The crystal structure of the C-terminal domain of a putative U32 peptidase from G. thermoleovorans is reported; it is one of the most tightly packed protein structures reported to date.
Keywords: solvent content, crystal packing, high resolution
Abstract
While small organic molecules generally crystallize forming tightly packed lattices with little solvent content, proteins form air-sensitive high-solvent-content crystals. Here, the crystallization and full structure analysis of a novel recombinant 10 kDa protein corresponding to the C-terminal domain of a putative U32 peptidase are reported. The orthorhombic crystal contained only 24.5% solvent and is therefore among the most tightly packed protein lattices ever reported.
1. Introduction
Unlike inorganic and organic crystals, protein crystals are very fragile (McPherson, 1999 ▶). This is a result of far fewer and much weaker interactions contributing to the crystalline architecture in proportion to the molecular mass of the molecule composing the crystal (Drenth & Haas, 1992 ▶). This also affects the kinetic parameters of crystal growth: whereas small-molecule crystals crystallize within minutes or a few days, proteins generally require a much longer time, sometimes weeks or months. The majority of these properties result from a single feature: the solvent content (McPherson, 1999 ▶). While metal and atomic crystals are completely anhydrous, inorganic ionic crystals may be anhydrous, like rock salt, or may contain a few water molecules per molecular unit. The same holds for organic small-molecule crystals, for which the solvent content is generally between 23 and 35% (Kitaigorodskii, 1973 ▶). In contrast, protein crystals have a high solvent content per volume. For most protein crystals, this varies from 40 to 60%; very few protein crystals have been reported with solvent contents of <30% or >90% (McPherson, 1999 ▶; Matthews, 1968 ▶). In addition, solvent molecules are generally ordered in all parts of organic and inorganic crystals; in contrast, macromolecular crystals have large channels of only partially ordered solvent. As a result, protein crystals cannot be dehydrated and prolonged exposure to air results in destruction of the crystalline order (Bernal & Crowfoot, 1934 ▶).
Here, we report the crystal structure determination and crystal-packing analysis of a 10 kDa protein, the C-terminal domain of the putative U32 peptidase from Geobacillus thermoleovorans, and analyze the findings in the general context of the Protein Data Bank.
2. Materials and methods
2.1. Protein production and purification
A gene coding for a hypothetical U32-type peptidase from G. thermoleovorans (previously G. lituanicus; Dinsdale et al., 2011 ▶; 422 residues; 47 928 Da; UniProt code G5DCB7; L2V mutation for cloning strategy) was cloned into a modified pET-28a vector using NcoI and SalI restriction sites and was verified by DNA sequencing. The protein was produced by heterologous overexpression in Escherichia coli BL21 (DE3) cells, which were grown at 310 K in Luria–Bertani medium supplemented with kanamycin to a final concentration of 30 µg ml−1. Cell cultures were induced at an OD550 of 0.8 with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM and growth was continued for 5 h at 310 K. The selenomethionine variant was obtained in the same way except that the cells were grown in minimal medium containing selenomethionine (Sigma) instead of methionine. After centrifugation at 7000g for 30 min at 277 K, the pellet was washed twice with buffer A (50 mM Tris–HCl, 250 mM NaCl, 20 mM β-mercaptoethanol pH 8.0) and resuspended in the same buffer containing EDTA-free protease-inhibitor cocktail tablets (Roche Diagnostics) and DNase I (Roche Diagnostics). The cells were lysed at 277 K using a cell disrupter (Constant Systems) at a pressure of 135 MPa, the cell debris was removed by centrifugation at 50 000g for 1 h at 277 K and the supernatant was filtered (0.22 µm pore size; Millipore). The protein was found to serendipitously bind nickel–nitrilotriacetic acid (Ni–NTA) resin despite the absence of a polyhistidine tag. As such, the sample was incubated with Ni–NTA resin (Invitrogen) previously equilibrated with buffer A and eluted using the same buffer plus 150 mM imidazole. The protein was subsequently purified by size-exclusion chromatography on a HiLoad 26/60 Superdex 75 column previously equilibrated with buffer B [20 mM Tris–HCl, 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), pH 8.0]. The protein identity and purity were assessed by Edman degradation, peptide mass fingerprinting and 15% Tricine–SDS–PAGE stained with Coomassie Blue. Fractions containing the ∼48 kDa protein were pooled, concentrated to 30 mg ml−1 by ultrafiltration using Vivaspin 15 filter devices with a 10 kDa cutoff (Sartorius Stedim Biotech) and incubated for 48 h at 310 K to test the stability of the protein over time. After incubation, the sample showed strong precipitation and was therefore centrifuged at 16 000g for 10 min at 277 K; the supernatant was subsequently filtered. SDS–PAGE analysis of this supernatant showed the presence of a major band at ∼10.4 kDa, which we attribute to reproducible heterologous cleavage by a contaminating peptidase. This protein species was purified by size-exclusion chromatography on a HiLoad 16/60 Superdex 75 column previously equilibrated with buffer C (20 mM Tris–HCl, 50 mM NaCl, 1 mM TCEP pH 7.5) and eluted as a monomer. Edman-degradation and mass-spectrometric analyses (10 392 Da; experimental molecular mass with internal calibration) revealed that this species corresponded to the C-terminal region of the U32-type peptidase spanning the segment Ser334–Asn422 (hereafter referred to as GT-U32-CTD; 89 residues; calculated molecular mass 10 395 Da). The protein was further concentrated to 75 mg ml−1 using Vivaspin 15 and 500 filter devices with a 5 kDa cutoff. The protein concentration was determined by measuring the absorbance at 280 nm using a spectrophotometer (NanoDrop) and a calculated absorption coefficient ∊0.1% of 0.82.
