Abstract
When cells change functions or activities (such as during differentiation, response to extracellular stimuli, or migration), gene expression undergoes large-scale reprogramming, in cell type- and function-specific manners. Large changes in gene regulation require changes in chromatin architecture, which involve recruitment of chromatin remodeling enzymes and epigenomic modification enzymes to specific genomic loci. Transcription factors must also be accurately assembled at these loci. SATB1 is a genome organizer protein that facilitates these processes, providing a nuclear architectural platform that anchors hundreds of genes, through its interaction with specific genomic sequences; this activity allows expression of all these genes to be regulated in parallel, and enables cells to thereby alter their function. We review and describe future perspectives on SATB1 function in higher-order chromatin structure and gene regulation, and its role in metastasis of breast cancer and other tumor types.
1. Introduction
When normal breast epithelial cells become malignant, disease progresses through a series of successive modifications that involve changes in the cells’ genetic and epigenetic status and interactions with the microenrivonment [1–3]. Metastasis is a late step in solid tumor progression and the primary cause of death for cancer patients[4, 5]. The ability to identify tumor cells that are likely to metastasize in patients with early-stage cancers could prolong their survival.
Gene expression profiling studies using DNA microarray technology for human breast carcinomas revealed that breast cancer, which is a highly heterogeneous disease, can be classified into at least four major molecular subtypes and a normal breast-like group[6–8]. These tumor subtypes might reflect different cell types within the breast or different stages of epithelial cell development[9]. Patients with these molecular subtypes of tumors have distinct outcomes, offering a basis for predicting response to treatment. It has been a challenge to translate differences in gene expression patterns into clinical practice, due to the heterogeneity of breast cancers[8]. Nevertheless, transcription profiling studies identified groups of genes whose specific expression patterns are associated with metastatic tumor cells (a poor prognosis signature); this expression profile could also be detected in some primary carcinomas[10–12]. Therefore, in contrast to a model in which metastatic tumor cells evolve from rare cell clones at only late stages of tumor development, primary carcinomas appear to already contain a large number of tumor cells with metastatic potential. Primary breast carcinomas can be further classified, based on the ‘poor prognosis signature’, which provides information about the likelihood of tumor metastasis[11]. Distant metastases not only have a poor prognosis signature, but also retain the expression profile of the primary tumor’s subtype from which they arose. Most subtypes of breast tumors therefore have the capacity to metastasize.
Although the host microenvironment influences tumor progression, tumor cells themselves must change their gene expression profile to become metastatic. An important question is, what are the molecular mechanisms by which tumor cells make the large changes in gene expression required to acquire metastatic features? Cells can alter gene expression patterns by accumulating genetic mutations, deletions, and amplifications. So does tumor progression proceed gradually, via randomly accumulation of genomic changes, until cells with the right combinations of changes can acquire an invasive, aggressive phenotype? Or is there a single turning point in which tumor cells to undergo a major change in gene expression that alters their phenotype?
Studies of the genome organizer protein special AT-rich binding protein, SATB1[13], have shown that breast cancer cells can make rapid, major changes in their gene expression pattern that alter their phenotype[14]. Breast cancer cells with sufficient levels of SATB1 undergo large changes in their gene expression profile and acquire a metastatic phenotype.
We review how SATB1 was identified and found to be a factor that promotes aggressive phenotypes of breast tumors. SATB1 regulates gene expression at the level of higher-order chromatin structure, so to understand SATB1 functions, it is necessary to briefly review chromatin folding in the mammalian genome and the specialized genomic sequences that are recognized by SATB1.
2. Higher-Order Chromatin Structures
The mammalian genome is organized into complex higher-order structures formed by hierarchical folding of DNA. In mammalian chromosomes, DNA is compacted ~10,000-fold[15]. These structures form when naked DNA is wrapped around octamers of core histone proteins to form nucleosomes. Nucleosomes are connected with linker DNA, forming ‘beads-on-a string’ chromatin fibers, which are 10 nm in thickness. The 10 nm chromatin fibers are further folded into fibers of increasing thickness (30 nm, and then into fibers of increased thickness). Even with the fiber of 30-nm diameter, its detailed structure is still under debate [16–19]. Beyond this point, it is not well understood how fibers are folded into higher-order chromatin structures.
However, it is known that chromosomes are organized into chromosome territories, largely separated from each other in the cell nucleus[20]. It has been proposed that chromatin, in chromosome territories, is folded into small-scale chromatin loops of ~50–200 kb[21]. In addition, giant loops of paired DNA, of several megabases, have been detected. These giant loops protrude from their original territory and intermingle with other chromosome territories[21]. Chromatin looping brings distal genomic loci into close spatial proximity. Long-range interactions between genomic sites within the same chromosome, and also from different chromosomes, have been detected by chromosomal conformation capture and related assays[22–33]. Chromatin fibers can fold into loops of varying sizes. Chromatin looping is not only important for chromatin compaction, but it is thought to be involved in gene regulation—a distal regulatory sequence could be brought to close proximity with a locus and thereby regulate it, or multiple co-inducible genes could be brought together to be co-regulated. This type of 3-dimensional chromatin architecture has been correlated with gene expression patterns [34, 35]. Therefore, eukaryotic gene expression is regulated at multiple hierarchical levels, from primary sequence to the 3-dimensional spatial organization of the genome[36]. In addition to gene regulation, the frequent and preferential juxtaposition of gene loci (e.g. Myc and Igh) in nuclei of B lymphocyte have been detected, which would predispose chromosomal translocation to take place [33, 37].
