Abstract
The age of most genes exceeds the longevity of their genomic and physiological associations by many orders of magnitude. Such transient contexts modulate the expression of ancient genes to produce currently appropriate and often highly distinct developmental and functional outcomes. The efficacy of such adaptive modulation is diminished by the high dimensionality of complex organisms and associated vast areas of neutrality in their genotypic and developmental networks (and, thus, weak natural selection). Here I explore whether epigenetic effects facilitate adaptive modulation of complex phenotypes by effectively reducing the dimensionality of their deterministic networks and thus delineating their developmental and evolutionary trajectories even under weak selection. Epigenetic effects that link unconnected or widely dispersed elements of genotype space in ecologically relevant time could account for the rapid appearance of functionally integrated adaptive modifications. On an organismal time scale, conceptually similar processes occur during recurrent epigenetic reprogramming of somatic stem cells to produce, recurrently and reversibly, a bewildering array of differentiated and persistent cell lineages, all sharing identical genomic sequences despite strongly distinct phenotypes. I discuss whether close dependency of onset, scope and duration of epigenetic effects on cellular and genomic context in stem cells could provide insights into contingent modulation of conserved genomic material on a much longer evolutionary time scale. I review potential empirical examples of epigenetic bridges that reduce phenotype dimensionality and accomplish rapid adaptive modulation in the evolution of novelties, expression of behavioural types, and stress-induced ossification schedules.
 |
Alexander Badyaev is a Professor of Evolutionary Biology at the University of Arizona. He works at the interface of evolutionary developmental biology and evolutionary ecology, with specific focus on the origin of adaptations. The central goal of his work is to understand the interplay of adaptation, contingency and randomness in the evolution of complex organismal forms and functions in vertebrates. Current research projects seek to reconcile adaptability and adaptations in physiological systems and variability and heritability in carotenoid-based colour displays.
Inheritance: historical scaling of a key evolutionary concept
Inheritance is a puzzling concept. On the one hand it is the central tenet of the theory of organismal evolution (reviewed in Jablonka, 2001; Jablonka & Lamb, 2006; Uller & Helanterä, 2014), on the other hand the concept of heredity or the topic of evolutionary retention is barely mentioned in major reviews in evolutionary genomics and developmental genetics among other major players in the evolutionary process (e.g. Davidson, 2006; Lynch, 2007; Koonin, 2011). Yet inheritance and, in particular, explicit separation of its genetic and epigenetic (‘above gene’) components is perceived to be of major importance for the modern theory of evolution (Helanterä & Uller, 2010; Day & Bonduriansky, 2011; Uller, 2013). Such dichotomy in the treatment of inheritance partly stems from the fact that inheritance presumes an existence of something to be inherited, which is usually an element of an organism's function, genomic sequence, epigenome, or development. And, it is in this context that inheritance, in an empirical sense, picks up elements of either natural selection, adaptation-specific or lineage-specific development, or tissue-specific modification of transcription that gives it its evolutionary importance; this explains why, over the history of evolutionary thought, the concept of inheritance has been repeatedly and interchangeably merged with either long-term natural selection or development (Badyaev, 2011).
Fundamentally, regardless of the transmission mode or level of organization, the phenomenon of inheritance refers to a limitation of variation that could be potentially expressed in the next generation. This principle applies to all types of inheritance, whether it is aggregation of genomic determinants of taxa-specific development, DNA imprinting that influences gene activity, epigenetic modulation of ontogenetic trajectories based on the environment of past generations, or cultural inheritance of a subset of local dialects. This principle raises two general questions: first, what determines the limits and duration of inheritance and its content, and second, how can inheritance, which consistently limits variability, be reconciled with evolutionary diversification? Darwin (1859) had a clear answer to the first question: in his view organismal functioning itself determined the limits of inheritance and generated heritable variation by disturbing existing adaptations (extended by Baldwin, 1902; Schmalhausen, 1969), such that the historical experience of a lineage determines the range of its heritable variation. The importance of historical contingency in determining the range of genetic inheritance was further developed by Dobzhansky (1974) and Schmalhausen (1938). A similar perspective is widely used in epigenetics literature where duration of genomic sequence modification (e.g. through DNA imprinting by methylation, histone or higher order chromatin modifications) is often either directly related to time-keeping since the inducing event (e.g. epigenetic marks are lost or ‘diluted’ passively as a function of cell division without epigenetic marks maintenance) or directly maintained by cells or tissues during functioning (Rando & Chang, 2012; Smith & Meissner, 2013).
