Abstract
Discoveries in cytogenetics, molecular biology, and genomics have revealed that genome change is an active cell-mediated physiological process. This is distinctly at variance with the pre-DNA assumption that genetic changes arise accidentally and sporadically. The discovery that DNA changes arise as the result of regulated cell biochemistry means that the genome is best modelled as a read–write (RW) data storage system rather than a read-only memory (ROM). The evidence behind this change in thinking and a consideration of some of its implications are the subjects of this article. Specific points include the following: cells protect themselves from accidental genome change with proofreading and DNA damage repair systems; localized point mutations result from the action of specialized trans-lesion mutator DNA polymerases; cells can join broken chromosomes and generate genome rearrangements by non-homologous end-joining (NHEJ) processes in specialized subnuclear repair centres; cells have a broad variety of natural genetic engineering (NGE) functions for transporting, diversifying and reorganizing DNA sequences in ways that generate many classes of genomic novelties; natural genetic engineering functions are regulated and subject to activation by a range of challenging life history events; cells can target the action of natural genetic engineering functions to particular genome locations by a range of well-established molecular interactions, including protein binding with regulatory factors and linkage to transcription; and genome changes in cancer can usefully be considered as consequences of the loss of homeostatic control over natural genetic engineering functions.
 |
James A. Shapiro, author of the 2011 book Evolution: A View fromthe 21st Century, is Professor of Microbiology at the University of Chicago. He has a BA in English Literature from Harvard (1964) and a PhD in Genetics from Cambridge (1968). During a postdoctoral at the Institut Pasteur in 1968, he established insertion mutations in bacteria. In 1969, he and colleagues at Harvard Medical School used in vivo genetic manipulations to clone and purify the lac operon. With Bukhari and Adhya in 1976, he organized the first conference on DNA insertion elements. In 1979, Shapiro formulated a molecular model for transposition. In 1984, he showed that selection stress triggers transposon action. Since 1992, he has been writing about the importance of biologically regulated natural genetic engineering.
Introduction
A major accomplishment of cytogenetics and molecular biology in the 20th century was the revelation that genome repair and genome change are active cell processes. Cells write their own genome modifications (Shapiro, 2011, 2013). When pre-DNA neo-Darwinian assumptions dictated that mutations had to be random and accidental, it did not make sense to discuss the physiology of genetic changes. But now that we know about the regulated molecular processes that proofread, repair and modify genomic DNA, we can discuss the physiology of how cells protect the genome and write new genomic structures when appropriately stimulated.
The goals of this review will be (i) to acquaint physiologists with the wide array of regulated biochemical systems we have come to recognize that underlie both genome stability and genome change, and (ii) to relate those systems to the processes of homeostatic regulation (McClintock, 1984, 1987).
Replication proofreading and mismatch repair
Cells actively protect themselves from mistakes by the replication apparatus. There are at least two levels for which we know details of the error-avoidance systems.
Exonuclease proofreading
Cellular DNA replication complexes contain exonuclease activities that come into play when an incorrect base has been incorporated onto the nascent DNA strand (Perrino & Loeb, 1989; Fazlieva et al. 2009; Ibarra et al. 2009). The polymerase function of a replication complex arrests when a mismatched duplex has been formed so that the exonuclease activity can remove the most recently incorporated nucleotides. Following exonuclease excision, polymerization resumes to extend the nascent strand without misincorporation. Studies with Escherichia coli indicate that exonuclease proofreading removes about 99.9% of the accidental misincorporations from the nascent strand (Kunkel & Bebenek, 2000).
Post-replication mismatch repair
For those misincorporations that escape exonuclease proofreading, cells have a backup mismatch repair system (Modrich & Lahue, 1996; Hays et al. 2005; Jiricny, 2006; Modrich, 2006; Fukui, 2010). The mismatch repair system employs duplex monitoring molecules named MutS (the E. coli model) or a eukaryotic homologue, such as MutSH1–6 for humans. When MutS detects a mismatch, it recruits MutL (E. coli) or one of its eukaryotic homologues plus an endonuclease activity to cleave the newly replicated strand on either side of the mismatch, a helicase to remove the error-containing oiligonucleotide and a repair polymerase plus ligase to produce an error-free duplex. In E. coli, discrimination of new and template strands occurs by detecting DNA hemimethylations (Modrich & Lahue, 1996). It is not yet clear how the strands are discriminated in eukaryotes. Mismatch repair in E. coli removes about 99% of the post-replication incorporation errors (Kunkel & Bebenek, 2000).
In summary, exonuclease proofreading plus mismatch repair can reduce error-driven mutations by five orders of magnitude in E. coli (and presumably by a similar degree in other organisms). These two physiological processes are homeostatic and respond to molecular sensing of double helix distortions.
DNA damage repair systems
Genomes are sensitive to damage by a number of physical and chemical agents, including the reactive products of oxidative metabolism in all aerobic organisms (Walker, 2000; Guetens et al. 2002). A variety of repair systems have evolved to deal with these external sources of genome change. The homeostatic nature of these systems was recognized as early as the 1930s.
McClintock and X-ray-induced ‘mutations’
The first demonstration of cell-mediated repair of induced genome change arose out of McClintock's cytogenetic investigations of X-ray-induced maize mutants in the 1930s (McClintock, 1932). She studied the mutant strains collected by Stadler at the University of Missouri. Rather than the presumed ‘gene mutations’, she found that the mutants carried chromosome rearrangements. Some mutants had an unstable ‘variegating’ phenotype. McClintock hypothesized that variegation resulted from mitotic instability of ring chromosomes formed by joining of X-ray-induced breaks near the telomeres of a single chromosome. Although colleagues ridiculed her hypothesis, she demonstrated the postulated ring chromomes cytologically (McClintock, 1932) and went on to demonstrate that two broken chromosome ends are joined together with great efficiency in maize embryos (McClintock, 1939, 1942).
It is worth quoting McClintock's comments on these experiments in her 1983 Nobel Prize lecture because she emphasized homeostasis, cell monitoring of its genome and informed physiological responses to unpredictable genome damage events:
‘The conclusion seems inescapable that cells are able to sense the presence in their nuclei of ruptured ends of chromosomes and then to activate a mechanism that will bring together and then unite these ends, one with another… The ability of a cell to sense these broken ends, to direct them toward each other, and then to unite them so that the union of the two DNA strands is correctly oriented, is a particularly revealing example of the sensitivity of cells to all that is going on within them… There must be numerous homeostatic adjustments required of cells. The sensing devices and the signals that initiate these adjustments are beyond our present ability to fathom. A goal for the future would be to determine the extent of knowledge the cell has of itself and how it utilizes this knowledge in a “thoughtful” manner when challenged’ (McClintock, 1984).
Damage-specific repair systems
Joining two duplexes with double-strand (DS) breaks at their ends is today called ‘non-homologous end joining’ (NHEJ), and cells have various biochemical systems for carrying it out (van Gent et al. 2001; Pitcher et al. 2005; Bowater & Doherty, 2006; van Gent & van der Burg, 2007; Bennardo et al. 2008; Brissett & Doherty, 2009). NHEJ is but one of a series of biochemical systems that have evolved to protect genomes from many kinds of physical and chemical damage. The partial list in Table 1 gives an idea of how extensive and sophisticated the physiology of DNA damage repair has evolved to be. We will return later to the significance of error-free and error-prone repair modalities.
Table 1.