Although examples are limited, architectural chromatin proteins that make chromatin fibers fold into loops have been identified. In mammalian nuclei, the homeobox protein SATB1 has been shown to fold chromatin into loops and regulate expression of large numbers of genes, in cell type- and cell function-specific manners[26, 38–42]. Methyl-CpG binding protein 2 (MeCP2), whose mutations cause RETT syndrome (RTT), an X-linked mental disorder[43]; MeCP2 actively forms transcriptionally-silent chromatin loop configurations at its target loci in brains of mice that are a model of RTT [24]. There is also much evidence to indicate that the zinc-finger protein CTCF (CCCTC-binding factor) acts as an insulator protein that blocks the effects of transcriptional enhancers, by promoting chromatin looping at specific loci [44, 45]. Further studies of proteins that function in chromatin looping will increase our knowledge about regulation of genes at the level of higher-order chromatin structure, and the role of these processes in disease pathogenesis[46].
3. SATB1 and Higher-Order Chromatin Structure
Some decades ago, a specialized ATC sequence context, comprising approximately 100–300 base pairs (bp), was identified in the mammalian genome; it conferred a strong unpairing propensity when placed under negative superhelical strain. The sequence is characterized by a cluster of specialized sequences (20–40 bp) with a complete bias in C and G distribution (1 strand consists of exclusively As, Ts, and Cs, referred to as ATC sequences)[13, 47, 48]. The extensive unpairing of genomic regions with ATC sequence clusters was originally identified by chemical probes (either chloro- or bromoacetaldehyde) that react specifically with unpaired adenine and cytosine bases[49]. Disruption of ATC sequence context by minimal mutagenesis eliminates the propensity toward unpairing.
Genomic regions characterized by the ATC sequence context are called base-unpairing regions (BURs); 1 is found, on average, in every 40,000 bp (unpublished data). Most genes have several major BURs within 100 kb 5′, in introns, and 100 kb 3′. BURs are well characterized because of their unique physical properties, which are distinct from those of the rest of the genome. Not all AT-rich sequences have properties of BURs. Many, but not all, sequences previously referred to as matrix attachment region (MAR) or scaffold attachment regions (SAR) contain a BUR. BURs are well-defined sequences with a specialized ATC sequence context that has unusual physical properties.
SATB1 was identified as a protein that binds exclusively to BURs when they are in the double-stranded DNA conformation (rather than an unpaired DNA structure); SATB1 recognizes the altered phosphate backbone structure[13]. Because SATB1 was cloned by virtue of its specificity for BURs[13], and BURs are distributed throughout mammalian genomes, researchers began to investigate whether SATB1 might be able to fold chromatin into loops. SATB1 was initially detected at a high level in thymocytes, in progenitor cells such as amyloblasts and osteoblasts, and at the basal layer of epidermis. Hundreds of BURs that bound to SATB1 in thymocytes were individually cloned and many were found to remain in the residual nuclei after extraction with high salt. However, in Satb1-null thymocytes, the BURs were found in DNA halos that surround the salt-extracted nuclei[38]. These data indicated that BURs anchor to SATB1 to form bases of chromatin loops. In nuclei of thymocytes, SATB1 is found exclusively in euchromatin, in a ‘chicken-wire’ or cage structure that surrounds heterochromatin. The SATB1-bound BURs are tethered to this nuclear architecture[40].
More information on the functions of SATB1 has come from chromatin conformation capture (3C) assays, combined with chromatin immunorecipitation (ChIP-3C or ChIP-loop), in studies of T-cell activation[26]. SATB1 was found to be induced upon activation of T-helper (Th)2 cells and bind to multiple BURs in a 200 kb cytokine gene cluster, bringing distal Il4, Il5, and Il13 loci together by chromatin looping. The SATB1-induced structure that is formed by the 200 kb region and contains the loops is loaded with genes that encode Th2-specific factors, such as GATA3, STAT6, and c-Maf, as well as the chromatin-remodeling enzyme Brg1 and RNA polyerase II. This transcriptionally-active chromatin structure enables activation of these cytokine genes in parallel[26]. Without SATB1, these looping events did not take place, and the cytokine genes were not activated. Therefore, the chromatin loops formed by SATB1 through its interaction with BURs were correlated with gene regulation, and SATB1 is required for Th2 cell activation and the cytokine production[26].
4. SATB1 and Epigenetic Regulation
SATB1 not only folds chromatin into loops via binding to BURs, but also provides a nuclear platform to recruit chromatin remodeling and modifying enzymes to loci around the BURs. Because chromatin folds to form complex higher-order structures, SATB1 tethering of BURs might provide core sites that facilitate assembly of chromatin remodeling or modifying factors. Evidence for this model came from the finding that SATB1 binds to chromatin-remodeling complexes, such as NURD and ACF/ISWI complexes[50]. At the gene level, SATB1 was shown to recruit the ACF/ISWI complex and histone deacetylase to the Il2Ra locus and repress it in thymocytes. SATB1 also promoted region-specific histone modification at specific loci, resulting in histone marks on chromatin that either activated or repressed transcription[50]. Induction of SATB1 upon activation of Th2 cells is required for the large increase in active histone marks that are observed at discrete loci over the 200 kb cytokine gene cluster region[26]. Therefore, SATB1 not only forms loops, but regulates epigenomic modifications and proper nucleosome positioning, by targeting chromatin remodeling and modifying enzymes to specific loci.