The second general question has been more difficult to address. During the formation of the Modern Synthesis, the debate centred on reconciling environmental contingency of development and functioning with long-term persistence of organismal features (Mayr & Provine, 1980). Such debate shifted the focus from the functional to transmission mode of inheritance, ultimately culminating in the view that each adaptive feature is genetically unique and a product of long-term acumination of small genetic differences, where genes can be viewed as ‘keepers of adaptations’, such that ‘the search for homologous genes is quite futile, except in very close relatives’ (Mayr, 1963). The extent to which this central assumption of the Modern Synthesis turned out to be empirically incorrect is striking: ‘The typical time of decay of genomic sequence similarity between homologous genes is comparable with the time of life's existence on Earth’ (Koonin, 2011). Recent discoveries from comparative genomics, particularly of the extremely ancient nature of many genes (i.e. their orthologous lineages) compared to the age of genomes in which they function (Tatusov 2003; Wolf et al. 2009), calls for a re-examination of the nature of inheritance in complex organisms and their highly specialized adaptations (Fig. 1). Another challenge comes from the realization that, on an organismal time scale, epigenetic reprogramming of cells with identical genomic material routinely produces the level of cell and tissue divergence comparable with those of extensive evolutionary radiations (e.g. mature neuron vs. epithelial cell) often in a highly context-specific manner. Ironically, if epigenetic effects are what facilitate adaptive modulation of ancient or identical genetic material on an ecological time scale, then Darwinian evolution by natural selection that requires the inheritance of context-specific gene expression might be, to a great extent, enabled by epigenetic effects and their inheritance (Oyama, 2000; Badyaev & Uller, 2009).
Figure 1. The discrepancy between the time scales of the longevity of genes, genomic associations, gene functions and gene expression states calls for examination of the place and duration of heredity phenomena (horizontal line).
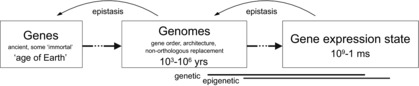
The placement of lines on the figure assumes that heredity occurs at the same time scale as a function (in the sense of phenotypic contribution to fitness). Distinct fitness consequences of the same gene combinations and expression in different contexts (epistasis: backward lines) maintains cohesiveness of the sequence despite widely distinct time scales on which its elements operate. Heredity, then, is enriched with assumptions of the past contexts (genetic or epigenetic) in which the gene functioned.
Although, implicitly, inheritance is often taken to mean genomic inheritance, in an empirical sense it is necessarily a combination of reliably transferred developmental resources needed to reconstruct, express and modify genetically and epigenetically inherited components in a lineage (Fig. 2A). The statistical framework of quantitative genetics can sometimes distinguish among some of these components in terms of their transgenerational stability, directionality and duration (Lynch & Walsh, 1998; Tal et al. 2010). Distinguishing between the most stably inherited epigenetic components of an adaptation, especially those associated with transcription machinery and its genomic components, is possible in systems where these components can be studied directly (e.g. Gerstein et al. 2012). In such systems, it is often found that epigenetic and genomic components form long-term compensatory interactions (e.g. as in sequence-driven methylation imprints Rando & Chang, 2012). When such associations escape decoupling over multiple generations, as is common in some taxa (e.g. plants), heritability of complex adaptations can be overwhelmingly due to inheritance of epigenetic components (e.g. Cortijo et al. 2014).
Figure 2. Inheritance in relation to its epigenetic and genetic components, functionality, pluripotency and organismal complexity.
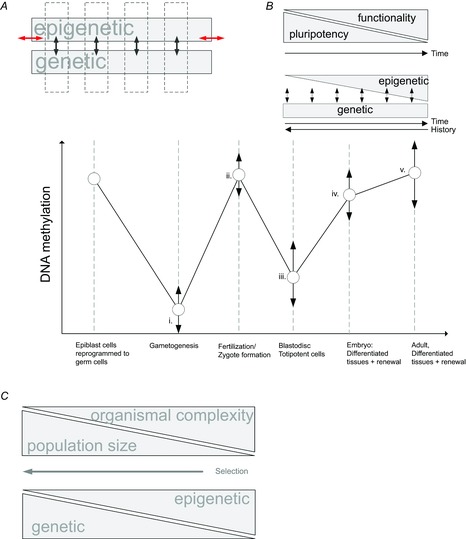
A, inherited ‘states’ (vertical arrows) (e.g. disease expression) arise as a combination and alignment (horizontal arrows) of genetic and epigenetic components, enabling evolution by natural selection. B, developmental and repair processes start with pluripotency that is reduced to accomplish functionality and specialization (upper inset). Greater and context-specific epigenetic modification of the genomic component accomplishes such specialization. Graph shows ontogeny of the setting and resetting of epigenetic marks using the example of DNA methylation (Smallwood & Kelsey, 2012; Smith & Meissner, 2013). Primordial germ cells undergo extensive demethylation during their migration from already differentiated epiblast tissues (e.g. in birds and mammals) to the site of future gonads; there germ cells acquire de novo methylation, followed by post-fertilization demethylation and subsequent tissue-specific methylation needed for normal cell differentiation. During recurrent regeneration of adult tissues, somatic stem cells undergo additional bouts of methylation. The rate and time of methylation setting and removal vary strongly (vertical arrows) depending on the cellular context in which it occurs (e.g. sex, tissue type, or age) providing extensive opportunity for epigenetic imprints to escape erasure in relation to factors of biological significance (e.g. sex-specific imprints, previous experience, or stress). For example, duration of de novo methylation of male and female gametes (i) differs by orders of magnitude (e.g. in mammals methylation of spermatogonia stops before birth, but methylation of oocytes continues until ovulation). Similarly, post-fertilization demethylation (ii), that is often under maternal control, is slow and typically passive (e.g. a consequence of cell division without imprint maintenance); extensive overlap with subsequent tissue-specific methylation (iii) enables many of the maternal imprints to escape erasure and to affect patterns of embryo tissue methylation. In contrast, the resetting of paternal imprints is an active and rapid process. During embryonic and adult stages (iv and v), the rate with which somatic stem cells acquire methylation varies widely with environmental impact on adult tissues (e.g. stress or injury), age and surrounding tissues. A greater context-dependency of epigenetic effects results in their increase throughout ontogeny, fewer shared contexts throughout the history of a lineage lead to their lesser importance over longer time spans. C, smaller population sizes and longer generation times lead to weaker purifying selection and thus greater organismal complexity. This raises a possibility that the overwhelming importance of epigenetic effects in the development and functioning of many organisms is an inevitable consequence of their complexity and associated necessity of producing thousands of different cell types from a single totipotent zygote. If organismal complexity has fewer dimensions in the epigenome than in the genome, then epigenetic modulation of a complex organism might be more effective over ecological time scales than genomic regulation, the phenomena well exemplified by stem cell dynamics.