Some repair processes for distinct kinds of DNA damage (from Shapiro, 2013, with references added)
| DNA damage type or agent | Error-free repair | Error-prone/mutagenic repair | References |
|---|---|---|---|
| Ionizing radiation, double-strand (DS) breakage, replication fork collapse | Homologous recombination (Rec) | Non-homologous end-joining (NHEJ) | (Cox, 2001; Lusetti & Cox, 2002; Pastwa & Blasiak, 2003; Frankenberg-Schwager et al. 2008; Fattah et al. 2010; Kass & Jasin, 2010; Grabarz et al. 2012; Kurosawa et al. 2013) |
| UV radiation (thymine dimers) | Nucleotide excision repair (NER) | Lesion bypass repair | (Petit & Sancar, 1999; Tang et al. 1999; Sutton et al. 2000; Walker, 2000; Goodman, 2002; Rechkunova & Lavrik, 2010) |
| Alkylation damage | Base excision repair (BER), NER, dealkylation | Lesion bypass repair | (Fromme & Verdine, 2004; Robertson et al. 2009; Kondo et al. 2010; Jacobs & Schar, 2012) |
| Large chemical adducts (e.g. benzpyrene damage) | Lesion bypass repair (DNA PolIV and V in E. coli) | (Wagner et al. 1999; Napolitano et al. 2000; Goodman, 2002) | |
| Oxidative damage | BER | Lesion bypass repair | (Lu et al. 2001; Fromme & Verdine, 2004; Yamada et al. 2006; Wirtz et al. 2010; Jacobs & Schar, 2012) |
| Cytosine deamination to uracil | BER (uracil-n-glycosylase) | BER (uracil-n-glycosylase) | (Jacobs & Schar, 2012; Perez-Duran et al. 2012; Li et al. 2013) |
DNA damage checkpoints
Critical to the operation of DNA repair systems is the ability cells have to delay progress through the cell cycle until repair is complete. The physiological process of monitoring each task in the cell cycle and controlling the overall cycle so that it remains synchronized is called a ‘checkpoint’ (Hartwell & Weinert, 1989; Elledge, 1996).
The first checkpoint to be explicitly recognized as such was the inhibition of entry into the mitotic cell division phase following DNA damage in budding yeast (Weinert & Hartwell, 1988, 1989). If division occurs before repair is complete, non-viable daughter cells form with incomplete genomes. A similar cell division control had long been known in the E. coli SOS response to ultraviolet (UV) irradiation (Huisman et al. 1984; Schoemaker et al. 1984; Freudl et al. 1987).
We know about checkpoint controls monitoring many different aspects of the cell cycle, including genome damage (Ishikawa et al. 2006), replication status (Weinert, 1992; Navas et al. 1995; Segurado & Tercero, 2009), chromosome positioning and alignment on the spindle pole (Hoyt, 2001; Taylor et al. 2004; Varetti & Musacchio, 2008; Nezi & Musacchio, 2009), and cell size (Rupes et al. 2001; Fang et al. 2006; Sabelli et al. 2013). The reason checkpoints are so important is that they exemplify at the molecular level the processes of self-evaluation and self-control that McClintock emphasized in her Nobel Prize lecture.
Apoptosis decisions in response to DNA damage
An evaluation of both intracellular and extracellular information occurs when cells have to decide between undergoing either genome repair or programmed cell death (apoptosis) in response to DNA damage (Zmasek et al. 2007; Schlereth et al. 2010; Tentner et al. 2012). There are apoptosis-promoting intercellular signals in both prokaryotes (Engelberg-Kulka et al. 2006; Kolodkin-Gal et al. 2007; Kolodkin-Gal & Engelberg-Kulka, 2008) and eukaryotes (Yin, 2000; Krieghoff-Henning & Hofmann, 2008; Walsh & Edinger, 2010). In mammalian cell culture, the presence of growth factors (such as insulin-like growth factor, IGF) protects against apoptosis and leads to a DNA repair response, while the presence of ‘death factors’ (such as tumour necrosis factor, TNF) induces an apoptotic response (Remacle-Bonnet et al. 2000; Danielsen & Maihle, 2002; Linseman et al. 2002; Janes et al. 2005; Gomez-Vicente et al. 2006; Song, 2007; Holoch & Griffith, 2009; Joza et al. 2009). The ability of cells to sense their biological environment and make life-or-death decisions based on intercellular signals is a prototypical homeostatic process (Loewer & Lahav, 2006). We will see other cell signal responses when we discuss the control of molecular systems to restructure the genome.
Mutator polymerases and point mutations
In the early 1950s, it became evident that UV-induced mutagenesis in E. coli is an active cell process, part of what we later came to call the ‘SOS’ DNA damage response (Witkin, 1975, 1991; Little & Mount, 1982). Jean Weigle, a Swiss physicist turned molecular biologist, performed the clarifying experiments (Weigle, 1953; Weigle & Bertani, 1953). He used bacterial virus ‘lambda’ as his test organism. The advantage of a virus was that the test DNA could be treated independently of the cells in which it replicated and mutated by irradiating cell-free suspensions of lambda prior to infection.
Weigle systematically tested all combinations of untreated and irradiated virus and host cells. He found that irradiated cells had much greater capacity both to repair lethal damage and also to induce mutations in the irradiated lambda genomes. To universal surprise, he also found augmented mutagenesis in untreated virus infecting irradiated cells. In other words, UV irradiation induced mutagenesis activity in the host cells, active even on unirradiated DNA. This activity was dubbed ‘error-prone’ repair (Witkin, 1973).
Error-free and error-prone repair
The notion that certain repair processes are precise and ‘error-free’ whereas others are imprecise and ‘error-prone’ (Table1) is based on considering repair as the chief evolved functionality. The distinction works readily for cases such as DS break repair. For broken duplexes, repair by the multi-step process of homologous recombination with an undamaged template is intrinsically error-free (Kowalczykowski et al. 1994; Kowalczykowski, 2000). NHEJ, by contrast, generally creates new sequence structures and is therefore inherently error-prone and mutagenic (Lieber, 2010; Symington & Gautier, 2011).
In other cases, the distinction between error-prone and dedicated mutator function is less clear. For example, as indicated in Table 1, base excision repair removal of uracil from DNA by uracil-n-glycosylase (Ung) activity can lead either to accurate repair, as in E. coli, or to mutagenic repair, as in activated B lymphocytes. In the immune system, Ung activity is essential to somatic hypermutation for antibody refinement (Perez-Duran et al. 2012). Similarly, the UV-inducible E. coli SOS functions DinB and UmuCD play key roles in mutagenesis, but their absence has little effect on bacterial repair capacity (Maenhaut-Michel, 1985; Brotcorne-Lannoye & Maenhaut-Michel, 1986). Does this mean that some SOS activities act more as mutator than repair functions (Goodman, 1998; Garcia-Diaz et al. 2003; McKenzie et al. 2003; Galhardo et al. 2009)?
Trans-lesion bypass ‘mutator’ polymerases
An ambiguous answer to the question about UmuCD and DinB mutator function comes from the discovery that they represent a new class of DNA polymerase activity. DinB encodes DNA polymerase IV, and UmuCD encodes DNA polymerase V (Reuven et al. 1999; Wagner et al. 1999).
PolIV and PolV typify a class of non-processive polymerases that have the capacity to elongate nascent DNA strands a few nucleotides opposite a damaged template strand (‘trans-lesion bypass’ polymerization) (Goodman, 2002). As would be expected from the ability to overcome template damage, trans-lesion bypass polymerases are highly prone to mutagenic misincorporations. Thus, they have also been called ‘mutator DNA polymerases’ (McKenzie & Rosenberg, 2001).
Detailed study of UV-induced mutagenesis in E. coli revealed that particular trans-lesion polymerases (or pairs thereof) are necessary for specific kinds of mutations to occur, such as −1 or −2 frameshifts or particular base substitutions (Napolitano et al. 2000; Fuchs et al. 2001). These unexpected results indicate that individual localized point mutations are the result of action by specific biochemical functions. Certainly, this conclusion fits with the demonstrations that PolIV and PolV account for the inducible mutagenic activity that Weigle discovered on unirradiated lambda DNA (Caillet-Fauquet & Maenhaut-Michel, 1988; Maenhaut-Michel & Caillet-Fauquet, 1990; Maenhaut-Michel et al. 1992). The involvement of identifiable enzymes with particular DNA changes is far more in agreement with the physiological view of mutation than with the random accident assumption.