5. SATB1 Regulation of Cell Phenotypes
When cells undergo phenotypic transitions, it is expected that global, but specific changes in gene expression take place. The ability of SATB1 to make many gene loci accessible to chromatin remodeling/modification and transcription factors allows it to control differentiation of specific cell lineages and cell functions. For example, SATB1 is required for development of thymocytes[39] and activation of Th2 cells[26]. Without SATB1, thymocyte development is blocked at CD4+CD8+ (double-positive) stage. Similarly, without SATB1, Th2 cells cannot become activated and produce cytokines. SATB1 is also functionally linked to Wnt–b-catenin signaling[51, 52], which is required for thymoctye development and lineage commitment of naïve T cells[53]. SATB1 implements the effects of Wnt by binding to b-catenin and recruiting it to SATB1 target loci, where b-catenin activates transcription. By this mechanism, SATB1 upregulates expression of GATA3[52], which induces Th2 lineage commitment of naïve T cells[54]. Of note, during Th2 activation, expression of another factor required for cytokine expression, c-Maf[55], also depends on SATB1[26]. Repression of SATB1 by Foxp3 in T regulatory (Treg) cells, which mediate self-tolerance and immune homeostasis, is required for maintenance of their suppressive function and inability to produce effector cytokines after activation[56]. Once SATB1 is released from Foxp3 repression in Treg cells, they differentiate into T effector (Teff) cells and induce production of effector cytokines[56]. SATB1 has been linked to acute myeloid leukemia (AML). Disruption of the distal enhancer of PU.1 reduces its expression, leading to AML in mice[57, 58]. A SNP which is frequently associated with human acute myeloid leukemia (AML) was identified within the distal enhancer of the PU.1 gene, and this mutation specifically inhibits SATB1 binding, causing reduced PU.1 expression in myeloid progenitor cells[59]. SATB1 also has an important role in X-chromosome inactivation, which is mediated by non-coding Xist RNA in specific developmental contexts[60]. Using a thymic lymphoma model, SATB1 was found to be necessary and sufficient for gene silencing by Xist, indicating that SATB1 is an important determinant for X inactivation[60].
SATB1’s role in cell differentiation is not limited to the T-cell lineage. It was recently shown that p63 directly regulates SATB1 during development of the epidermis and that SATB1 regulates many epidermis-specific genes to control epidermal differentiation[61]. Furthermore, differentiation of mouse embryonic stem cells is impaired in the absence of Satb1[62]. These data indicated that SATB1 and SATB2, which have 98% homology at the amino acid level, might have antagonistic activities and that the relative levels of these proteins may regulate the balance of self-renewal and differentiation[62].
Satb1 also has a role in brain function. Satb1 is expressed in post-natal neurons and regulates temporal expression of immediate early genes during cortical development. Disruption of Satb1 reduces dendritic spine density in the cerebral cortex[63]. SATB1 is therefore an important regulator of many different cell types and functions.
6. SATB1 in Breast Cancer Metastasis
A role for SATB1 in breast cancer metastasis was first proposed because SATB1 is expressed in aggressive cancer cell lines, though it is absent or undetectable in normal and immortalized human mammary epithelial cells[14]. Immunohistochemical analysis of breast carcinoma specimens from patients showed that some specimens contained tumor cells with SATB1 in their nuclei, whereas other specimens had no tumor cells with detectable levels of SATB1. Most breast carcinomas expressing SATB1 were poorly differentiated, whereas adjacent normal tissues had no detectable expression of SATB1. This led to a model in which expression of SATB1 by breast cancer cells alters their expression profile to promote acquisition of an aggressive phenotype.
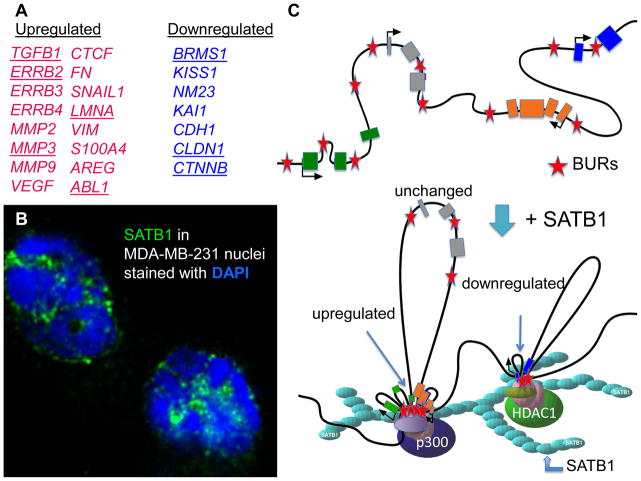
Studies of human breast cancer cells in culture and in mice (as xenograft tumors) revealed that SATB1 can function as a determinant for breast cancer metastasis[14]. SATB1 promoted tumor metastasis by regulating expression of ~1000 genes, predominantly those that control cell adhesion, signaling, extracellular matrix (ECM) formation, and the cell cycle[14]. In cells with increased levels of SATB1, genes that promote cancer progression and metastasis were upregulated, whereas those that suppressed metastasis were repressed. The genes regulated by SATB1 were identified by comparing the gene expression profiles of an aggressive breast cancer cell line (MDA-MB-231 cells) and MDA-MB-231 cells that were depleted of SATB1. SATB1 was found to upregulate the epidermal growth factor receptor (EGFR) genes ERBB1, ERBB2, ERBB3, and ERBB4, as well as the EGFR ligands NRG and AREG. ERBB2 (HER-2 or NEU) is the most oncogenic member of the ERBB2 family. Other genes upregulated by SATB1 and known to promote metastasis included those encoding metastasin (S100A4), vascular endothelial growth factor B (VEGFB), matrix metalloproteases (MMP2, 3, and 9), transforming growth factor-B1 (TGFB1), and connective tissue growth factor (CTGF). Genes found to be downregulated by SATB1 included suppressors of metastasis, such as BRMS1, KAI1, KISS1, NME1 (NM23), and E-cadherin (CDH1) (Figure 1).
Figure 1. SATB1 provides a nuclear architecture for large-scale gene regulation. This scheme illustrates that SATB1-target genes, but not non-target genes, are anchored to the SATB1 ‘cage-like’ structure via BUR sequences in each of these loci. The anchored gene loci will then be assembled with chromatin remodeling and modifying enzymes as well as transcription factors.