Further, recent discoveries from developmental genetics, in particular work on somatic (‘adult’) stem cells that produce recurrent within-generation regeneration of functionalized tissues, emphasize that plasticity and totipotency is an ancestral state in organismal development and function and that specialized adaptations and context-dependent functionalization and differentiation of cells with identical genomic sequence is produced by narrowing and modulating such pluripotency (Fig. 2B), largely by epigenetic reprogramming (Nakaki et al. 2013; Smith & Meissner, 2013; Obokata et al. 2014a). Most tissues in adult animals harbour a population of somatic stem cells that retain their tissue-specific pluripotency and recurrently and reversibly produce lineages of highly phenotypically distinct and highly persistent cells. In many systems, the transition from totipotent embryonic stem cells to pluripotent somatic stem cells to specialized cells can be bi-directional and experimentally induced by ecologically relevant cues (Gafni et al. 2013; Rais et al. 2013; Obokata et al. 2014b). Thus, epigenetic modulation enables these cells to repeat ontogenetic development from a pluripotent state to a highly functionalized state during organismal life (Fig. 2B), such that a replacement of a particular bone, or an element of beak, or a feather modification that, at the phenotypic level, is a precise contemporary adaptation, is accomplished repeatedly during the organism's lifetime by the setting and resetting of epigenetic imprints on an identical DNA sequence in either somatic stem cells or differentiated cells (Ito et al. 2007; Conrad et al. 2008; Kim et al. 2010; Obokata et al. 2014b). Some elements of epigenetic reprogramming, such as DNA methylation and demethylation, can be under genomic control or activated by transcription itself (Smallwood & Kelsey, 2012; Kelsey & Feil, 2013). In either case, the mechanisms by which an interplay between genomic and epigenetic elements reliably recreates context-specific modification of the phenotype within a generation is of great interest to evolutionary biology in general and to our understanding of the mechanics of inheritance in particular (Fig. 2B). Particularly relevant in this context are findings that such reprogramming can be directly linked with an organism's experience of local ecological conditions or age (e.g. Adkins et al. 2011; Teschendorff et al. 2013), or guided by other elements of the phenotype, such as by reciprocal interactions between adjacent tissues and traits that provide each other's ‘environments’ during development, in a process that might be akin to ‘developmental epistasis’ (e.g. Newman, 2012; Badyaev & Walsh, 2014).
Interplay between universal rules and transient contingency in the evolution of complex traits
The current biological function of genes arises from the complementary interplay between universal rules that guide gene and genome evolution (e.g. cost of replication, regulation, protein robustness) and their contingent modulation by the transient contexts of phenotypes and genotypes. Retention of past contingencies in newly formed genomes, and thus accumulated complexity of organisms, is thought to be proportional to the efficacy of purifying selection exerted by both new contexts and the universal rules (Lynch, 2010; Lynch & Abegg, 2010), such that increasing complexity of structures constrains their optimization for current functions resulting in a ‘complexity catastrophe’ or ‘curse of dimensionality’ (Kauffman & Levin, 1987). The constraint emerges from an increase in neutrality of vast areas of genotypic and fitness landscapes that themselves are a consequence of their dimensionality (many genotypes having an identical phenotype) (Gavrilets, 2004). Although greater areas of neutrality can sustain greater explorative evolution without modifying the existing phenotype, they retard effective modification of the phenotype by decreasing the probability of encounter of evolutionary innovation by chance on time scales that are most relevant to natural, often small, populations (Kauffman, 1969; Gavrilets, 2004).