2.2. Crystallization and X-ray diffraction data collection
Crystallization assays for GT-U32-CTD were carried out by the sitting-drop vapour-diffusion method using 96 × 2-well MRC plates (Innovadyne) and a Cartesian nanodrop robot (Genomic Solutions) at the IBMB/IRB crystallization service. Crystallization plates were stored in Bruker steady-temperature crystal farms at 277 and 293 K. Successful hits were scaled up to the microlitre range in 24-well Cryschem crystallization plates (Hampton Research). The best crystals of native GT-U32-CTD appeared at 293 K using protein solution at 75 mg ml−1 in buffer C containing 1 mM phenylmethylsulfonyl fluoride and reservoir solution consisting of 100 mM sodium acetate, 200 mM ammonium acetate, 30%(w/v) polyethylene glycol 4000 pH 4.6 in microlitre plates. The best crystals of the selenomethionine-derivatized protein were obtained from microlitre drops at 293 K using protein solution at 75 mg ml−1 in buffer C and reservoir solution consisting of 100 mM sodium citrate, 2 M ammonium sulfate pH 5.5. Crystals were cryoprotected by successive passages through reservoir solution containing increasing amounts of glycerol [up to 20–25%(v/v)]. Complete diffraction data sets were collected at 100 K from liquid-nitrogen flash-cryocooled crystals (Oxford Cryosystems 700 series cryostream) using a PILATUS 6M pixel detector (Dectris) on beamline ID29 of the European Synchrotron Radiation Facility (ESRF) Grenoble, France within the Block Allocation Group ‘BAG Barcelona’ (native protein) and using an ADSC Q315R CCD detector on beamline PROXIMA 1 of synchrotron SOLEIL, Paris, France (selenomethionine-derivatized protein). The crystals of native and selenomethionine-derivatized protein were orthorhombic (maximal resolution 1.10 Å) and trigonal (maximal resolution 1.15 Å), respectively, with one molecule per asymmetric unit. Mass-spectrometric analysis of carefully washed crystals revealed the presence of full-length GT-U32-CTD (approximately 60%) and three shorter forms that lacked two, three and five N-terminal residues, respectively (approximately 40% in total). Diffraction data were integrated, scaled, merged and reduced using the programs XDS (Kabsch, 2010 ▶) and SCALA (Evans, 2006 ▶) within the CCP4 suite (Winn et al., 2011 ▶; Table 1 ▶).
Table 1. Crystallographic data.
Values in parentheses are for the outermost resolution shell.
