Migr. 2010;4:299–312. doi: 10.4161/cam.4.2.11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.