How constraining the dimensionality of genomic networks is for adaptive modifications depends on several factors (Gavrilets, 2004; Wagner, 2011). First is the size of the smallest evolutionary step (e.g. mutational step) that can reach genotype areas conferring different fitness without leaving the current phenotypically invariant network (e.g. the step that enables both exploratory evolutionary search for innovation and preservation of the existing phenotype) (Waxman & Peck, 1998; Wagner et al. 2008). Central to this is the distribution and connectivity of genotype areas conferring different fitness (Maynard Smith, 1970; Gavrilets, 2004; Carneiro & Hartl, 2010; Draghi et al. 2010). When such areas form a connected network that can be reached by the smallest mutational steps in ecologically relevant time, the time and speed of adaptive evolution is accelerated. Second is the retention of previous adaptive solutions within such a connected network, such as when a population experiences distinct, but partially overlapping environments over evolutionary time (‘the ghosts of environments past’). Such exaptations could act as stepping stones in adaptive evolution (Chetverikov, 1926; Stebbins & Hartl, 1988; Badyaev, 2007; Wagner, 2011). Third is the possibility of functional integration between novel elements and an existing well-adapted phenotype, a feature accomplished by robustness of underlying deterministic networks (Waddington, 1953; Siegal & Bergman, 2002; Draghi et al. 2010) and features of organismal homeostasis (West-Eberhard, 2005; Badyaev, 2013). Taken together, such constraints result in a majority of potential evolutionary pathways being interrupted by areas of very low fitness thus significantly reducing the evolutionary dimensionality of the phenotype, that is, the evolutionary pathways available for evolutionary change (Poelwijk et al. 2007; Breen et al. 2012).
Here I propose that, by acting at a different level of organization, epigenetic modulation of genomic networks can act as short-term bridges across areas of low fitness or over absent (or not accessible) genomic connectivity and thus can extend the time available for adaptive evolution and increase its speed, partially overcoming constraints imposed by the curse of dimensionality (Fig. 3A). This can be accomplished when epigenetic effects (1) change the size of phenotypically invariant networks and therefore increase the speed and gait of the smallest step available to encounter phenotypic innovation (see also Geoghegan & Spencer, 2013a; Klironomos et al. 2013; Furrow & Feldman, 2014), (2) provide a buffer of phenotypic plasticity that gives populations time to cross low-fitness gaps (by either finding previous solutions or forming new ones in ecologically relevant time) (Feinberg & Irizarry, 2010; Espinosa-Soto et al. 2011; Roux et al. 2011; de Vos et al. 2013), (3) expose links between genomic elements or developmental stages that were either not available or not accessible before (e.g. expression of previously methylated sequences during resetting of epigenetic imprints or in compensatory interactions between epigenetic and genetic elements (Rando & Chang, 2012), (4) lower ‘barriers’ that separate the pathways of stem cell differentiation emerging from gene network connectivity by exposing newly available pathways of differentiation or reversing their directionality (Kauffman, 1969; Huang et al. 2009), or (5) accomplish functional integration among newly encountered elements by combining exaptations from different environments (Geoghegan & Spencer, 2013b) or environments of different generations (Cowley & Atchley, 1992; Badyaev, 2008). Overall, such effects predict that epigenetic modulations should produce phenotypically invariant networks that are larger, but have lesser dimensionality compared to their genomic counterparts (Badyaev & Walsh, 2014). If so, then epigenetically delineated evolutionary pathways should enable rapid and drastic short-term modulation of genotypes, such as seen in maternal effects, developmental polymorphisms and phenotypic plasticity.
Figure 3. Epigenetic reduction of dimensionality of lower lever deterministic networks.
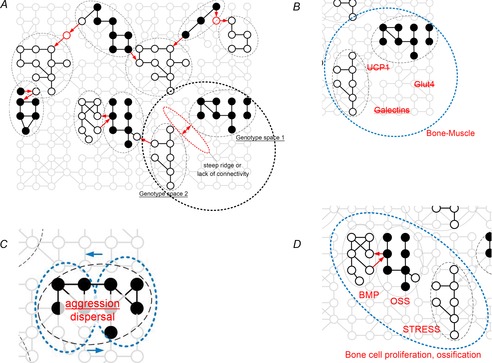
A, epigenetic bridges can cross unconnected or widely dispersed elements of deterministic networks of lower level (e.g. genomics networks) thereby facilitating adaptive evolution on an ecological time scale. B, epigenetic effects (large ellipsoid) of muscle–ossification interface encompass previously unconnected genomic areas to compensate for successive losses of genes under the thermogenesis hypothesis for the origin of birds. UCP1, uncoupling protein 1; Glut4, insulin-responsive glucose transporter. C, epigenetic effects ‘subsample’ elements of distinct behavioural strategies to produce rapid matching of morphs to prevalent contexts without eroding the genetic integration of components of complex behavioural phenotypes in bluebirds. D, epigenetic recruitment and integration of novel pathways of calcium synthesis under stress-induced growth in house finches. BMP, bone morphogenetic protein; OSS, ossification.