NHEJ, chromosome rearrangements and DS break repair centres
As McClintock observed over the many years that she studied the fate of broken chromosome ends, NHEJ is the source of many kinds of chromosome rearrangements. In the prior citation from her Nobel Prize lecture, she pointed out that cells must have the ability to bring broken chromosome ends together. Analysis of DS break repair in budding yeast has thrown some light on this process (Lisby et al. 2001, 2003a,b; Lisby & Rothstein, 2004, 2005).
When DS breaks occur in eukaryotic cells, specialized chromatin, containing the exceptional histones H2A (yeast) or H2AX (mammals), forms at the broken ends. By using immunofluorescence and fluorescent protein tags to detect H2A, it is possible to see that broken ends, homologous recombination proteins and NHEJ proteins localize to subnuclear foci (‘repair centres’) in a cell cycle-dependent manner (Lisby & Rothstein, 2004). By differential labelling of nearby DNA, the presence of broken ends from two different chromosomes can be observed in a single repair centre (Lisby et al. 2003b), where the ends can be joined by NHEJ to form a translocation or other rearrangement structure.
For recombination and NHEJ to take place, motor proteins are required for two distinct tasks: (i) chromatin remodelling of the damaged DNA (Giglia-Mari et al. 2011; Xu et al. 2012; Bennett et al. 2013; North et al. 2013; Seeber et al. 2013) and (ii) powering the DNA movements needed for repair centre localization and execution of strand exchanges (Mazin et al. 2010; Ceballos & Heyer, 2011; Burgess et al. 2013). The nature of control over chromosome movements towards the repair centres remains unclear, but the physiological nature of DS break repair is unmistakable insofar as its cell biology is concerned.
Natural genetic engineering (NGE) functions
In addition to systems used both for DNA repair and for genome change, there are a number of evolved complex molecular systems that appear dedicated to generating novel genome structures. Table 2 lists a number of categories for these genome innovation systems.
Table 2.
Genome restructuring natural genetic engineering (NGE) systems
| Function | System | References |
|---|---|---|
| Horizontal nucleic acid transfer between cells | DNA import and export complexes | (Zupan et al. 2000; Chen & Dubnau, 2004; Cehovin et al. 2013) |
| Plasmids and other conjugative elements | (Hayes, 1968; Sonea, 1987; Smillie et al. 2010; Leclercq et al. 2012) | |
| Viruses | (Forterre, 2006; Comeau et al. 2008; Forterre, 2010; Krupovic et al. 2011) | |
| Virus-like ‘gene transfer agents’ (GTAs) | (Lang & Beatty, 2007; Stanton, 2007; Zhao et al. 2009; Leung et al. 2010) | |
| Generate copies or variants of genome coding sequences | Reverse transcription and genome insertion of processed RNAs | (Brosius, 2003; Baertsch et al. 2008) |
| Mobilize DNA segments within and between molecules | Site-specific recombination systems | (Landy, 1989; Hall & Stokes, 1993; Grindley et al. 2006; Cambray et al. 2010; Hallet & Sherratt, 2010) |
| DNA transposons | (Curcio & Derbyshire, 2003) | |
| Retroviruses and retroviral-like LTR retrotransposons | (McDonald et al. 1997; Rho et al. 2007; Novikova, 2009) | |
| Non-LTR retrotransposons | (Schmidt, 1999; Han, 2010) | |
| Inteins | (Elleuche & Poggeler, 2010) | |
| Retrosplicing introns | (Lambowitz & Zimmerly, 2011) | |
| Generate diversity in protein structure or expression | Silent cassette conversion into expression sites | (Plasterk et al. 1985; Barry & McCulloch, 2001; Horn, 2004; Barbour et al. 2006; Cahoon & Seifert, 2011) |
| Shufflons and invertons | (Komano, 1999; Hallet & Sherratt, 2010) | |
| Diversity-generating retroelements | (Medhekar & Miller, 2007) | |
| Variable lymphocyte receptor diversification | (Rogozin et al. 2007; Boehm et al. 2012) | |
| VDJ joining | (Bassing et al. 2002; Alt et al. 2013) | |
| Somatic hypermutation | (Honjo et al. 2002; Longerich et al. 2006; Peled et al. 2008) | |
| Isotype class switching | (Honjo et al. 2002; Longerich et al. 2006; Bothmer et al. 2011; Bothmer et al. 2013) | |
| Genome immunity acquisition | Clustered regular interspersed palindromic repeats (CRISPRs) | (Barrangou, 2013; Sorek et al. 2013) |
| piRNA loci | (Brennecke et al. 2007; Handler et al. 2013) | |
| Developmental chromatin diminution | (Muller & Tobler, 2000; Bachmann-Waldmann et al. 2004) | |
| Ciliate macronucleus development | Excision of germline-specific sequences, unscrambling of jumbled coding sequences, telomere capping of short multicopy minichromosomes | (Juranek & Lipps, 2007; Nowacki et al. 2011) |
Abbreviations: LTR, long terminal repeat; piRNA, piwi-binding RNA; VDJ, variable, diversity and join cassettes of immunoglobulin-encoding DNA sequences.
From the list of diverse DNA transfer and rearrangement systems in Table 2, it is evident that cells have complex physiologies for assembling and modifying their genomes. The range of molecular functionalities enables living organisms to accomplish the following adaptive genome writing tasks:
Import and integrate extensive DNA segments encoding adaptive functions (horizontal transfer)
The ability of cells to acquire DNA by direct uptake from the environment, virus infection, conjugal transfer, parasite vectors (Houck et al. 1991) or other as yet unknown means has made it possible for genomes to acquire coding capacity for new biochemical activities. The importance of horizontal DNA transfer in the spread of antibiotic resistance and specialized adaptations in bacteria and archaea is widely recognized (Grassi et al. 2012) (http://shapiro.bsd.uchicago.edu/ExtraRefs.AntibioticResistanceAndHorizontalTransfer.shtml). Less well known are a rapidly growing number of examples where eukaryotic microbes and multicellular eukaryotes have acquired adaptive functions from prokaryotes or other eukaryotes by horizontal DNA transfer (Kondrashov et al. 2006; Keeling & Palmer, 2008; Keeling, 2009; Whitaker et al. 2009; Danchin et al. 2010; Oliver et al. 2010; Acuna et al. 2012; Danchin & Rosso, 2012; Xi et al. 2012).
Diversify protein structures
DNA rearrangements serve the antagonistic cells providing adaptive immunity (http://shapiro.bsd.uchicago.edu/ExtraRefs.ImmuneSystemChanges.shtml) and their infectious microbial targets, which undergo surface antigen variation to escape immune system defences (Wisniewski-Dye & Vial, 2008; Deitsch et al. 2009; Cahoon & Seifert, 2011). In addition, site-specific recombination ‘shufflons’ serve to expand pilus attachment specificity for plasmid transfer (Gyohda et al. 2004), and ‘diversity-generating retroelements’ extend bacteriophage host range (Doulatov et al. 2004).
Alter the regulation of existing functions
Insertion of a mobile genetic element in or near a particular genetic locus is among the most common ways of altering regulation of an existing genetic locus (Nakayashiki, 2011) (http://shapiro.bsd.uchicago.edu/Table4B(4).MoreFormattingofInsertionTargetsbyMobileGeneticElements.html). The regulatory changes include transcription factor-, micro RNA- and RNA-directed epigenetic controls. There are numerous examples of transcription factor regulatory sites mobilized by transposons, retrotransposons and retroviruses (http://shapiro.bsd.uchicago.edu/Table5C-1.MobileElementsFoundtobeExaptedascis-RegulatoryControlSitesinAnimals.html), and epigenetic imprinting is often linked to the presence of DNA from mobile elements (Youngson et al. 2005; Kinoshita et al. 2007; Suzuki et al. 2007; Fujimoto et al. 2008; Gehring et al. 2009; Pask et al. 2009; Cowley et al. 2011).