A) Representative list of genes upregulated (red) and downregulated (blue) by SATB1 expression in breast cancer cells is shown. Underlined genes are confirmed to be directly bound to SATB1 via BURs.
B) MDA-MB-231 cells grown on 3-dimensional matrix were immunostained for SATB1 (green) and DAPI (blue), exhibiting the ‘cage-like’ nuclear distribution of the SATB1 network.
C) SATB1 (light blue) binds to BUR sequences (red stars) within specific target genes (upregulated genes: orange and green; downregulated genes: dark blue; boxes represent exons) to create chromatin loops, which bring distal loci closer in proximity. This intricate organization of the chromatin, together with SATB1 recruitment of histone acetylase p300 to activated gene loci or histone deacetylase HDAC1 to repressed gene loci, results in the regulation of a multitude of genes in parallel. Some genes whose expression is SATB1-independent (gray) remained unbound to SATB1 even though they contain BURs.
Claudin family members, such as CLDN1, 3, 10 and 11, are repressed by SATB1[14]. The claudins are tight junction proteins that are either lost or mislocalized in invasive tumors[64, 65]. A more recently identified subgroup of breast tumors express low levels of claudin (claudin-low, 5%–10% of breast tumors)[66]. These tumors are characterized by their high grade, undifferentiated status (stem cell-like), features of the epithelial–mesenchymal transition (EMT), and frequent immune-cell infiltrate [66]. These are also features of tumors with SATB1-expressing cells. The relationship between SATB1 expression and the claudin-low subtype of tumors requires further investigation.
The expression profiling studies of SATB1-regulated genes revealed their overlap with those in the Rossetta poor-prognosis signature[10]: 231 genes that change in expression during bone[67] or lung metastasis[68]. But the most interesting information from the profiling studies of SATB1 was that SATB1 binds to and regulates a specific group of genes in breast cancer cells by anchoring BURs associated with each of their loci, leaving non-target genes unbound[14]. A significant portion of the large cohort of genes, if not all the genes, is expected to be directly tethered to the SATB1 nuclear architecture. In a study of randomly chosen loci that included SATB1-upregulated and -downregulated genes, all bound in vivo to SATB1 at the predicted BUR sites in their loci, whereas all genes whose expression did not depend on SATB1 remained unbound to SATB1, even though there were multiple BURs in each of their loci. Fig. 1 shows how genes are anchored onto the SATB1 nuclear architecture, mediated by BURs. Further studies are required to determine how this selectivity is achieved.
SATB1 binds not only to BURs in target genes in breast cancer cells, but recruits histone-modifying enzymes (such as p300 and HDAC1) to the BURs to establish region-specific epigenomic modification. p300 is recruited to genes upreguated by SATB1, to sustain a transcriptionally active histone mark (such as histone H3 K9/14 acetylation); when SATB1 is removed, the same sites are occupied by HDAC1, and acetylation of H3K9 is greatly reduced. Conversely, HDAC1 is recruited to their BURs of genes that are downregulated by SATB1. When SATB1 is removed, these BURs are occupied by p300, and acetylation of H3K9 is greatly increased. The mechanisms by which SATB1 determines the epigenomic status of specific genes is therefore similar to its gene-organizing activity in T cells.
Consistent with the activity of SATB1 in establishing a gene expression profile that promotes breast cancer progression, ectopic expression of SATB1 in non-aggressive breast cancer cells (e.g. SKBR3 and Hs578T cells) causes them to acquire aggressive phenotypes. Ectopic expression of SATB1 in these cells caused them to form large undifferentiated tumors in mammary fat pads, invade blood vessels, and survive in the circulation. They also acquired the ability to form lung metastases when injected into tail veins of mice.
Conversely, knockdown of SATB1 with small hairpin RNA in highly metastatic human breast cancer cells (e.g. MDA-MB-231 and BT549) reverses their morphology and causes them to form acinus-like structures in culture; these cells lose their invasiveness and ability to undergo anchorage-independent growth. Surprisingly, knockdown of SATB1 from MDA-MB-231 cells not only prevented them from forming lung metastases in mice, but also prevented tumor formation from cells injected into mammary fat pads, indicating a complete reversal of their aggressive phenotype. This complete reversal of metastatic and tumorigenic abilities was unexpected, because these cells have aberrant chromosomes (translocation and amplification) and mutations[69]. The accumulation of these mutations, over time, was previously believed to cause their aggressive phenotype. However, knockdown of a single protein, SATB1, was sufficient to reverse this phenotype in the model system used within the experimental time period observed.
Based on ability of SATB1 to promote metastasis of cancer cell lines, it was expected that expression of SATB1 by human breast tumors would be associated with poor prognosis. Analysis of 5–10 year follow-up records from 985 patients with ductal breast carcinomas showed that those with a large percentage of tumor cells with high levels of SATB1 in their nuclei (~6% of patient samples examined) had (shorter overall survival times. On the other hand, patients with no tumor cells that expressed SATB1 had better outcomes. Using 3-level scoring, based on the SATB1 levels and the percentage of tumor cells that express SATB1, SATB1 levels were correlated with patient survival times, independent of lymph node status[14]. A multivariate analysis of 1318 specimens, including all breast tumor types, showed that SATB1 is an independent prognostic factor for patients with breast cancer[14]. Although these results require validation with different antibodies and independent, large cohorts of patient samples, SATB1 has potential to be a useful prognostic marker, and might be used to predict the likelihood of tumor progression to metastasis in patients with breast cancer.