| Data set | Native | Selenomethionine (absorption peak)† |
|---|---|---|
| Space group | P212121 | P3221 |
| Unit-cell parameters () | a = 28.40, b = 36.30, c = 65.67 | a = b = 45.55, c = 60.90 |
| Wavelength () | 0.9724 | 0.9791 |
| No. of measurements | 200643 | 131989 |
| No. of unique reflections | 27941 | 26349 |
| Resolution range () | 32.81.10 (1.161.10) | 39.51.15 (1.211.15) |
| Completeness (%) | 98.8 (96.0) | 99.7 (98.8) |
| Anomalous completeness (%) | 97.2 (91.0) | |
| R merge ‡ | 0.116 (0.159) | 0.036 (0.348) |
| R r.i.m. (= R meas)‡ | 0.125 (0.172) | 0.045 (0.459) |
| R p.i.m. ‡ | 0.046 (0.066) | 0.026 (0.296) |
| Average intensity {[I/(I)]} | 13.7 (8.6) | 19.0 (3.3) |
| B factor (Wilson) (2) | 6.6 | 11.4 |
| Average multiplicity | 7.2 (6.3) | 5.0 (3.7) |
| Resolution range used for refinement () | 1.10 | 1.15 |
| No. of reflections used | 27158 | 25557 |
| No. of reflections used for test set | 734 | 792 |
| Crystallographic R factor‡ | 0.123 | 0.141 |
| Free R factor‡ | 0.163 | 0.170 |
| No. of protein atoms | 760 | 692 |
| No. of solvent molecules | 108 | 80 |
| No. of ligands | 1 acetate, 1 cation | 1 sulfate |
| R.m.s.d. from target values, bonds () | 0.015 | 0.014 |
| R.m.s.d. from target values, angles () | 2.33 | 2.44 |
| Average B factor for protein atoms (2) | 10.4 | 18.3 |
| Main-chain conformational angle analysis§ | ||
| Residues in favoured regions | 85 | 78 |
| Outliers | 0 | 0 |
| All residues | 87 | 80 |
2.3. Structure determination and refinement
The structure was determined by single-wavelength anomalous diffraction using the selenomethionine derivative and the program SHELXD (Sheldrick, 2010 ▶). Diffraction data were collected from a crystal at the selenium absorption-peak wavelength, as inferred from a previous XANES fluorescence scan, and enabled the program to identify the two selenium sites of the monomer present in the asymmetric unit. Subsequent phasing with SHELXE (Sheldrick, 2010 ▶), including implementation of the ‘free-lunch’ algorithm and auto-tracing, resolved the twofold ambiguity intrinsic to a SAD experiment owing to the difference in the values of the pseudo-free correlation coefficients of the two possible hands (79.2% versus 49.8%), which confirmed P3221 as the correct space group. An initial model was built into the experimental electron density with the program TURBO-FRODO (Carranza et al., 1999 ▶) on a Silicon Graphics Octane2 workstation and was refined with PHENIX (Adams et al., 2002 ▶), BUSTER (Blanc et al., 2004 ▶) and SHELXL (Sheldrick, 2008 ▶). This model was employed to solve the native structure (true space group P212121) by molecular replacement (Huber, 1965 ▶) using Phaser (McCoy, 2007 ▶), which rendered one unambiguous solution with Z-scores of 9.7 and 13.8 for the rotation and translation functions, respectively. Model building and refinement, which included TLS refinement and refinement of anisotropic thermal displacement parameters, proceeded as described above. The final native model comprised all of the residues of the protein (Ser334–Asn422) plus one oxygen-pentacoordinated cation, an acetate ion and 108 solvent molecules. Table 1 ▶ provides a summary of data collection and final model refinement.
2.4. Solvent-content analysis
A solvent-content distribution for all X-ray crystal structures deposited in the Protein Data Bank (PDB) before September 2012 was calculated for a nonredundant set of unique crystal forms. To obtain this data set, structures that share equivalent asymmetric units were clustered together. The crystal structures in each cluster possessed the same polypeptide composition, were determined in the same space group and showed differences of up to 5% in unit-cell volume. The working pipeline was composed of the following steps. Firstly, to determine the polypeptide composition, each protein structure was labelled with a formula describing the number of polypeptides and their sequence-cluster identifiers. The pre-calculated sequence clusters had been generated by BLAST (Altschul et al., 1990 ▶) using a 90% identity threshold (obtained from ftp://resources.rcsb.org/sequence/clusters/). At this stage, the data set contained clusters of all possible protein assemblies. Sequences shorter than 20 amino acids, split entries, virus capsids, synthetic constructs, complexes with nucleic acids, polysaccharides and d-polypeptides were excluded from the analysis. Structures missing 20% or more of the residues of the associated sequence were likewise rejected. Next, the aforementioned clusters were subdivided based on the crystal space group and unit-cell volume. The latter were calculated based on the unit-cell dimensions defined in the mmCIF files using the MATTHEWS_COEF program from the CCP4 suite (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). The molecular weight of the polypeptides in the asymmetric unit was calculated from the sequences reported by the authors using the ProtParam methodology (Gasteiger et al., 2005 ▶). Representatives of each final cluster were chosen based on a simple quality factor defined as 1/resolution − R value. For the resulting data set, the solvent content of each protein crystal structure was determined using MATTHEWS_COEF (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶). Subsequently, manual curation of the final data set enabled the removal of special cases in which the structures were the result of ensemble refinement (PDB entries 1cwq and 1gtv), desiccated crystals and those obtained by phase transition (PDB entries 1c0c, 1xek, 1xej and 1v7t), crystals with ‘order–disorder’ (Pletnev et al., 2009 ▶; PDB entry 3h1r) and other nonstandard protocols for structure determination (PDB entries 3gi0 and 2xge). Cases for which the solvent content was artificially low owing to long missing N- and C-terminal fragments were not considered either (PDB entries 2xnq, 2duy, 2axo, 3bqh, 2f9l, 3m4s, 3nzl, 2xjx, 4eti, 2ds8 and 1vfq). The final resulting data set consisted of 35 656 X-ray structures.