In addition to mobile elements, integrated viruses also change the regulatory configuration of the genome (Kokosar & Kordis, 2013). Viruses provide sequences for non-coding (ncRNAs) (Frias-Lasserre, 2012), sites for transcriptional control (Peaston et al. 2004; Dunn et al. 2005; Maksakova et al. 2006; Conley et al. 2008; Jern & Coffin, 2008; Cohen et al. 2009; Beyer et al. 2011) and epigenetic regulation (Brunmeir et al. 2010; Macfarlan et al. 2011; Conley & Jordan, 2012; Ward et al. 2013).
Shuffle exons to generate novel multi-domain protein activities
It has become apparent from genome sequencing that many new protein functionalities arise from the accretion and rearrangement of functional ‘domains’ shared by many different proteins (Doolittle & Bork, 1993; Doolittle, 1995; Lander et al. 2001; Toll-Riera & Alba, 2013). There is direct evidence in several species that mobile genetic elements have mediated exon shuffling in the past and can do so experimentally in real time (Moran et al. 1999; Hiller et al. 2000; Ejima & Yang, 2003; Liu & Grigoriev, 2004; Morgante et al. 2005; Damert et al. 2009; Hancks et al. 2009; Elrouby & Bureau, 2010) (http://shapiro.bsd.uchicago.edu/Table5A.ExamplesofDocumentedExonShufflingbyMobileGeneticElements.html).
Create new polynucleotide coding sequences
One of the outstanding problems in protein evolution is the origin of novel domain and whole protein coding sequences. This problem has been partially solved by discovering (i) that new exons frequently arise post-insertion by utilization of splice signals inside mobile elements (‘exonization’) (Sorek, 2007; Burns & Boeke, 2008; Toll-Riera et al. 2009; Wissler et al. 2013) (http://shapiro.bsd.uchicago.edu/Origin_of_New_Protein_Domains.html) and (ii) that ‘neogene’ formation occurs by insertion of reversed transcribed sequences into genetic loci, often generating chimeric coding regions (Long, 2001; Betrán et al. 2002; Piriyapongsa et al. 2007; Chen et al. 2010; Fu et al. 2010; Schmitz & Brosius, 2011; Mandal et al. 2013) (http://shapiro.bsd.uchicago.edu/Table5B.Reportsofretrogenesinplantandanimalgenomes.html).
New coding sequences also arise by the reverse transcription and insertion of ‘edited’ (sequence altered) mRNAs or ncRNAs (Lev-Maor et al. 2007; Xie et al. 2012; Mandal et al. 2013). In addition to mobile elements, fragments of viral and organelle sequences have also undergone exonization to generate novel proteins and domains in nuclear genomes (Taylor & Bruenn, 2009; Koonin, 2010; Liu et al. 2010, 2011a,b; Chiba et al. 2011; Lloyd & Timmis, 2011; Woehle et al. 2011; Rousseau-Gueutin et al. 2012).
Enhance protein diversity by alternative transcription and splicing signals
When a mobile element inserts into an intron, it can generate transcript and protein diversity by providing both alternative transcription initiation and termination sites as well as alternative splice signals (Han et al. 2004; Han & Boeke, 2005; Wheelan et al. 2005; Zemojtel et al. 2007; Burns & Boeke, 2008; Gogvadze & Buzdin, 2009; Kaer et al. 2011). These additional regulatory sites lead to the production of messages truncated either at the 5′ start or at the 3′ tail and encoding proteins with altered combinations of exons.
Rapidly disperse common regulatory sequences to distant genetic loci to generate coordinately controlled genome networks
The ability of mobile elements to move rapidly to many genome locations allows them to solve the problem of generating functionally coordinated networks without having to alter each locus independently (Feschotte, 2008; Kunarso et al. 2010; Lynch et al. 2011) (http://shapiro.bsd.uchicago.edu/Table5C-1.MobileElementsFoundtobeExaptedascis-RegulatoryControlSitesinAnimals.html). Single events can activate mobile element activity that results in multiple genome modifications. This is of particular importance after interspecific hybridization and whole genome duplication (WGD), which create redundant copies of entire networks (Teichmann & Babu, 2004). It is noteworthy that such WGD events correspond to major transitions in the evolutionary record, such as the emergence of vertebrates (Kasahara, 2007).
Viruses as part of the distributed RW genome
Listing viruses as agents of genome transfer may appear surprising to many readers accustomed to think of these subcellular infectious agents as parasites. But it is also useful to consider viruses as both extracellular and intracellular extensions of the collective read–write (RW) genome of living cells. This view was implicit in the work of Lwoff and his students Wollman and Jacob on temperate bacteriophages, which could enter and exit from the genomes of bacteria (Lwoff, 1954, 1957, 1966; Jacob & Wollman, 1961; Wollman & Jacob, 1961). Sonea and Panisset explicitly articulated the collective genome concept for prokaryotes in the 1970s and 1980s (Sonea, 1971; Sonea & Panisset, 1983; Sonea & Mathieu, 2001).
Sonea and Panisset viewed the prokaryotic genome as an ecology-wide distributed system containing DNA sequences encoding all manner of adaptive functions. When a particular ecological niche became available, the requisite functions could be assembled in one cell by horizontal DNA transfer, including viral infections, to produce an organism that could exploit the new niche. In accordance with this view, the results of environmental metagenomics show that the virosphere provides a significant reservoir of DNA encoding a wide range of cellular functions (Williamson et al. 2008; Kristensen et al. 2010; Alperovitch-Lavy et al. 2011; Sharon et al. 2011; Breitbart, 2012; Hurwitz et al. 2013; Roux et al. 2013; Schoenfeld et al. 2013).
Moreover, sequencing the genomes of recently discovered giant DNA viruses that infect amoebae, algae and other protists has revealed a realm of genome mixing between the three domains of life. The megabase-range genomes of these viruses contain mixtures of viral, archaeal, bacterial and eukaryotic sequences (Boyer et al. 2009; Wilson et al. 2009; Colson & Raoult, 2010; Fischer et al. 2010; Yutin et al. 2013). Large DNA viruses have extended host ranges, and the protists they infect harbour bacterial symbionts that also infect plants and animals (Huws et al. 2008; Bozzaro & Eichinger, 2011; Steinert, 2011; Yousuf et al. 2013). There is even evidence of conjugal transfer within amoebae between animal and plant pathogenic bacteria (Saisongkorh et al. 2010). Thus, there exist multiple biological paths for distributing novelties originating within the lower eukaryote-cum-large DNA virus ‘melting pot’ for genome sequence innovation to all kinds of cells (Boyer et al. 2009; Moliner et al. 2010).
Activation of NGE
One of the major features of cell-mediated genome change is regulation over the biochemical activities involved. This means physiologically that genome change functions are facultative and responsive to external and internal stimuli (http://shapiro.bsd.uchicago.edu/TableII.7.shtml). The range of activating NGE stimuli extend from DNA damage and other stress events (infection, hybridization, starvation) to evolved responses to intercellular signals (pheromones, lymphokines).
My personal experience with physiological activation of NGE came from studying the action of a transposable element in mediating the fusion of the araB and lacZ coding sequences to direct synthesis of a hybrid protein (Shapiro, 1984, 1997). Fusions were not detectable in over 3 × 1010 plated bacteria following normal growth (i.e. no colonies within the first 3–4 days), but after several days of additional incubation colonies began to sprout in ever greater numbers, and the frequency rapidly increased to at least one fusion per 105 viable bacteria on the selection plates. Fusions were dependent on transposase activity (Shapiro & Leach, 1990). The key parameter inducing this particular DNA restructuring process was aerobic starvation, independent of the initial carbon source (Maenhaut-Michel & Shapiro, 1994), and a number of regulatory proteases and transcription factors were necessary for fusion activation (Shapiro, 1993; Gomez-Gomez et al. 1997; Lamrani et al. 1999).