The role of SATB1 in breast cancer has been supported by independent studies. Highly invasive, multidrug-resistant (MDR) breast cancer cells were also found to have increased levels of SATB1 compared with non-resistant parental cells and SATB1 depletion partially reversed the MDR phenotype [70]. Another study reported roles of SATB1 in chemotherapy-induced EMT and progression of malignant tumors. Chemotherapy downregulates levels of microRNA (miR)-448, which was shown to target SATB1 mRNA. Therefore, suppression of miR-448 by chemotherapy could increase levels of SATB1 to promote the EMT[71]. In one study, higher levels of SATB1 mRNA were correlated with more advanced-stage breast carcinomas[72].
There have been some successful approaches to reduce the levels of SATB1 in breast tumors, with aims of reversing the malignant phenotype. In one report, specific DNA sequences that specifically bind SATB1 greatly reduced the invasive and metastatic capacity of MDA-MB-231 cells[73]. Another study found that overexpression of FOXP3 (which directly represses SATB1 in Treg cells) in an aggressive breast cancer cell line, BT549, not only directly repressed SATB1 but also induced miR-7 and miR-155, which target the 3′UTR of SATB1 to repress its expression[74]. These findings provide many new and interesting therapeutic approaches for breast cancer.
7. SATB1 Regulates Different Genes in Different Cell Types
SATB1 roles vary among cell types and processes. The genes regulated by SATB1 during epidermal differentiation are expected to differ from those involved in breast cancer metastasis, because of great differences in these cell types and their environments. Bioinformatic analyses of SATB1-dependent expression have been performed to determine differences and similarities among different systems.
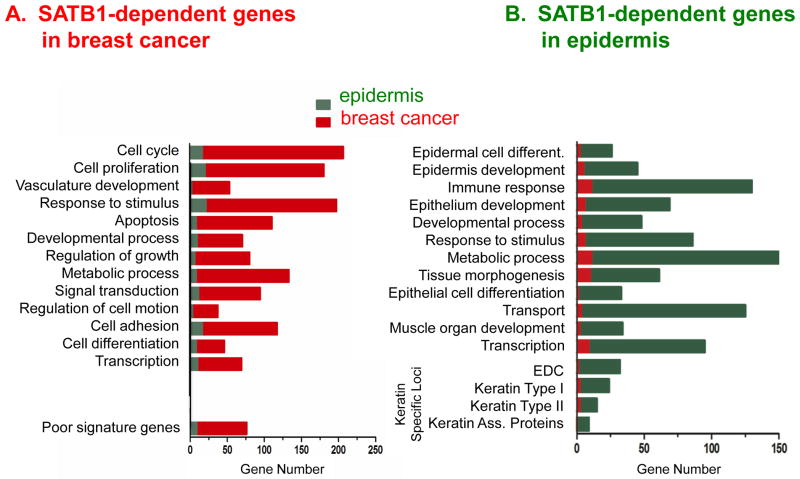
The David functional annotation tool and in-house R script were used to analyze gene ontology (GO) functional enrichment for published expression profiles of breast cancer cells and mouse primary keratinocytes with SATB1 knockout[61]. For each cell type, genes that changed at least 2-fold in expression level when SATB1 was lost, were selected for further analysis. Genes were annotated to GO biological process terms using the David tool, and R scripts were then used to filter redundant ontologies by combining hierarchical related and highly overlapped functional categories and to assign enrichment probability for functional groups. The final sets include 13 functional categories for SATB1-dependant genes in breast cancer cells and 12 for SATB1-dependent genes in epidermis development.
The top statistical-enriched ontologies of genes regulated by SATB1 in breast cancer cells were totally different from those in epidermal differentiation (Figure 2). In breast cancer cells, SATB1 regulated expression of genes involved in proliferation, the cell cycle, cell proliferation, vasculature development, cell adhesion—all of which are relevant to metastasis. In each of these categories, genes regulated by SATB1 during epidermis differentiation were greatly under-represented. In addition, genes whose expression was altered by silencing of SATB1 in breast cancer cells were significantly enriched in combined set of poor-prognosis signature genes[14], but were not among the genes altered in primary keratinocytes. On the other hand, during epidermis differentiation, genes regulated by SATB1 encoded factors involved primarily in differentiation and development. Those genes were found to be highly over-represented among keratin-specific loci (epidermis differentiation complex, keratin types I and II, and keratin -associated proteins). In all categories of SATB1-regulated genes involved in epidermis differentiation, those associated with cancer metastasis were greatly under-represented. Even under the GO category of transcription, those 2 sets of genes had minimal overlap. This bioinformatic analysis revealed that SATB1 regulates specific groups of genes in different cell types, and that these genes are associated with the particular activities of the cell. Therefore, the role of SATB1 appears to be to provide a nuclear architecture to which specific genes are tethered and assembled with transcriptional complexes, to mediate specific phenotypic changes in cells. Studies are needed to determine how SATB1 regulates specific sets of genes (and processes) in different cell types.
Figure 2.
The limited overlap in genes targeted by SATB1 between breast cancer and epidermis suggests that SATB1 regulates distinct sets of genes depending on the cell type. Published gene expression profiles in breast cancer (A, red) and in epidermis (B, green) were used to identify SATB1-dependent genes, which were then grouped according to basic GO functional groups. The top most represented groups for each cell type are shown. Note the difference in GO groups that were highly represented for each cell type and the lack of common SATB1-dependent genes with in each GO group between breast cancer and epidermis.