2.5. Miscellaneous
Figures were prepared with TURBO-FRODO and UCSF Chimera (Pettersen et al., 2004 ▶). The total interaction surface (taken as half of the surface area buried at a complex interface; probe radius 1.4 Å) and close contacts (defined as atoms separated by less than 4 Å) were determined using CNS (Brünger et al., 1998 ▶). Pairwise interaction surfaces were calculated with the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel & Henrick, 2005 ▶). Surface shape complementarity was calculated with the program SC (Lawrence & Colman, 1993 ▶) within CCP4 with a probe radius of 1.4 Å. Structure similarities were investigated with DALI (Holm & Rosenström, 2010 ▶). Model validation was performed with MolProbity (Chen et al., 2010 ▶) and the WHAT_CHECK routine of WHAT IF (Vriend, 1990 ▶). The final coordinates of native and selenomethione-derivatized GT-U32-CTD have been deposited in the PDB (http://www.pdb.org; accession codes 4he5 and 4he6).
3. Results and discussion
3.1. Crystallization and structure determination
We studied the crystal structure of the C-terminal domain of a putative peptidase from G. thermoleovorans belonging to MEROPS (http://merops.sanger.ac.uk; Rawlings et al., 2012 ▶) family U32 (GT-U32-CTD), which was obtained by spontaneous proteolytic fragmentation of the recombinant full-length protein. Native GT-U32-CTD crystallized in space group P212121 using 100 mM sodium acetate, 200 mM ammonium acetate, 30%(w/v) polyethylene glycol 4000 pH 4.6 as the reservoir solution and the crystals diffracted to 1.10 Å resolution (see §2 and Table 1 ▶). The structure was determined by single-wavelength anomalous diffraction of a selenomethione-derivatized variant of the protein, which crystallized in space group P3221 using 100 mM sodium citrate, 2 M ammonium acetate pH 5.5 as the reservoir solution; the crystals diffracted to 1.15 Å resolution. The resulting molecular model was employed to solve the structure of the orthorhombic crystal form, which revealed all of the residues of the only chain present in the asymmetric unit plus one (hypothetical) sodium cation, an acetate ion and 108 solvent molecules. Table 1 ▶ provides a summary of the crystallographic data-collection and final model-refinement parameters.
3.2. Architecture of GT-U32-CTD
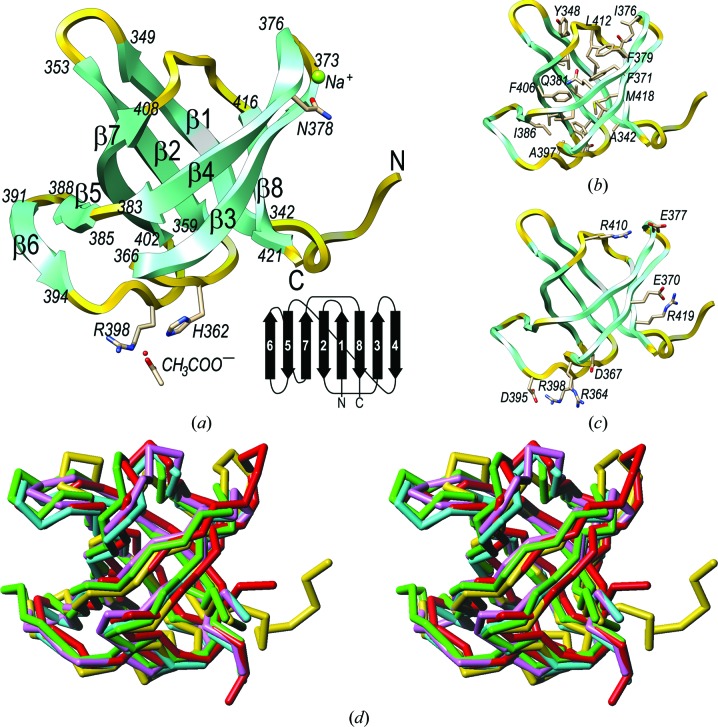
The structure shows a compact distorted open β-barrel made up of eight β-strands (β1–β8), into which the first eight residues (Ser334–Phe341) and the last single residue (Asn422) are inserted (Fig. 1 ▶ a). The first six residues are present with a refined occupancy of 60%, which is in accordance with mass-spectrometric analysis of carefully washed crystals (see §2). The barrel is actually arranged as a strongly twisted, curled and arched antiparallel β-sheet (β6–β5–β7–β2–β1–β8–β3–β4; connectivity −1, +3, +1, −6, −1, +2, +3); the flanking strands, β4 and β6, do not interact with each other through their main chains. In addition, β4 and β7 are close to giving rise to a perfect barrel with β1–β3 and β8 (Fig. 1 ▶ a), but they do not contact each other through