Origin and evolution of epigenetic effects
The view of epigenetic effects as bridges reducing distances and dimensionalities in genomic or other deterministic networks calls for explicit discussion of their origin – one of the most neglected topics of epigenetics. Are current epigenetic effects emergent properties of organismal complexity, such as aggregation of components of exaptations that retain their environmental sensitivities and thus can acquire function in some environments? Have epigenetic modulations evolved in an entirely different context? (e.g. for the silencing of transposable elements or gene copies, or the necessity to produce distinct tissues from a single cell in multicellular organisms) that is only secondarily coopted for other functions, such as maintenance of contemporary adaptations? What is the evolutionary future of epigenetic effects that escape ‘resetting episodes’ during germ cell formation and post-fertilization (Fig. 2B)? Are such effects eventually replaced in organismal organization by genomic effects once those are encountered or have time to evolve? Is the persistence of epigenetic effects through periods of resetting and reprogramming increased once they form functional associations with the underlying gene network or additional phenotypic elements of an organism?
If a population genetic framework is applicable to the evolutionary dynamics of epigenetic effects, especially in relation to the efficacy of natural selection, then we can predict that in complex organisms or small populations epigenetic effects and associations would be easier to gain than to lose, despite transgenerational resetting (Fig. 2C), making their evolutionary accumulation likely. That should, in turn, lead to selection for their homeostatic accommodation. Further, larger neutral networks accomplished by epigenetic effects can shield genomic elements from selection, whereas changes in the intensity of natural selection driven by fluctuations in population sizes can lead to alternation of neutral and adaptive evolution and thus contribute to the ‘resetting’ of epigenetic effects between selective environments or contexts in which they are expressed.
Hypothetical examples of epigenetic resolution of the ‘curse of dimensionality’
Rescue of gene loss effects by epigenetic networks
The thermogenetic muscle hypothesis (Newman, 2011) seeks to explain extraordinary hyperplasia and diversification of avian musculature and skeleton. It proposes that modern birds originated from an ancestral population that underwent successive episodes of loss of genes associated with thermogenesis, myogenesis and skeletogenesis. Some of these losses show phylogenetic signatures of newly disconnected genotype spaces (e.g. most genomic elements are present but no longer form a functional pathway) (Mezentseva et al. 2008), with each loss setting the stage for strong selection for rescue effects. Thus the loss of the gene for uncoupling protein 1 (UCP1), responsible for the generation of heat in brown adipose tissue, leads to the shift of avian thermogenesis to muscle tissues and associated muscle hyperplasia. Muscle hyperplasia, in turn, is partially caused by the loss of the insulin-responsive glucose transporter Glut4 enabling birds to repurpose insulin and glucose as muscle growth factors. In turn, muscle expansion was associated with the massive loss of genes in the galectin family resulting in the loss of redundant regulation of skeletogenesis and corresponding exceptional diversification of the avian skeleton in response to external stress exerted by muscle hyperplasia (Newman et al. 2013). The remarkable effect of muscle activity on patterns of avian ossification, a common epigenetic effect (Newman & Müller, 2005), can be demonstrated empirically, where the extent of development of bird-specific skeletal elements is proportional to the extent of muscle paralysis during ontogeny (reviewed in Newman et al. 2013). One potential explanation for the observed pattern is that each successive episode of gene loss could have been followed by compensatory epigenetic rescue effects directly capitalizing on the ossification–muscle growth interface (Fig. 3B) when strong fitness consequences of a novel mode of muscle thermogenesis could have favoured novel linkages among its contributors, eventually encompassing formerly unconnected genomic areas (e.g. those associated with glucose metabolism and skeletal formation).
Adaptive behavioural integration
Western bluebird (Sialia mexicana) males have two distinct behavioural phenotypes (‘morphs’) within a population (Duckworth, 2008). One morph shows high aggression, long natal dispersal, and limited parental behaviour, whereas the other is non-aggressive, does not disperse, and can raise nestlings in cooperation with relatives thus tolerating high population densities. Colonization of new environments is accomplished by the aggressive and dispersive morph; however, within a few generations, the population frequency of this morph declines and it is replaced by an increasing non-dispersing and non-aggressive morph (Duckworth & Badyaev, 2007). Eventually, a population runs out of breeding resources and the dispersing morph again increases in frequency and a population establishes in a new location.
The entire cycle is driven by the frequency of natural forest fires that create available successional habitats and takes less than 15–20 bluebird generations. Behavioural components of each morph phenotype are strongly genetically correlated (Duckworth & Kruuk, 2009), but the mechanism by which the frequency of the highly distinct and integrated complex phenotype is matched to the most appropriate conditions at such a short time scale is not known. Recent experimental work showed that maternal experience with nest site competition and associated elevation of maternal corticosterone during oogenesis affects hormonal allocation into growing oocytes, which in turn influences both their ovulation order (and therefore position in the hatching hierarchy) and behaviour of produced juveniles (Duckworth, 2009). Such effects of differential hormonal allocation can be caused by either induced oocyte selection before ovulation (e.g. Rando & Chang, 2012; West et al. 2013) or hormonal modification of oocyte DNA imprinting, as is found in other systems (Kelsey & Feil, 2013). How can epigenetic effects in this case enable phenotypic integration of only some elements of integrated phenotypes (Fig. 3C), accomplishing abrupt changes in the frequency of such phenotypes and their adaptive matching to the environment? Three factors make the involvement of epigenetic effects likely. First is the ubiquity of age- and experience-dependency of establishment and maintenance of DNA methylation in animals (Adkins et al. 2011; Teschendorff et al. 2013), such that variable allocation of hormones into growing oocytes depending on maternal age and experience with competition for nesting resources can have variable effects on patterns of DNA methylation during oocyte growth and maturation. Second is strong differences between the sexes in methylation and demethylation of their germ cells, gametes and embryo tissues (Smallwood & Kelsey, 2012) and, consequently, potential cycles of prevalence of maternally- versus paternally-set imprints depending on the demographic composition of a population (e.g. mostly young dispersing males and local females in the beginning of the cycle). Third is a pronounced cyclical change in genetic relatedness in such populations driven by patterns of dispersal (e.g. genetic relatedness of females to local males progressively increases as more male relatives are recruited into the population at later stages of the cycle) that could set a stage for alternation of strong epigenetic modulation (e.g. by DNA methylation imprinting) and its effective erasure during fertilization and development.