The araB–lacZ fusion system was the first example of a more general phenomenon that has come to be called ‘adaptive mutation’ (Foster, 1993; Shapiro, 1997; Hall, 1998; Rosenberg, 2001) (http://shapiro.bsd.uchicago.edu/TableII.7.shtml). Although the phenomenon and the term ‘adaptive’ have proved controversial (Roth et al. 2006), I argue that the term is appropriate in two senses: (i) increased mutability occurs as an adaptive response to starvation conditions, and (ii) among the resulting mutations, there are invariably some that provide adaptation to the selective conditions.
The stresses or deviations from the normal genomic state that trigger mutability in eukaryotes correlate with stimuli that alter epigenetic formatting (http://shapiro.bsd.uchicago.edu/TableII.10.shtml). It therefore appears that a major mode of NGE activation is disruption of epigenetic controls known to inhibit the activity of mobile elements and possibly other DNA restructuring functions (Hollister & Gaut, 2009; Handler et al. 2013; Nuthikattu et al. 2013). In addition to destabilizing NGE controls, perturbations of various outside inputs ranging from nutrition to maternal care lead to trans-generational epigenetic changes without altering the underlying DNA sequences (http://shapiro.bsd.uchicago.edu/Transgenerational_Epigenetic_Effects.html). How and why various stresses and stimuli affect epigenetic silencing is an active topic of research concerning physiological impacts on the genome.
As expected from physiological processes, there are a number of diverse but specific effects of intercellular signalling molecules on NGE activities. In bacteria, pheromones and quorum sensing signals stimulate transfer of conjugative elements and uptake of extracellular DNA (Fuqua & Winans, 1994; Auchtung et al. 2005; Meibom et al. 2005; Kozlowicz et al. 2006a,b; Dunny, 2007; Christie-Oleza et al. 2009; Suckow et al. 2011; Lo Scrudato & Blokesch, 2012; Cook & Federle, 2013). In budding yeast, retrotransposition of the Ty3 and Ty5 elements is coordinated with mating events by sex pheromone induction (Kinsey & Sandmeyer, 1995; Ke et al. 1997). In rodents, steroid hormones induce reproduction and retrotransposition of mouse mammary tumour virus (Truss et al. 1992). In the mouse and human immune systems, lymphokine molecules determine the sites of DS breaks and NHEJ for isotype switching (Kinoshita et al. 1998; Dunnick et al. 2011).
Cell- and tissue-type specific NGE processes as part of the normal life cycle
The DNA changes associated with antigen receptor synthesis in lymphocytes are the most thoroughly documented case of a tissue-specific NGE process (http://shapiro.bsd.uchicago.edu/ExtraRefs.ImmuneSystemChanges.shtml). But there are other cases, which show that the immune system is far from exceptional. In Drosophila and nematodes, for example, the actions of P factors and other mobile genetic elements are specific to germ-line tissues (Laski et al. 1986; Siebel & Rio, 1990; Moerman et al. 1991; Takeda et al. 2007; Koga et al. 2008; Keng et al. 2009). The same germ-line restriction is true of a retrotransposon in mice (Dupressoir & Heidmann, 1996).
One of the more intriguing tissue-specific regulatory phenomena is the activation of long interspersed element (LINE) retrotransposition in mammalian neural cells (Muotri et al. 2005, 2010; Coufal et al. 2009; Thomas et al. 2012). This observation has led to the hypothesis that LINE-induced genome diversity is a contributing factor to neural network architecture in the mammalian nervous system (Muotri & Gage, 2006; Muotri et al. 2007). There is also the potential for deleterious changes, and a recent report links LINE retrotransposition to schizophrenia (Bundo et al. 2014).
Targeting of NGE
The ability of cells to regulate NGE functions is not limited to turning them on and off. Cells employ a range of different molecular mechanisms to target NGE activity both towards and away from particular regions of the genome (http://shapiro.bsd.uchicago.edu/TableII.11.shtml). The targeting mechanisms involve familiar molecular interactions: protein–DNA sequence binding, protein–protein binding, nucleic acid sequence homologies and coupling to transcription. What has made recognition of this widespread targeting capacity difficult for many biologists to accept is simply the unsupported ad hoc assertion that such targeting within the genome is not possible. A very partial discussion of targeting examples follows.
Homologous recombination
In principle, homologous recombination can occur between any two DNA duplexes that have the same sequence. But the real in vivo recombination process inevitably displays non-uniformity across the genome (‘hotspots’ and ‘coldspots’) (Smith, 1994; Huang & Keil, 1995; Amundsen & Smith, 2007; Cromie et al. 2007; Grey et al. 2009; Brunschwig et al. 2012; Steiner & Steiner, 2012; Martin-Castellanos et al. 2013). The non-uniformity reflects the biochemical complexity of the homologous exchange process (Kowalczykowski et al. 1994; Kowalczykowski, 2000; San Filippo et al. 2008), which provides opportunities for regulation, as well as the need for specific protein–DNA interactions to carry out the actual physiological recombination steps (Krejci et al. 2012; Baudat et al. 2013).
An example of sequence specificity is provided by the ‘chi’ sites regulating motor protein behaviour and exonuclease specificity of proteins needed to process broken duplex ends to initiate bacterial recombination (Spies et al. 2003; Amundsen & Smith, 2007). This feature of the prokaryotic recombination process has evolved independently at least twice in prokaryotes given that gram-negative and gram-positive bacteria have distinct sets of chi sites and cognate exonuclease proteins (El Karoui et al. 2000).
An example of targeting homologous exchange away from particular regions of the genome comes from studies of mice, where recombination events might disrupt evolved combinations of transcriptional regulatory signals (Brick et al. 2012). A particular protein has evolved to recognize those combinations and suppress recombination events (Segurel et al. 2011). This same protein also protects the integrity of its own coding sequences (Jeffreys et al. 2013). Chromatin formatting plays an important role in regulating the distribution of homologous recombination sites (Yamada et al. 2013), and the same recombination suppressing protein also influences this epigenetic regulation (Grey et al. 2011).
Targeting mobile element insertions towards adaptively useful locations (and away from harmful regions)
Protein–DNA and protein–protein interactions frequently target mobile element insertions to advantageous positions. Here we will look at only two of many examples.
The bacterial transposon Tn7 encoding antibiotic resistances provides a good prokaryotic example (Craig, 1991). When horizontal transfer introduces Tn7 into a new cell, DNA recognition by the multi-protein integration complex occurs at a specific chromosomal location where insertion has no deleterious effect on bacterial physiology. However, when a plasmid without Tn7 is leaving a cell that carries a chromosomal copy of the element, a different targeting protein becomes part of the transposition/integration complex. This particular protein binds to a DNA replication factor and targets new insertions in a sequence-independent manner to actively replicating DNA (Peters & Craig, 2001; Parks et al. 2009). As replication in the donor cell powers plasmid transfer events, this protein–protein interaction targets Tn7 insertions to mobilizing plasmids and facilitates dispersal of the transposon to new cells (Wolkow et al. 1996; Parks & Peters, 2009).
In yeast cells, there are a number of retroviral-like mobile elements that display distinctive insertion patterns determined by protein–protein interactions that keep them from disrupting expressed coding sequences but allow them to alter coding sequence expression (Levin & Moran, 2011):
In budding yeast, retrotransposons Ty1 and Ty3 both insert upstream of RNA PolIII transcription start sites. However, the precise molecular mechanism is specific to each element. For Ty1, the integrase protein interacts with nucleosome histone markers at the initiation site (Mou et al. 2006; Mularoni et al. 2012), while the Ty3 integrase interacts with two PolIII transcription initiation factors (Kirchner et al. 1995).
The budding yeast retrotransposon Ty5 inserts preferentially into unexpressed chromatin regions by binding of a phosphorylated domain of the integrase to the Sir4 silencing factor (Xie et al. 2001; Brady et al. 2008). However, under stress conditions, the phosphate is lost from the integrase domain, and insertion becomes untargeted, perhaps because insertions into expressed regions of the genome may prove useful (Dai et al. 2007).
In fission yeast, the Tf1 retrotransposon inserts upstream of RNA PolII transcription start sites due to interaction of the integrase protein with 5′-binding transcriptional activators (Behrens et al. 2000; Leem et al. 2008).