their main chains either. Instead, the gap is closed by the side chains of Phe379, Gln381, Lys409 and Arg410 (Fig. 1 ▶ b). The sheet wraps around a central hydrophobic core formed by the side chains of residues Ala342, Val345, Tyr348, Ala355, Val357, Ala359, Phe363, Val369, Phe371, Ile376, Phe379, Ile383, Gln381, Ala397, Val404, Phe406, Val408, Leu412, Asn416 and Met418. In contrast, the surface is mainly hydrophilic and shows seven lysines, five arginines, five aspartates and ten glutamates, which contribute to four salt bridges (Asp367–Arg364, Glu370–Arg419, Glu377–Arg410 and Asp395–Arg398; Fig. 1 ▶ c). Furthermore, a potential sodium cation was tentatively assigned on the surface based on very short binding distances to five liganding O atoms (Asn378 Oδ1, 2.35 Å; Gly373 O, 2.38 Å; Ile376 O, 2.28 Å; a solvent molecule, 2.32 Å; a symmetry-related Asp395 Oδ2 atom, 2.38 Å) arranged in a trigonal bipyramidal coordination sphere. Finally, an acetate ion was likewise found bound to the protein surface anchored to His362 N∊2 (2.90 Å), Arg398 Nη2 (2.85 Å) and a solvent molecule (2.64 Å) through one of its carboxylate O atoms. The other O atom is bound by Arg398 N∊ (2.89 Å), a solvent molecule (2.74 Å) and a symmetry-related Gln358 N∊2 atom (3.20 Å).
Figure 1.
(a) Ribbon-type plot of GT-U32-CTD with the eight β-strands labelled and marked with their flanking residues. Residues participating in ion binding are also shown and labelled. Inset (lower right): topology scheme of the protein. (b) Cartoon showing the side chains participating in the central hydrophobic core. (c) As (b) but showing the charged residues of the protein engaged in salt bridges. (d) Cα-type plot in stereo showing the superposed structures of GT-U32-CTD (yellow), LepA (green), eRF3 (lilac), EF-1a (cyan) and EF-Tu (red).
3.3. Structural relatives
Structure-similarity searches with GT-U32-CTD identified the LepA protein (PDB entry 3cb4; Evans et al., 2008 ▶), release factor eRF3 (PDB entry 3e20; Cheng et al., 2009 ▶), elongation factor 1a (EF-1a: PDB entry 1skq; Vitagliano et al., 2004 ▶) and elongation factor Tu (EF-Tu; PDB entry 2c77; Parmeggiani et al., 2006 ▶) as the most similar structures, with Z-scores of 10.9–11.0, r.m.s.d. values of 1.7–2.0 Å and aligned stretches of 78–80 residues. Superposition of these structures onto GT-U32-CTD (Fig. 1 ▶ d) revealed equivalent architectures, topologies and connectivities despite negligible sequence identity (8–19%). These structural relatives are all found in proteins that further comprise an additional N-terminal guanine-nucleotide binding domain and they participate in ribosomal protein translation elongation or termination (Andersen et al., 2003 ▶). For the elongation factors, it has been shown that the GT-U32-CTD-like domains interact with aminoacyl-loaded tRNAs and with antibiotics that target the protein-synthesis machinery (Andersen et al., 2003 ▶; Parmeggiani et al., 2006 ▶). Accordingly, a similar function to a protein engaged in binding is conceivable for the C-terminal domain of the putative U32-type peptidase of G. thermoleovorans.