Stress-induced cooption of calcium signalling
The main source of nestling mortality in Sonoran Desert house finches (Haemorhous mexicanus) is exposure to nest mites (Badyaev et al. 2006; Hamstra & Badyaev, 2009). Breeding females accumulate mites when collecting nest material and infect their future nest site. During a breeding attempt that coincides with the infestation period, nestlings have a distinct ontogeny, growing their long bones up to 50% faster and earlier than nestlings of the same breeding pair during other times of the year, which enable these nestlings to leave infested nests earlier and minimize their exposure to mites (Badyaev et al. 2006). Such distinct growth trajectories are evident at the earliest embryonic stages, up to 2 weeks prior to hatching (and thus the nestlings’ first direct exposure to mites). We showed experimentally that chronic elevation of maternal baseline corticosterone resulted in its greater transfer to developing oocytes where, in turn, it triggered earlier activity of bone morphogenetic protein (BMP) genes, and lead to faster and earlier ossification. Comparison of RNA-seq profiles of transcribed and non-transcribed genes associated with the maternal stress-induced growth of the offspring revealed that faster ossification results mostly from recruitment of novel genetic pathways involved in Ca2+ signalling and only partially from upregulation of calcium synthesis in gene pathways associated with normal ossification (A. V. Badyaev, R. L. Young, K. P. Oh, E. A. Landeen, unpublished observations). Epigenetic effects in this case can recruit, expose, or integrate calcium biosynthesis from novel genetic pathways responding to corticosterone-mediated stress and enable faster ‘emergency’ growth (Fig. 3D).
Although speculative and requiring confirmatory tests of assumptions, these empirical examples nevertheless suggest that when epigenetic effects operate at a different scale of organization (spatial or temporal) from the genomic elements whose phenotypic outcomes they modify, such epigenetic effects can strongly facilitate modification of complex phenotypes to fluctuations in contemporary natural selection.
Acknowledgments
I thank Denis Noble, Gerd Müller, Eva Jablonka, Mike Joyner and Stig Omholt for their invitation to contribute and the editors and anonymous reviewers for comments and suggestions. I am also grateful to Rufus Johnstone, Ido Pen and Bram Kuijper for their invitation to participate in the symposium on ‘Non-genetic inheritance in evolution’, where some of the ideas outlined here were presented and discussed.
Additional information
Competing interests
None declared.
References
- Adkins RM, Thomas F, Tylavsky FA, Krushkal J. Parental ages and levels of DNA methylation in the newborn are correlated. BMC Med Genet. 2011;12:47. doi: 10.1186/1471-2350-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV. Evolvability and robustness in colour displays: Bridging the gap between theory and data. Evol Biol. 2007;34:61–71. [Google Scholar]
- Badyaev AV. Maternal effects as generators of evolutionary change: A reassessment. Ann N Y Acad Sci. 2008;1133:151–161. doi: 10.1196/annals.1438.009. [DOI] [PubMed] [Google Scholar]
- Badyaev AV. Origin of the fittest: Link between emergent variation and evolutionary change as a critical question in evolutionary biology. Proc Biol Sci. 2011;278:1921–1929. doi: 10.1098/rspb.2011.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV. ‘Homeostatic hitchhiking’: a mechanism for the evolutionary retention of complex adaptations. Integr Comp Biol. 2013;53:913–922. doi: 10.1093/icb/ict084. [DOI] [PubMed] [Google Scholar]
- Badyaev AV, Hamstra TL, Oh KP, Acevedo Seaman D. Sex-biased maternal effects reduce ectoparasite-induced mortality in a passerine bird. Proc Natl Acad Sci U S A. 2006;103:14406–14411. doi: 10.1073/pnas.0602452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV, Uller T. Parental effects in ecology and evolution: Mechanisms, processes, and implications. Philos Trans R Soc Lond B Biol Sci. 2009;364:1169–1177. doi: 10.1098/rstb.2008.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV, Walsh JB. Epigenetic processes and genetic architecture in character origination and evolution. In: Charmantier A, Garant D, Kruuk LEB, editors. Quantitative Genetics in the Wild. Oxford University Press; 2014. pp. 177–189. [Google Scholar]
- Baldwin JM. Development and Evolution. New York: Macmillan; 1902. [Google Scholar]
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 2012;490:535–538. doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Hartl DL. Adaptive landscapes and protein evolution. Proc Natl Acad Sci U S A. 2010;107:1747–1751. doi: 10.1073/pnas.0906192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverikov SS. On certain aspects of the evolutionary process from the standpoint of modern genetics. J Exp Biol A. 1926;2:1–40. [Google Scholar]
- Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, Aicher W, Bühring H, Mattheus U, Mack A, Wagner HH, Minger S, Matzkies M, Reppel M, Hescheler J, Sievert K, Stenzl A, Skutella T. Generation of pluripotent stem cells from adult human testis. Nature. 2008;456:344–349. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- Cortijo S, Wardenaar R, Colome-Tatche M, Gilly A, Etcheverry M, Labadie K, Caillieux E, Hospital F, Aury JM, Wincker P, Roudier F, Jansen RC, Colot V, Johannes F. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- Cowley DE, Atchley WR. Quantitative genetic models for development, epigenetic selection and phenotypic evolution. Evolution. 1992;46:495–518. doi: 10.1111/j.1558-5646.1992.tb02054.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. The Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press; 2006. [Google Scholar]
- Day T, Bonduriansky R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat. 2011;178:E18–E36. doi: 10.1086/660911. [DOI] [PubMed] [Google Scholar]
- de Vos MGJ, Poelwijk FJ, Battich N, Ndika JDT, Tans SJ. Environmental dependence of genetic constraint. PLoS Genet. 2013;9:e1003580. doi: 10.1371/journal.pgen.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Chance and creativity in evolution. In: Ayala F, Dobzhansky T, editors. Studies in the Philosophy of Biology. London: McMillan; 1974. pp. 307–338. [Google Scholar]
- Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth RA. Adaptive dispersal strategies and the dynamics of a range expansion. Am Nat. 2008;172:S4–S17. doi: 10.1086/588289. [DOI] [PubMed] [Google Scholar]
- Duckworth RA. Maternal effects and range expansion: A key factor in a dynamic process? Philos Trans R Soc Lond B Biol Sci. 2009;364:1075–1086. doi: 10.1098/rstb.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth RA, Badyaev AV. Coupling of dispersal and aggression facilitates the rapid range expansion of a passerine bird. Proc Natl Acad Sci U S A. 2007;104:15017–15022. doi: 10.1073/pnas.0706174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth RA, Kruuk LEB. Evolution of genetic integration between dispersal and colonization ability in a bird. Evolution. 2009;63:968–977. doi: 10.1111/j.1558-5646.2009.00625.x. [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Martin OC, Wagner A. Phenotypic plasticity can facilitate adaptive evolution in gene regulatory circuits. BMC Evol Biol. 2011;11:5. doi: 10.1186/1471-2148-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl. 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrow RE, Feldman MW. Genetic variation and the evolution of epigenetic regulation. Evolution. 2014;68:673–683. doi: 10.1111/evo.12225. [DOI] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton, NJ, USA: Princeton University Press; 2004. [Google Scholar]
- Geoghegan JL, Spencer HG. The adaptive invasion of epialleles in a heterogeneous environment. Theor Popul Biol. 2013a;88:1–8. doi: 10.1016/j.tpb.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Spencer HG. Exploring epiallele stability in a population-epigenetic model. Theor Popul Biol. 2013b;83:136–144. doi: 10.1016/j.tpb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamstra TL, Badyaev AV. Comprehensive investigation of ectoparasite community and abundance across life history stages of avian host. J Zool. 2009;278:91–99. [Google Scholar]
- Helanterä H, Uller T. The Price equation and extended inheritance. Philos Theor Biol. 2010;2:1–17. [Google Scholar]
- Huang S, Ernberg I, Kauffman S. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin Cell Dev Biol. 2009;20:869–876. doi: 10.1016/j.semcdb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar S, Costsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jablonka E. The systems of inheritance. In: Oyama S, Griffiths PE, Gray RD, editors. Cycles of Contigency: Developmental Systems and Evolution. Cambridge, MA, USA: MIT Press; 2001. pp. 99–116. [Google Scholar]
- Jablonka E, Lamb MJ. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. A Bradford Book; 2006. [Google Scholar]
- Kauffman S, Levin S. Towards a general theory of adaptive walks on rugged landscapes. J Theor Biol. 1987;128:11–45. doi: 10.1016/s0022-5193(87)80029-2. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos FD, Berg J, Collins S. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays. 2013;35:571–578. doi: 10.1002/bies.201200169. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press Science; 2011. [Google Scholar]
- Lynch M. The Origins of Genome Architecture. Sunderland, MA, USA: Sinauer Associates; 2007. [Google Scholar]
- Lynch M. Scaling expectations for the time to establishment of complex adaptations. Proc Natl Acad Sci U S A. 2010;107:16577–16582. doi: 10.1073/pnas.1010836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Abegg A. The rate of establishment of complex adaptations. Mol Biol Evol. 2010;27:1404–1414. doi: 10.1093/molbev/msq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA, USA: Sinauer Associates; 1998. [Google Scholar]