These distinct yeast retrotransposon targeting mechanisms with remarkably similar specificities appear to have evolved independently for adaptive utility.
Targeting isotype class switching by intercellular signalling
One example of NGE targeting that deserves emphasis is class switching in the DNA encoding immunoglobulins. This process occurs in activated B cells once VDJ joining and somatic hypermutation have generated and refined the antigen binding capacity of an immunoglobulin molecule (Bothmer et al. 2011).
Immunoglobulins originate as cell-bound IgM molecules because the constant region of the heavy protein chain is encoded by a μ exon (Fig. 1). Directing the antigen specificity of the immunoglobulin towards different locations in the body occurs by a change in the constant region of the heavy chain (HC). This requires a ‘class switch’ of the encoding HC exon from μ to one encoding another Ig class or ‘isotype’ (Fig. 1).
Figure 1. Mechanism of class switch recombination that allows isotype switching in activated B cells.
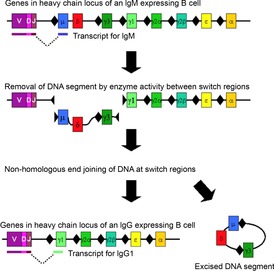
The boxes indicate exon cassettes and black diamonds indicate switch regions. Copied from Wikipedia and used according to Wikimedia Free Commons.
The isotype switch occurs by inducing DS breaks at special ‘switch’ regions (S) preceding each HC exon at the immunoglobulin heavy chain (IgH) locus (black diamonds in Fig. 1) (Bothmer et al. 2013). The DS break requires transcription at Sμ (already transcribed) and a second switch region. The downstream switch regions each have a promoter responsive to a combination of lymphokine signalling molecules. As only activated and transcribed switch regions undergo breaks followed by end-joining, the cells of the immune system effectively instruct the activated B cell which class/isotype antibody to synthesize.
The connection of transcription and DNA rearrangement is a powerful tool for directing NGE activities to any functionally regulated region of the genome.
Genome homeostasis and cancer
One implication of the RW genome concept is that cells have elaborate regulatory circuits controlling NGE activities so the genome is not disrupted during normal reproduction.
We observe homeostatic control of highly evolved NGE systems where complex DNA rearrangement processes occur only in certain tissues, such as vertebrate adaptive immune systems, and where specific DNA changes occur only at defined stages of the life cycle, such as bacterial sporulation (Stragier et al. 1989; Kunkel et al. 1990), cyanobacterial heterocyst differentiation (Carrasco & Golden, 1995; Golden & Yoon, 1998), yeast mating-type switches following spore germination (Haber, 2012) and massive post-mating genome restructuring in ciliated protozoa (Prescott, 2000) (http://shapiro.bsd.uchicago.edu/Ciliate_macronuclear_development.html).
The RW genome perspective provides us with a new way of thinking about the high levels of genome change in cancer cells (Richards, 2001; Davies, 2013). Instead of considering stochastic mutations as causes of cell reprogramming in cancer, we can rather think of particular NGE system activations as consequences of oncogenic changes in cell regulatory networks.
Activation of specific NGE functions may help to explain some of the recurrent DNA rearrangement patterns observed in particular kinds of tumours, exemplified by the Philadelphia chromosome in chronic myelogenous leukaemia (CML) (Rowley, 1973, 2008). In the case of lymphomas due to aberrations of immune system rearrangements, the identity of the specific NGE operator can be determined (Nambiar & Raghavan, 2011; Rocha et al. 2012; Rocha & Skok, 2013).
It is notable that genome changes become more extensive as cancer progresses (Jeggo, 2005; Alexandrov et al. 2013). Tumours exemplify the extreme kinds of genome changes cells can produce when normal regulation of NGE radically fails. This failure can lead to activation of genome change programmes that generate novel cell proliferation phenotypes (Davies, 2013). The phenomenon of ‘chromothripsis’ (literally, chromosome shattering) is particularly interesting (Kloosterman et al. 2011; Chen et al. 2012; Korbel & Campbell, 2013). In chromothripsis, individual chromosomes are fragmented and rearranged, sometimes in a new structure or sometimes with pieces transposed to other chromosomes. Such extreme genome rearrangements resemble some of the chromosome scrambling observed when related genomes are compared, such as human and mouse (Waterston et al. 2002; Armengol et al. 2003). Perhaps the rapid occurrence of similar scrambling in cancer has lessons for evolutionary biology.
Another lesson of chromothripsis is that NGE events can often occur multiple times in spatially restricted domains, such as a single chromosome. One aspect of RW genome changes is that regulatory interactions can intersect with NGE functions (Lin et al. 2009). This possibility has been suggested for the chromosome looping effect of androgen receptors on the location of recurrent rearrangement breakpoints in prostate cancer (Wu et al. 2011; Barbieri et al. 2012; Martin et al. 2013). We have seen transcription factor-mediated NGE targeting with yeast retrotransposons, and cancer genomics may well provide further examples.
A physiological view of the RW genome, its maintenance and differentiation
The preceding sections have attempted to make two major arguments: (i) the concept of an RW genome altered by cell action is more compatible with the discoveries of molecular genetics than the pre-DNA idea of a read-only memory (ROM) subject to accidental change; (ii) the concepts of physiological regulation can be applied to the control of the NGE operators that alter DNA sequences and genome structure in non-random and controlled ways.
With respect to genome maintenance, the parallels appear to be straightforward between homeostatic regulation of organismal physiology and the phenomenology of genome proofreading, DNA repair and inhibition of NGE functions during normal reproduction. However, the same parallel is applicable to a ROM memory view of the genome.
Where the idea of an RW genome diverges from conventional thinking in both genetics and physiology is in the realm of genome differentiation. In this case, regulation is for change rather than for stability. Genomes can change to take on specialized tasks that occur repeatedly in the life cycle or for adaptive innovation in the face of evolutionary challenges.
We know that the genome changes as part of the normal life cycle fit into organismal physiology in the same way as cellular, tissue and morphological differentiation do in multicellular development. Normal life-cycle genome changes occur under well-defined conditions and have characteristic features, even if the functional purpose is to generate diversity, as in the adaptive immune system and its antagonist, antigenic variation of infectious organisms (http://shapiro.bsd.uchicago.edu/ExtraRefs.NaturalGeneticEngineeringPartNormalLifeCycle.shtml). The existence of highly evolved and tightly regulated NGE processes that generate genome changes in a predictable fashion is an empirical reality incompatible with the ROM genome view.
The major challenge we face in understanding non-random genome change is its success in generating complex adaptive evolutionary innovations (Shapiro, 2011). It is not difficult to see how horizontal DNA transfer, domain shuffling and mobile element dispersal of regulatory signals expedite the search of functional genome space and thereby facilitate the evolution of novel biochemical activities and coordinately regulated networks.
Nonetheless, constructing the genomic basis for a major adaptive evolutionary innovation, such as a novel form of mimicry or a four-chambered heart, comprises coordinated changes in multiple developmental processes. How could that be accomplished? We know from studies of activation and targeting of NGE activities that genome change operators and cell regulatory circuits interact in multiple ways. A major 21st century research challenge for the life sciences (including physiology, molecular genetics and genomics) will be to explore the depth of control circuit–NGE interactions and learn how ‘informed’ the process of genome rewriting may be. I predict that big surprises are in store for all of us.
Acknowledgments
I thank Denis Noble for the invitation to speak at the International Union of Physiology Societies Congress in July 2013, and for the opportunity to write this article.
Glossary
- DS
double-strand
- HC
immunoglobulin heavy chain constant region
- IgH
immunoglobulin heavy chain
- LINE
long interspersed nucleotide element
- ncRNA
non-coding RNA
- NGE
natural genetic engineering
- NHEJ
non-homologous end-joining
- ROM
read-only memory
- RW
read–write (memory)
- S
switch region
- Sμ
switch region upstream of the heavy chain mu exon
- Ung
uracil-n-glycosylase
- UV
ultraviolet
- WGD
whole genome duplication
Additional information
Competing interests
The author has no competing interests.