3.4. Crystal-packing analysis
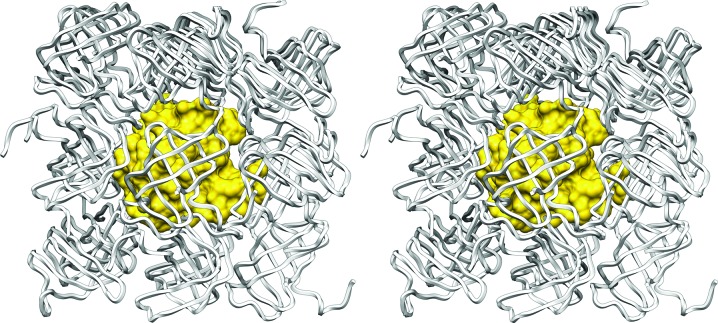
Notwithstanding the interest of the abovementioned results, the most striking feature of the present study is that the native protein crystallized in an orthorhombic crystal form with an extremely low solvent content of 24.5% (Matthews parameter V M of 1.63 Å3 Da−1; Matthews, 1968 ▶), which may at least partially account for its strong diffraction power; crystals with less solvent tend to diffract better (Kantardjieff & Rupp, 2003 ▶). In contrast, the selenomethionine-derivatized protein, which only differs from the former by the replacement of two S atoms by selenium within the 10 kDa molecule, crystallized in a trigonal (i.e. higher symmetry) space group with 30.0% solvent content (V M = 1.75 Å3 Da−1). In addition, none of the abovementioned structural relatives packed with less than 39% solvent. Detailed inspection of the native crystal lattice (Fig. 2 ▶) confirmed very tight binding of the protein molecules within the crystal, with almost no solvent channels and only ordered solvent molecules ‘gluing’ the protein molecules together. Each protein molecule interacts directly with its surrounding crystallographic neighbours through a total of 136 close contacts and a total contact surface of 3480 Å2, which is in good agreement with the value obtained by adding the binary interaction surfaces with each neighbour when calculated by another approach (3590 Å2; see §2). The total contact surface represents 65% of the total surface of a monomer, which is far larger than generally reported for crystal contact surfaces (15–49%; Carugo & Argos, 1997 ▶). In comparison, tight protein–protein complexes, which exist not only in the crystalline state but also in solution, such as proteinase–inhibitor and antibody–antigen complexes, interact through surfaces of 600–1000 Å2, which correspond to 5–20% of their total surfaces (Janin & Chothia, 1990 ▶). The shape complementarity between the surface of one GT-U32-CTD protomer and its surrounding crystal relatives is 0.69. This falls within the ranges of values for proteinase–inhibitor complexes (0.70–0.76) and antibody–antigen complexes (0.64–0.68) (Lawrence & Colman, 1993 ▶), which is noteworthy taking into account the overall size of the total contact surface. Taken together, all of these features explain the very tightly packed crystal lattice.
Figure 2.
Cartoon in stereo depicting the packing of one GT-U32-CTD molecule (shown with its Connolly surface in yellow) surrounded by its crystallographic symmetry equivalents (shown as white ropes).
3.5. Low-solvent crystal structures in the PDB
To assess the uniqueness of the present packing in the general context of the PDB, we investigated the cluster of tightly packed protein crystal structures that have been deposited. Detailed studies of the solvent-content distribution have been published based on 116 (Matthews, 1968 ▶), 15 641 (Kantardjieff & Rupp, 2003 ▶) and 9081 (Chruszcz et al., 2008 ▶) macromolecular crystal structures. These studies concluded that ∼0.5% of nonredundant protein structures contain 25% solvent or less (Kantardjieff & Rupp, 2003 ▶), which is consistent with recent estimates that less than 1% of proteins pack with less than 25–26% solvent (C. Weichenberger & B. Rupp, personal communication). As the annotation of PDB entries with respect to solvent content is inaccurate (Andersson & Hovmöller, 2000 ▶), we decided to perform a comprehensive search for low-solvent-content crystal structures (see §2). We calculated the solvent contents from the molecular masses of the annotated protein sequences of 35 656 nonredundant protein crystal forms from the PDB determined using X-ray crystallography (Berman et al., 2000 ▶), excluding virus capsids, protein–DNA and protein–RNA complexes, synthetic constructs and peptides (defined as comprising 20 residues or less). Artificially reduced values such as those obtained by crystal desiccation or treatment with organic solvents were also omitted. These calculations revealed only ten nonredundant structures (∼0.028%) with a solvent content equal to or less than 26% (see Table 2 ▶). These are human CD59 (79 residues; PDB entry 2uwr; Leath et al., 2007 ▶), human matrix metalloproteinase 8 (163 residues; PDB entry 1zp5; Campestre et al., 2006 ▶), the SH3 domain of chicken α-spectrin (62 residues; PDB entry 2f2v; Casares et al., 2007 ▶), two structures of cypoviral polyhedrines from Wiseana signata NPV (243 residues; PDB entry 3jvb; Coulibaly et al., 2007 ▶, 2009 ▶) and Bombyx mori (248 residues; PDB entry 2oh7; Coulibaly et al., 2007 ▶), a five-stranded phenylalanine zipper (56 residues; PDB entry 2guv; Liu et al., 2006 ▶), the CLP-protease adaptor protein (176 residues; PDB entry 3gw1; Román-Hernández et al., 2009 ▶), a small antifungal protein from Ecommia ulmoides (41 residues; PDB entry 1p9g; Xiang et al., 2004 ▶) and the mating pheromone Er-1 from Euplotes raikovi (40 residues; PDB entry 2erl; Anderson et al., 1996 ▶), with five and three disulfide bridges, respectively, and effector protein AVR3A11 from Phytophthora capsici (65 residues; PDB entry 3zr8; Boutemy et al., 2011 ▶). Interestingly, four of the ten abovementioned structures crystallized in space group P212121, which is also the presently reported space group (see Table 1 ▶), and the other three in P21. This correlates well with the observation by Chruszcz and coworkers that these two space groups, together with P1, show the lowest mean solvent content in general (see Table 3 in Chruszcz et al., 2008 ▶).