- Maynard Smith J. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Cambridge, MA, USA: Harvard University Press; 1963. [Google Scholar]
- Mayr E, Provine WB, editors. The Evolutionary Synthesis: Perspectives on the Unification of Biology. Cambridge, MA, USA: Harvard University Press; 1980. [Google Scholar]
- Mezentseva NV, Kumaratilake JS, Newman SA. The brown adipocyte differentiation pathway in birds: an evolutionary road not taken. BMC Biol. 2008;6:17. doi: 10.1186/1741-7007-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Newman SA. Thermogenesis, muscle hyperplasia, and the origin of birds. Bioessays. 2011;33:653–656. doi: 10.1002/bies.201100061. [DOI] [PubMed] [Google Scholar]
- Newman SA. Physico-genetic determinants in the evolution of development. Science. 2012;338:217–219. doi: 10.1126/science.1222003. [DOI] [PubMed] [Google Scholar]
- Newman SA, Mezentseva N, Badyaev AV. Gene loss, thermogenesis and the origin of birds. Ann N Y Acad Sci. 2013;1289:36–47. doi: 10.1111/nyas.12090. [DOI] [PubMed] [Google Scholar]
- Newman SA, Müller GB. Origination and innovation in the vertebrate limb skeleton: an epigenetic perspective. J Exp Zool B Mol Dev Evol. 2005;304:593–609. doi: 10.1002/jez.b.21066. [DOI] [PubMed] [Google Scholar]
- Obokata H, Sasai Y, Niwa H, Kadota M, Andrabi M, Takata N, Tokoro M, Terashita Y, Yonemura S, Vacanti CA, Wakayama T. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature. 2014a;505:676–680. doi: 10.1038/nature12969. [DOI] [PubMed] [Google Scholar]
- Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, Yamato M, Vacanti CA. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014b;505:641–647. doi: 10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- Oyama S. The Ontogeny of Information: Developmental Systems and Evolution. Durham: Duke University Press; 2000. [Google Scholar]
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, Mor N, Baran D, Weinberger L, Jaitin DA, Lara-Astiaso D, Blecher-Gonen R, Shipony Z, Mukamel Z, Hagai T, Gilad S, Amann-Zalcenstein D, Tanay A, Amit I, Novershtern N, Hanna JH. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Rando Thomas A, Chang Howard Y. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Colome-Tatche M, Edelist C, Wardenaar R, Guerche P, Hospital F, Colot V, Jansen RC, Johannes F. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics. 2011;188:1015–1017. doi: 10.1534/genetics.111.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhausen II. Organism as a Whole in Individual Development and History. Leningrad: Academy of Sciences, USSR; 1938. [Google Scholar]
- Schmalhausen II. Problems of Darwinism. Leningrad: Nauka; 1969. [Google Scholar]
- Siegal ML, Bergman A. Waddington's canalization revisited: Developmental stability and evolution. Proc Natl Acad Sci U S A. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stebbins GL, Hartl DL. Comparative evolution: Latent potentials for anagenetic advance. Proc Natl Acad Sci U S A. 1988;85:5141–5145. doi: 10.1073/pnas.85.14.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal O, Kisdi E, Jablonka E. Epigenetic contribution to covariance between relatives. Genetics. 2010;184:1037–1050. doi: 10.1534/genetics.109.112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift. Hum Mol Genet. 2013;22:R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uller T. Non-genetic inheritance and evolution. In: Kampourakis K, editor. Philosophy of Biology: A Companion for Educators. Springer Verlag; 2013. pp. 267–287. [Google Scholar]
- Uller T, Helanterä H. Heredity and evolutionary theory. In: Walsh H, Huneman, editors. Challenges to Evolutionary Theory. Oxford University Press; 2014. In press. [Google Scholar]
- Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:119–127. [Google Scholar]
- Wagner A. The Origins of Evolutionary Innovations: A Theory of Transformative Change in Living Systems. Oxford University Press; 2011. [Google Scholar]
- Wagner GP, Kenney-Hunt JP, Pavlicev M, Peck JR, Waxman D, Cheverud JM. Pleiotropic scaling of gene effects and the ‘cost of complexity’. Nature. 2008;452:470–472. doi: 10.1038/nature06756. [DOI] [PubMed] [Google Scholar]
- Waxman D, Peck JR. Pleiotropy and the preservation of perfection. Science. 1998;279:1210–1213. [PubMed] [Google Scholar]
- West-Eberhard MJ. Phenotypic accommodation: adaptive innovation due to developmental plasticity. J Exp Zool B Mol Dev Evol. 2005;304:610–618. doi: 10.1002/jez.b.21071. [DOI] [PubMed] [Google Scholar]
- West J, Widschwendter M, Teschendorff AE. Distinctive topology of age-associated epigenetic drift in the human interactome. Proc Natl Acad Sci U S A. 2013;110:14138–14143. doi: 10.1073/pnas.1307242110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 2009;106:7273–7280. doi: 10.1073/pnas.0901808106. [DOI] [PMC free article] [PubMed] [Google Scholar]