8. Functions in Different Tumor Types
It will be important to determine if SATB1 promotes metastasis of other tumor types. SATB1 is expressed in many different adult progenitor cells, as well as in embryonic stem cells. From 2010 to 2012, a number of papers reported roles for SATB1 in different types of cancer, including laryngeal squamous cell carcinoma (LSCC)[75], endometrioid endometrial cancer (EEC)[76], hepatocellular carcinoma (HCC)[77, 78], rectal cancer[79], cutaneous malignant melanoma (CMM)[80], and gastric cancer[81, 82]. Based on immunohistochemical analyses of tumor samples, the level of SATB1 was found to have prognostic and clinicopathological significance for CMM (based on 97 tissue samples—47 primary, 15 metastatic, and controls) and gastric cancer (based on 118 tumor samples—66 with lymph node metastasis and 16 with distant metastasis). High levels of SATB1 in tumor cells correlated with metastasis of CMM and gastric cancer. Kaplan-Meier analysis associated high levels of SATB1 with reduced survival times of patients with CMM[80] or gastric cancer[82]. Mutivariate analysis showed that SATB1 is an independent prognostic marker for CMM[80] and for gastric cancer[82], as it is for breast cancer[14]. For gastric cancer, high levels of SATB1 mRNA were also shown to be well correlated with high metastatic potential and shorter survival of patients[81] in agreement with results from immunohistochemical studies.
Similar results were observed for rectal cancer (based on 93 paired samples of rectal cancer vs normal tissues) [79] and LSCC (based on 80 samples of LSCC and 25 of control mucosa)[75]. Immunohistochemical studies correlated levels of SATB1 protein with depth of rectal tumor invasion and metastasis [79]. SATB1 was detected in LSCC, but not in control tissues, where protein and mRNA levels were below the limit of detection[75]. High levels of SATB1 mRNA and protein were detected in HCC tissues (based on 45 pairs of tumor and non-tumor adjacent tissue samples) and in liver cancer cell lines with high metastatic potential[77]. SATB1 was also shown to promote tumor growth and metastasis when expressed in liver cancer cells. SATB1-mediated regulation (either directly or indirectly) of genes involved in cell cycle progression, inhibition of apoptosis, and induction of the EMT have been reported, and many overlap with those identified for breast cancer [77]. SATB1 mRNA expression was detected in the majority of liver tumor samples, but not healthy liver tissues, from patients with hepatitis B virus-related HCC[78]. Laser-capture microdissection followed by expression microarray of EEC samples, which allows for precise assessment of homogeneous cell populations, identified SATB1 as a gene whose transcript level was significantly upregulated during endometrial carcinogenesis[76].
Although studies in rectal tumors, LSCC, HCC, and EEC correlated expression of SATB1 with tumor development, studies in lung tumor tissues have reported contradicting results. One study reported increased levels of SATB1 mRNA in non-small cell lung cancers (NSCLCs), compared with healthy lung tissues, and even higher levels in metastatic NSCLCs[83]. Another correlated loss of SATB1 with NSCLC and reduced survival times of patients[84]. Further research is required to solve this discrepancy.
SATB2, which is a homolog of SATB1, might serve as a diagnostic marker for colorectal cancer (CRC)—especially when its level is combined with that of cytokeratin 20, based on immunohistochemical analysis of 1882 tumor specimens[85]. Two studies have reported that SATB2 might be a prognostic factor for patients with CRC (1 study of 146 samples[86] and another of 527 samples[87], based on immunohistochemical analyses). In contrast to SATB1, these studies associated reduced levels of SATB2 with poor prognosis[86] and consistently high levels of SATB2 with good prognosis[87] for patients with CRC. As it was suggested previously in embryonic stem cells [62], SATB1 and SATB2 may have antagonistic activities in colorectal cancer. This is an important area for further research, along with the roles of SATB1 in SATB2-low CRC cells.
Some studies have reported no association between SATB1 and breast cancer progression or prognosis[88, 89]. However, these findings were based on transcription analyses of whole breast tumor samples. Given that expression of SATB1 is not limited to tumor cells, studies of only transcripts (mRNAs) isolated from whole tumor specimens would not necessarily identify SATB1 as a prognostic marker. Activated lymphocytes, fibroblasts and macrophages, which can be found in some tumor regions, also express SATB1. Therefore, immunohistochemistry must be used to determine protein levels of SATB1 in the cell nuclei of each sample, and then each sample scored for the number of SATB1-positive tumor cells. This might be of particular importance for breast carcinomas; whereas, in some other tumor types such as gastric cancer, results from SATB1 mRNA and immunohistochemical analysis agreed well and correlated with the aggressive phenotypes of cancer and the outcome of patients [81, 82].
Based on an analysis of transcription data available online, Iorns et al. reported that levels of SATB1 mRNA did not change in human breast cancer cell lines, compared with non-tumorigenic cell lines[88], in contrast to the findings of Han et al[14]. However, large differences have been reproducibly detected in levels of SATB1 protein or mRNA between non-tumorigenic and aggressive breast cancer cell lines [90, 91]. Due to the high degree of homology between SATB1 and SATB2, it is important to validate the specificity of oligonucleotide probes and antibodies for SATB1 vs SATB2. Furthermore, phenotypes of cultured cells can change during passage and with freezing and thawing. This drift in cell phenotypes, which results from the selection of different cell subpopulations during culture, might account for some of the discrepancies among laboratories.
9. Future Directions
SATB1, as a genome organizer, reprograms the cell’s gene expression profiles and thereby allows it to rapidly change phenotypes. SATB1 acts like gene glue, providing a nuclear architectural platform for anchoring loci that bind to specialized genomic marks, or BURs. Amazingly, the genes regulated by SATB1 vary among cell types and cell functions, so there is much to learn about the mechanisms by which SATB1 anchors specific groups of genes. There is much evidence that SATB1 promotes progression of breast cancer and other tumor types, and that its level has prognostic significance. Interestingly, many genes whose expression is regulated by SATB1 are shared between breast and liver tumors[14, 77].
In breast tumor specimens, some but not all tumor cells express SATB1. It is important to determine whether the cells that express the high levels of SATB1 in primary carcinomas are those that are destined to metastasize. This would be an intriguing hypothesis, based on studies of human breast cancer cells that overexpress SATB1 in nude mice[14]. If this is the case, it might be possible to therapeutically target SATB1-expressing cells in early-stage tumors.