References
- Acuna R, Padilla BE, Florez-Ramos CP, Rubio JD, Herrera JC, Benavides P, Lee SJ, Yeats TH, Egan AN, Doyle JJ, Rose JK. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc Natl Acad Sci U S A. 2012;109:4197–4202. doi: 10.1073/pnas.1121190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Andrew Futreal P, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alperovitch-Lavy A, Sharon I, Rohwer F, Aro EM, Glaser F, Milo R, Nelson N, Beja O. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I and photosystem-II gene suites inferred from oceanic metagenomic datasets. Environ Microbiol. 2011;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Smith GR. Chi hotspot activity in Escherichia coli without RecBCD exonuclease activity: implications for the mechanism of recombination. Genetics. 2007;175:41–54. doi: 10.1534/genetics.106.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Waldmann C, Jentsch S, Tobler H, Muller F. Chromatin diminution leads to rapid evolutionary changes in the organization of the germ line genomes of the parasitic nematodes A. suum and P. univalens. Mol Biochem Parasitol. 2004;134:53–64. doi: 10.1016/j.molbiopara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Dai Q, Restrepo BI, Stoenner HG, Frank SA. Pathogen escape from host immunity by a genome program for antigenic variation. Proc Natl Acad Sci U S A. 2006;103:18290–18295. doi: 10.1073/pnas.0605302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Behrens R, Hayles J, Nurse P. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 2000;28:4709–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G, Papamichos-Chronakis M, Peterson CL. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat Commun. 2013;4:2084. doi: 10.1038/ncomms3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci U S A. 2011;108:3624–3629. doi: 10.1073/pnas.1016201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Rommel PC, Gazumyan A, Polato F, Reczek CR, Muellenbeck MF, Schaetzlein S, Edelmann W, Chen PL, Brosh RM, Jr, Casellas R, Ludwig T, Baer R, Nussenzweig A, Nussenzweig MC, Robbiani DF. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210:115–123. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R, Doherty AJ. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzaro S, Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011;12:942–954. doi: 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady TL, Schmidt CL, Voytas DF. Targeting integration of the Saccharomyces Ty5 retrotransposon. Methods Mol Biol. 2008;435:153–163. doi: 10.1007/978-1-59745-232-8_11. [DOI] [PubMed] [Google Scholar]
- Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissett NC, Doherty AJ. Repairing DNA double-strand breaks by the prokaryotic non-homologous end-joining pathway. Biochem Soc Trans. 2009;37:539–545. doi: 10.1042/BST0370539. [DOI] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- Brotcorne-Lannoye A, Maenhaut-Michel G. Role of RecA protein in untargeted UV mutagenesis of bacteriophage lambda: evidence for the requirement for the dinB gene. Proc Natl Acad Sci U S A. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Simboeck E, Sawicka A, Egger G, Hagelkruys A, Zhang Y, Matthias P, Miller WJ, Seiser C. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 2010;6:e1000927. doi: 10.1371/journal.pgen.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunschwig H, Levi L, Ben-David E, Williams RW, Yakir B, Shifman S. Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics. 2012;191:757–764. doi: 10.1534/genetics.112.141036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, Kato M, Kasai K, Kishimoto T, Nawa H, Okano H, Yoshikawa T, Kato T, Iwamoto K. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:396–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Sebesta M, Sisakova A, Marini VP, Lisby M, Damborsky J, Klein H, Rothstein R, Krejci L. The PCNA interaction protein box sequence in Rad54 is an integral part of its ATPase domain and is required for efficient DNA repair and recombination. PLoS One. 2013;8:e82630. doi: 10.1371/journal.pone.0082630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. Great exaptations. J Biol. 2008;7:5. doi: 10.1186/jbiol66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Seifert HS. Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria. Mol Microbiol. 2011;81:1136–1143. doi: 10.1111/j.1365-2958.2011.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Fauquet P, Maenhaut-Michel G. Nature of the SOS mutator activity: genetic characterization of untargeted mutagenesis in Escherichia coli. Mol Gen Genet. 1988;213:491–498. doi: 10.1007/BF00339621. [DOI] [PubMed] [Google Scholar]
- Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- Carrasco CD, Golden JW. Two heterocyst-specific DNA rearrangements of nif operons in Anabaena cylindrica and Nostoc sp. strain Mac. Microbiology. 1995;141:2479–2487. doi: 10.1099/13500872-141-10-2479. [DOI] [PubMed] [Google Scholar]
- Ceballos SJ, Heyer WD. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci U S A. 2013;110:3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res. 2012;750:52–59. doi: 10.1016/j.mrrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza JA, Lanfranconi MP, Nogales B, Lalucat J, Bosch R. Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J Bacteriol. 2009;191:1239–1247. doi: 10.1128/JB.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Colson P, Raoult D. Gene repertoire of amoeba-associated giant viruses. Intervirology. 2010;53:330–343. doi: 10.1159/000312918. [DOI] [PubMed] [Google Scholar]
- Comeau AM, Hatfull GF, Krisch HM, Lindell D, Mann NH, Prangishvili D. Exploring the prokaryotic virosphere. Res Microbiol. 2008;159:306–313. doi: 10.1016/j.resmic.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Conley AB, Jordan IK. Endogenous retroviruses and the epigenome. In: Witzany G, editor. Viruses: Essential Agents of Life. Dordrecht: Springer; 2012. pp. 309–323. [Google Scholar]
- Conley AB, Piriyapongsa J, Jordan IK. Retroviral promoters in the human genome. Bioinformatics. 2008;24:1563–1567. doi: 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev. 2013 doi: 10.1111/1574-6976.12046. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, de Burca A, McCole RB, Chahal M, Saadat G, Oakey RJ, Schulz R. Short interspersed element (SINE) depletion and long interspersed element (LINE) abundance are not features universally required for imprinting. PLoS One. 2011;6:e18953. doi: 10.1371/journal.pone.0018953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu Rev Genet. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]
- Craig NL. Tn7: a target site-specific transposon. Mol Microbiol. 1991;5:2569–2573. doi: 10.1111/j.1365-2958.1991.tb01964.x. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EG, Rosso MN. Lateral gene transfers have polished animal genomes: lessons from nematodes. Front Cell Infect Microbiol. 2012;2:27. doi: 10.3389/fcimb.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EG, Rosso MN, Vieira P, de Almeida-Engler J, Coutinho PM, Henrissat B, Abad P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc Natl Acad Sci U S A. 2010;107:17651–17656. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen AJ, Maihle NJ. The EGF/ErbB receptor family and apoptosis. Growth Factors. 2002;20:1–15. doi: 10.1080/08977190290022185. [DOI] [PubMed] [Google Scholar]
- Davies PCW. Implications of read-write genomics for cancer biology. Phys Life Rev. 2013;10:338–340. doi: 10.1016/j.plrev.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Bork P. Evolutionarily mobile modules in proteins. Sci Am. 1993;269:50–56. doi: 10.1038/scientificamerican1093-50. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate β3GAL-T5. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Holden V, Fontaine C, Collins JT. The role of germline promoters and I exons in cytokine-induced gene-specific class switch recombination. J Immunol. 2011;186:350–358. doi: 10.4049/jimmunol.1003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol Cell Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet. 2003;12:1321–1328. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- El Karoui M, Schaeffer M, Biaudet V, Bolotin A, Sorokin A, Gruss A. Orientation specificity of the Lactococcus lactis Chi site. Genes Cells. 2000;5:453–461. doi: 10.1046/j.1365-2443.2000.00342.x. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Poggeler S. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl Microbiol Biotechnol. 2010;87:479–489. doi: 10.1007/s00253-010-2628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Bureau TE. Bs1, a new chimeric gene formed by retrotransposon-mediated exon shuffling in maize. Plant Physiol. 2010;153:1413–1424. doi: 10.1104/pp.110.157420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, de los Reyes C, Umen JG. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2006;2:e167. doi: 10.1371/journal.pgen.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazlieva R, Spittle CS, Morrissey D, Hayashi H, Yan H, Matsumoto Y. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37:2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Forterre P. Defining life: the virus viewpoint. Orig Life Evol Biosph. 2010;40:151–160. doi: 10.1007/s11084-010-9194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation: the uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg-Schwager M, Becker M, Garg I, Pralle E, Wolf H, Frankenberg D. The role of nonhomologous DNA end joining, conservative homologous recombination, and single-strand annealing in the cell cycle-dependent repair of DNA double-strand breaks induced by H2O2 in mammalian cells. Radiat Res. 2008;170:784–793. doi: 10.1667/RR1375.1. [DOI] [PubMed] [Google Scholar]