Table 2. Crystal structures with 25% solvent content.
| PDB code | Solvent content (%) | Space group | Cell volume (3) | Theoretical sequence length (amino acids) | No. of residues actually observed | Quality factor† |
|---|---|---|---|---|---|---|
| 3jvb | 21.32 | I23 | 1074906 | 243 | 204 | 0.2968 |
| 2uwr | 21.77 | P212121 | 57793 | 79 | 79 | 0.5393 |
| 1zp5 | 22.53 | P212121 | 114865 | 163 | 163 | 0.3366 |
| 2f2v | 22.64 | P212121 | 45796 | 62 | 62 | 0.3653 |
| 2oh7 | 22.72 | I23 | 1084536 | 248 | 247 | 0.2862 |
| 2guv | 24.12 | P1211 | 109779 | 280 | 280 | 0.5156 |
| 1p9g | 24.29 | P1211 | 13536 | 41 | 40 | 1.1221 |
| 3gw1 | 24.48 | P1211 | 66477 | 176 | 168 | 0.1520 |
| 2erl | 24.50 | C121 | 28766 | 40 | 40 | 0.8710 |
| 3zr8 | 24.86 | P212121 | 48935 | 65 | 65 | 0.9844 |
Defined as 1/resolution R value.
4. Conclusion
Accordingly, the present orthorhombic crystal form of GT-U32-CTD represents one of the most tightly packed protein structures reported to date; it shows a solvent content that is more similar to those attributable to crystals of small organic compounds than to a 10 kDa protein.
Supplementary Material
PDB reference: C-terminal domain of putative U32 peptidase, native, 4he6
PDB reference: selenomethionine variant, 4he5
Acknowledgments
We are particularly indebted to Bernhard Rupp for his interest, assistance and helpful discussion. We also thank Tibisay Guevara and the IBMB/IRB crystallization service for assistance during crystallization experiments. This study was supported in part by grants from the American, European, Lithuanian, Spanish and Catalan agencies (FP7-HEALTH-F3-2009-223101 ‘AntiPathoGN’, FP7-HEALTH-2010-261460 ‘Gums & Joints’, FP7-PEOPLE-2011-ITN-290246 ‘RAPID’, 31V-151 ‘COSMETIZYM’, BIO2009-10334, BFU2009-07134/BMC, BFU2012-32862, BFU2012-33516, CSD2006-00015, a JAE postdoctoral contract from CSIC, an FPU PhD fellowship from the Spanish Ministry for Science and Technology and Fundació ‘La Marató de TV3’ grants 2009-100732 and 2009SGR1036). We acknowledge the help provided by the ESRF synchrotron local contacts and, in particular, the excellent assistance and outstanding dedication of Andrew Thompson of SOLEIL beamline PROXIMA 1. Funding for travelling and data collection was provided in part by the ESRF and SOLEIL.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Andersen, G. R., Nissen, P. & Nyborg, J. (2003). Trends Biochem. Sci. 28, 434–441. [DOI] [PubMed]
- Anderson, D. H., Weiss, M. S. & Eisenberg, D. (1996). Acta Cryst. D52, 469–480. [DOI] [PubMed]
- Andersson, K. M. & Hovmöller, S. (2000). Acta Cryst. D56, 789–790. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bernal, J. D. & Crowfoot, D. (1934). Nature (London), 133, 794–795.
- Blanc, E., Roversi, P., Vonrhein, C., Flensburg, C., Lea, S. M. & Bricogne, G. (2004). Acta Cryst. D60, 2210–2221. [DOI] [PubMed]
- Boutemy, L. S., King, S. R., Win, J., Hughes, R. K., Clarke, T. A., Blumenschein, T. M., Kamoun, S. & Banfield, M. J. (2011). J. Biol. Chem. 286, 35834–35842. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Campestre, C., Agamennone, M., Tortorella, P., Preziuso, S., Biasone, A., Gavuzzo, E., Pochetti, G., Mazza, F., Hiller, O., Tschesche, H., Consalvi, V. & Gallina, C. (2006). Bioorg. Med. Chem. Lett. 16, 20–24. [DOI] [PubMed]
- Carranza, C., Inisan, A.-G., Mouthuy-Knoops, E., Cambillau, C. & Roussel, A. (1999). AFMB Activity Report 1996–1999, pp. 89–90. Marseille: CNRS-UPR 9039.