The nuclear distribution patterns and genes regulated by SATB1 change, depending on culture conditions (such as 2-dimensional vs 3-dimensional), indicating that SATB1 responds to signals from microenvironment. It will be interesting to study whether and how SATB1-expressing tumor cells influence the stromal cells to upregulate expression of SATB1, or vice versa. More research is needed to understand what controls the mRNA and protein levels of SATB1, along with its post-translational modification and activities in nuclear architecture formation. MicroRNAs have been reported to regulate levels of SATB1[56, 71, 74, 92, 93], and some post-translational modifications of SATB1, with biological significance, have been identified[51, 94]. Further studies are needed to examine levels of SATB1 in different subtypes of breast carcinomas, to see if these have additional prognostic significance.
Acknowledgments
We thank Kris Novak for critical reading of the manuscript. This work was supported by National Cancer Institute grants R37CA039681 and R01 CA146444 to T. K-S, NRSA fellowship(F32CA138109) to E.O. and CIRM Scholarship to H-J. Han. The work was also supported by Low Dose Radiation Research Program, US Department of Energy (DE-AC02-05CH11231).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–46. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tysnes BB, Bjerkvig R. Cancer initiation and progression: involvement of stem cells and the microenvironment. Biochim Biophys Acta. 2007;1775:283–97. doi: 10.1016/j.bbcan.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–63. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 5.Parker B, Sukumar S. Distant metastasis in breast cancer: molecular mechanisms and therapeutic targets. Cancer Biol Ther. 2003;2:14–21. doi: 10.4161/cbt.188. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2012;9:58–64. doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- 9.Prat A, Perou CM. Mammary development meets cancer genomics. Nat Med. 2009;15:842–4. doi: 10.1038/nm0809-842. [DOI] [PubMed] [Google Scholar]
- 10.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–8. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–45. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 14.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–93. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 15.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–8. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghirlando R, Felsenfeld G. Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J Mol Biol. 2008;376:1417–25. doi: 10.1016/j.jmb.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rydberg B, Holley WR, Mian IS, Chatterjee A. Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J Mol Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- 20.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 21.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 23.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 24.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–31. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–88. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 28.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 29.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, et al. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–83. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 35.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–15. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 36.van Driel R, Fransz PF, Verschure PJ. The eukaryotic genome: a system regulated at different hierarchical levels. J Cell Sci. 2003;116:4067–75. doi: 10.1242/jcs.00779. [DOI] [PubMed] [Google Scholar]
- 37.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–48. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–35. [PMC free article] [PubMed] [Google Scholar]
- 40.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 41.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, et al. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Di LJ, Lv X, Zheng W, Xue Z, Guo ZC, et al. Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster. PLoS One. 2009;4:e4629. doi: 10.1371/journal.pone.0004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amir RE, Zoghbi HY. Rett syndrome: methyl-CpG-binding protein 2 mutations and phenotype-genotype correlations. Am J Med Genet. 2000;97:147–52. doi: 10.1002/1096-8628(200022)97:2<147::aid-ajmg6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 44.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–60. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohwi-Shigematsu T, Kohwi Y. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 1990;29:9551–60. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- 48.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, et al. Biological significance of unwinding capability of nuclear matrix- associating DNAs. Science. 1992;255:195–7. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 49.Kohwi-Shigematsu T, Kohwi Y. Detection of non-B-DNA structures at specific sites in supercoiled plasmid DNA and chromatin with haloacetaldehyde and diethyl pyrocarbonate. Methods Enzymol. 1992;212:155–80. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- 50.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–5. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 51.Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, Galande S. Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol. 2009;29:1321–37. doi: 10.1128/MCB.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, et al. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 2010;8:e1000296. doi: 10.1371/journal.pbio.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gattinoni L, Ji Y, Restifo NP. Wnt/beta-catenin signaling in T-cell immunity and cancer immunotherapy. Clin Cancer Res. 2010;16:4695–701. doi: 10.1158/1078-0432.CCR-10-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 55.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–83. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 56.Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–30. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 58.Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38:1269–77. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- 59.Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117:2611–20. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, et al. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–16. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. 2011;194:825–39. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–38. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balamotis MA, Tamberg N, Woo YJ, Li J, Davy B, Kohwi-Shigematsu T, et al. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol Cell Biol. 2012;32:333–47. doi: 10.1128/MCB.05917-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang K, Yao HP, Wang MH. Activation of RON differentially regulates claudin expression and localization: role of claudin-1 in RON-mediated epithelial cell motility. Carcinogenesis. 2008;29:552–9. doi: 10.1093/carcin/bgn003. [DOI] [PubMed] [Google Scholar]
- 65.Tokes AM, Kulka J, Paku S, Szik A, Paska C, Novak PK, et al. Claudin-1, -3 and -4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 2005;7:R296–305. doi: 10.1186/bcr983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 68.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crepin M, Salle V, Raux H, Berger R, Hamelin R, Brouty-Boye D, et al. Steroid hormone receptors and tumorigenicity of sublines from breast tumor metastatic MDA-MB 231 cell line. Anticancer Res. 1990;10:1661–6. [PubMed] [Google Scholar]
- 70.Li QQ, Chen ZQ, Xu JD, Cao XX, Chen Q, Liu XP, et al. Overexpression and involvement of special AT-rich sequence binding protein 1 in multidrug resistance in human breast carcinoma cells. Cancer Sci. 2010;101:80–6. doi: 10.1111/j.1349-7006.2009.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen YY, et al. Involvement of NF-kappaB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ. 2011;18:16–25. doi: 10.1038/cdd.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patani N, Jiang W, Mansel R, Newbold R, Mokbel K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009;9:18. doi: 10.1186/1475-2867-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamayoshi A, Yasuhara M, Galande S, Kobori A, Murakami A. Decoy-DNA against special AT-rich sequence binding protein 1 inhibits the growth and invasive ability of human breast cancer. Oligonucleotides. 2011;21:115–21. doi: 10.1089/oli.2010.0277. [DOI] [PubMed] [Google Scholar]
- 74.McInnes N, Sadlon TJ, Brown CY, Pederson S, Beyer M, Schultze JL, et al. FOXP3 and FOXP3-regulated microRNAs suppress SATB1 in breast cancer cells. Oncogene. 2012;31:1045–54. doi: 10.1038/onc.2011.293. [DOI] [PubMed] [Google Scholar]
- 75.Zhao XD, Ji WY, Zhang W, He LX, Yang J, Liang HJ, et al. Overexpression of SATB1 in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2010;72:1–5. doi: 10.1159/000264777. [DOI] [PubMed] [Google Scholar]
- 76.Mokhtar NM, Ramzi NH, Yin-Ling W, Rose IM, Hatta Mohd Dali AZ, Jamal R. Laser capture microdissection with genome-wide expression profiling displayed gene expression signatures in endometrioid endometrial cancer. Cancer Invest. 2012;30:156–64. doi: 10.3109/07357907.2011.633290. [DOI] [PubMed] [Google Scholar]
- 77.Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, et al. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012 doi: 10.1111/j.1478-3231.2012.02815.x. [DOI] [PubMed] [Google Scholar]
- 78.Huang YK, Fan XG, Qiu F, Wang ZM. Genomics of hepatitis B virus-related hepatocellular carcinoma and adjacent noncancerous tissues with cDNA microarray. Chin Med J (Engl) 2011;124:2057–64. [PubMed] [Google Scholar]
- 79.Meng WJ, Yan H, Zhou B, Zhang W, Kong XH, Wang R, et al. Correlation of SATB1 overexpression with the progression of human rectal cancer. Int J Colorectal Dis. 2012;27:143–50. doi: 10.1007/s00384-011-1302-9. [DOI] [PubMed] [Google Scholar]
- 80.Chen H, Takahara M, Oba J, Xie L, Chiba T, Takeuchi S, et al. Clinicopathologic and prognostic significance of SATB1 in cutaneous malignant melanoma. J Dermatol Sci. 2011;64:39–44. doi: 10.1016/j.jdermsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Lu X, Cheng C, Zhu S, Yang Y, Zheng L, Wang G, et al. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep. 2010;24:981–7. doi: 10.3892/or.2010.981. [DOI] [PubMed] [Google Scholar]
- 82.Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu S, et al. Expression of SATB1 and heparanase in gastric cancer and its relationship to clinicopathologic features. APMIS. 2010;118:855–63. doi: 10.1111/j.1600-0463.2010.02673.x. [DOI] [PubMed] [Google Scholar]
- 83.Zhou LY, Liu F, Tong J, Chen QQ, Zhang FW. Expression of special AT-rich sequence-binding protein mRNA and its clinicopathological significance in non-small cell lung cancer. Nan Fang Yi Ke Da Xue Xue Bao. 2009;29:534–7. [PubMed] [Google Scholar]
- 84.Selinger CI, Cooper WA, Al-Sohaily S, Mladenova DN, Pangon L, Kennedy CW, et al. Loss of special AT-rich binding protein 1 expression is a marker of poor survival in lung cancer. J Thorac Oncol. 2011;6:1179–89. doi: 10.1097/JTO.0b013e31821b4ce0. [DOI] [PubMed] [Google Scholar]
- 85.Magnusson K, de Wit M, Brennan DJ, Johnson LB, McGee SF, Lundberg E, et al. SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol. 2011;35:937–48. doi: 10.1097/PAS.0b013e31821c3dae. [DOI] [PubMed] [Google Scholar]
- 86.Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM, et al. Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol. 2009;219:114–22. doi: 10.1002/path.2575. [DOI] [PubMed] [Google Scholar]
- 87.Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlen M, Ericson Lindquist K, et al. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931–8. doi: 10.1038/bjc.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iorns E, Hnatyszyn HJ, Seo P, Clarke J, Ward T, Lippman M. The role of SATB1 in breast cancer pathogenesis. J Natl Cancer Inst. 2010;102:1284–96. doi: 10.1093/jnci/djq243. [DOI] [PubMed] [Google Scholar]
- 89.Hanker LC, Karn T, Mavrova-Risteska L, Ruckhaberle E, Gaetje R, Holtrich U, et al. SATB1 gene expression and breast cancer prognosis. Breast. 2011;20:309–13. doi: 10.1016/j.breast.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 90.Kohwi-Shigematsu T, Han HJ, Russo J, Kohwi Y. Re: The role of SATB1 in breast cancer pathogenesis. J Natl Cancer Inst. 2010;102:1879–80. doi: 10.1093/jnci/djq440. author reply 80-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ordinario E, Han H-J, Furuta S, Heiser LM, Jakkula LR, Rodier F, Paul T, Spellman PT, Campisi J, Gray JW, Bissell MJ, Kohwi Y, Kohwi-Shigematsu T. ATM suppresses SATB1-induced malignant progression in breast epithelial cells. doi: 10.1371/journal.pone.0051786. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lena AM, Mancini M, Rivetti di Val Cervo P, Saintigny G, Mahe C, Melino G, et al. MicroRNA-191 triggers keratinocytes senescence by SATB1 and CDK6 downregulation. Biochem Biophys Res Commun. 2012 doi: 10.1016/j.bbrc.2012.05.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang S, Banerjee S, Freitas A, Cui H, Xie N, Abraham E, et al. miR-21 regulates chronic hypoxia-induced pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2012;302:L521–9. doi: 10.1152/ajplung.00316.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, et al. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–43. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]