- Carugo, O. & Argos, P. (1997). Protein Sci. 6, 2261–2263. [DOI] [PMC free article] [PubMed]
- Casares, S., López-Mayorga, O., Vega, M. C., Cámara-Artigas, A. & Conejero-Lara, F. (2007). Proteins, 67, 531–547. [DOI] [PubMed]
- Cheng, Z., Saito, K., Pisarev, A. V., Wada, M., Pisareva, V. P., Pestova, T. V., Gajda, M., Round, A., Kong, C., Lim, M., Nakamura, Y., Svergun, D. I., Ito, K. & Song, H. (2009). Genes Dev. 23, 1106–1118. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chruszcz, M., Potrzebowski, W., Zimmerman, M. D., Grabowski, M., Zheng, H., Lasota, P. & Minor, W. (2008). Protein Sci. 17, 623–632. [DOI] [PMC free article] [PubMed]
- Coulibaly, F., Chiu, E., Gutmann, S., Rajendran, C., Haebel, P. W., Ikeda, K., Mori, H., Ward, V. K., Schulze-Briese, C. & Metcalf, P. (2009). Proc. Natl Acad. Sci. USA, 106, 22205–22210. [DOI] [PMC free article] [PubMed]
- Coulibaly, F., Chiu, E., Ikeda, K., Gutmann, S., Haebel, P. W., Schulze-Briese, C., Mori, H. & Metcalf, P. (2007). Nature (London), 446, 97–101. [DOI] [PubMed]
- Dinsdale, A. E., Halket, G., Coorevits, A., Van Landschoot, A., Busse, H. J., De Vos, P. & Logan, N. A. (2011). Int. J. Syst. Evol. Microbiol. 61, 1802–1810. [DOI] [PubMed]
- Drenth, J. & Haas, C. (1992). J. Cryst. Growth, 122, 107–109.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, R. N., Blaha, G., Bailey, S. & Steitz, T. A. (2008). Proc. Natl Acad. Sci. USA, 105, 4673–4678. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Huber, R. (1965). Acta Cryst. 19, 353–356.
- Janin, J. & Chothia, C. (1990). J. Biol. Chem. 265, 16027–16030. [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kitaigorodskii, A. I. (1973). Molecular Crystals and Molecules. London: Academic Press.
- Krissinel, E. & Henrick, K. (2005). CompLife 2005, edited by M. R. Berthold, R. Glen, K. Diederichs, O. Kohlbacher & I. Fischer, pp. 163–174. Berlin, Heidelberg: Springer.
- Lawrence, M. C. & Colman, P. M. (1993). J. Mol. Biol. 234, 946–950. [DOI] [PubMed]
- Leath, K. J., Johnson, S., Roversi, P., Hughes, T. R., Smith, R. A. G., Mackenzie, L., Morgan, B. P. & Lea, S. M. (2007). Acta Cryst. F63, 648–652. [DOI] [PMC free article] [PubMed]
- Liu, J., Zheng, Q., Deng, Y., Kallenbach, N. R. & Lu, M. (2006). J. Mol. Biol. 361, 168–179. [DOI] [PubMed]
- Mallorquí-Fernández, N., Manandhar, S. P., Mallorquí-Fernández, G., Usón, I., Wawrzonek, K., Kantyka, T., Solà, M., Thøgersen, I. B., Enghild, J. J., Potempa, J. & Gomis-Rüth, F. X. (2008). J. Biol. Chem. 283, 2871–2882. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- McPherson, A. (1999). Crystallization of Biological Macromolecules. New York: Cold Spring Harbor Laboratory Press.
- Parmeggiani, A., Krab, I. M., Okamura, S., Nielsen, R. C., Nyborg, J. & Nissen, P. (2006). Biochemistry, 45, 6846–6857. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Pletnev, S., Morozova, K. S., Verkhusha, V. V. & Dauter, Z. (2009). Acta Cryst. D65, 906–912. [DOI] [PMC free article] [PubMed]
- Rawlings, N. D., Barrett, A. J. & Bateman, A. (2012). Nucleic Acids Res. 40, D343–D350. [DOI] [PMC free article] [PubMed]
- Román-Hernández, G., Grant, R. A., Sauer, R. T. & Baker, T. A. (2009). Proc. Natl Acad. Sci. USA, 106, 8888–8893. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Vitagliano, L., Ruggiero, A., Masullo, M., Cantiello, P., Arcari, P. & Zagari, A. (2004). Biochemistry, 43, 6630–6636. [DOI] [PubMed]
- Vriend, G. (1990). J. Mol. Graph. 8, 52–56. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xiang, Y., Huang, R.-H., Liu, X.-Z., Zhang, Y. & Wang, D.-C. (2004). J. Struct. Biol. 148, 86–97. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C-terminal domain of putative U32 peptidase, native, 4he6
PDB reference: selenomethionine variant, 4he5