- Freudl R, Braun G, Honore N, Cole ST. Evolution of the enterobacterial sulA gene: a component of the SOS system encoding an inhibitor of cell division. Gene. 1987;52:31–40. doi: 10.1016/0378-1119(87)90392-1. [DOI] [PubMed] [Google Scholar]
- Frias-Lasserre D. Non coding RNAs and viruses in the framework of the phylogeny of the genes, epigenesis and heredity. Int J Mol Sci. 2012;13:477–490. doi: 10.3390/ijms13010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- Fu B, Chen M, Zou M, Long M, He S. The rapid generation of chimerical genes expanding protein diversity in zebrafish. BMC Genomics. 2010;11:657. doi: 10.1186/1471-2164-11-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RP, Koffel-Schwartz N, Pelet S, Janel-Bintz R, Napolitano R, Becherel OJ, Broschard TH, Burnouf DY, Wagner J. DNA polymerases II and V mediate respectively mutagenic (-2 frameshift) and error-free bypass of a single N-2-acetylaminofluorene adduct. Biochem Soc Trans. 2001;29:191–195. doi: 10.1042/0300-5127:0290191. [DOI] [PubMed] [Google Scholar]
- Fujimoto R, Kinoshita Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Nasrallah ME, Shimizu KK, Kudoh H, Kakutani T. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 2008;4:e1000048. doi: 10.1371/journal.pgen.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K. DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/260512. doi: 10.4061/2010/260512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Ruiz JF, Juarez R, Terrados G, Blanco L. Are there mutator polymerases? ScientificWorldJournal. 2003;3:422–431. doi: 10.1100/tsw.2003.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze E, Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci. 2009;66:3727–3742. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Yoon HS. Heterocyst formation in Anabaena. Curr Opin Microbiol. 1998;1:623–629. doi: 10.1016/s1369-5274(98)80106-9. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez JM, Blazquez J, Baquero F, Martinez JL. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J Bacteriol. 1997;179:4620–4622. doi: 10.1128/jb.179.14.4620-4622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Vicente V, Doonan F, Donovan M, Cotter TG. Induction of BIMEL following growth factor withdrawal is a key event in caspase-dependent apoptosis of 661W photoreceptor cells. Eur J Neurosci. 2006;24:981–990. doi: 10.1111/j.1460-9568.2006.04990.x. [DOI] [PubMed] [Google Scholar]
- Goodman M. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Ann Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Goodman MF. Purposeful mutations. Nature. 1998;395:221–223. doi: 10.1038/26111. [DOI] [PubMed] [Google Scholar]
- Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- Grassi L, Grilli J, Lagomarsino MC. Large-scale dynamics of horizontal transfers. Mob Genet Elements. 2012;2:163–167. doi: 10.4161/mge.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Baudat F, de Massy B. Genome-wide control of the distribution of meiotic recombination. PLoS Biol. 2009;7:e35. doi: 10.1371/journal.pbio.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Guetens G, De Boeck G, Highley M, van Oosterom AT, de Bruijn EA. Oxidative DNA damage: biological significance and methods of analysis. Crit Rev Clin Lab Sci. 2002;39:331–457. doi: 10.1080/10408360290795547. [DOI] [PubMed] [Google Scholar]
- Gyohda A, Furuya N, Ishiwa A, Zhu S, Komano T. Structure and function of the shufflon in plasmid R64. Adv Biophys. 2004;38:183–213. [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- Hall RM, Stokes HW. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- Hallet B, Sherratt DJ. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 2010;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Han JS. Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA. 2010;1:15. doi: 10.1186/1759-8753-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH., Jr Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–1991. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. The genetic makeup of the Drosophila piRNA pathway. Mol Cell. 2013;50:762–777. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hayes W. The Genetics of Bacteria and their Viruses. 2nd edn. London: Blackwell; 1968. [Google Scholar]
- Hays JB, Hoffman PD, Wang H. Discrimination and versatility in mismatch repair. DNA Repair (Amst) 2005;4:1463–1474. doi: 10.1016/j.dnarep.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Hiller R, Hetzer M, Schweyen RJ, Mueller MW. Transposition and exon shuffling by group II intron RNA molecules in pieces. J Mol Biol. 2000;297:301–308. doi: 10.1006/jmbi.2000.3582. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr Mol Med. 2004;4:563–576. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- Houck MA, Clark JB, Peterson KR, Kidwell MG. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991;253:1125–1128. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. A new view of the spindle checkpoint. J Cell Biol. 2001;154:909–911. doi: 10.1083/jcb.200108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GS, Keil RL. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O, D'Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013;14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huws SA, Morley RJ, Jones MV, Brown MR, Smith AW. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiol Lett. 2008;282:258–265. doi: 10.1111/j.1574-6968.2008.01123.x. [DOI] [PubMed] [Google Scholar]
- Ibarra B, Chemla YR, Plyasunov S, Smith SB, Lazaro JM, Salas M, Bustamante C. Proofreading dynamics of a processive DNA polymerase. EMBO J. 2009;28:2794–2802. doi: 10.1038/emboj.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Ishii H, Saito T. DNA damage-dependent cell cycle checkpoints and genomic stability. DNA Cell Biol. 2006;25:406–411. doi: 10.1089/dna.2006.25.406. [DOI] [PubMed] [Google Scholar]
- Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Wollman EL. Viruses and genes. Sci Am. 1961;204:92–110. [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Cotton VE, Neumann R, Lam KW. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proc Natl Acad Sci U S A. 2013;110:600–605. doi: 10.1073/pnas.1220813110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA. Genomic instability in cancer development. Adv Exp Med Biol. 2005;570:175–197. doi: 10.1007/1-4020-3764-3_6. [DOI] [PubMed] [Google Scholar]
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Joza N, Pospisilik JA, Hangen E, Hanada T, Modjtahedi N, Penninger JM, Kroemer G. AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci. 2009;1171:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- Juranek SA, Lipps HJ. New insights into the macronuclear development in ciliates. Int Rev Cytol. 2007;262:219–251. doi: 10.1016/S0074-7696(07)62005-1. [DOI] [PubMed] [Google Scholar]
- Kaer K, Branovets J, Hallikma A, Nigumann P, Speek M. Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation. PLoS One. 2011;6:e26099. doi: 10.1371/journal.pone.0026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. The 2R hypothesis: an update. Curr Opin Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke N, Irwin PA, Voytas DF. The pheromone response pathway activates transcription of Ty5 retrotransposons located within silent chromatin of Saccharomyces cerevisiae. EMBO J. 1997;16:6272–6280. doi: 10.1093/emboj/16.20.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids. Methods Mol Biol. 2009;532:501–515. doi: 10.1007/978-1-60327-853-9_29. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Keng VW, Ryan BJ, Wangensteen KJ, Balciunas D, Schmedt C, Ekker SC, Largaespada DA. Efficient transposition of Tol2 in the mouse germline. Genetics. 2009;183:1565–1573. doi: 10.1534/genetics.109.100768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Tashiro J, Tomita S, Lee CG, Honjo T. Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequences of S regions. Immunity. 1998;9:849–858. doi: 10.1016/s1074-7613(00)80650-0. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJ, Koornneef M, Kakutani T. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007;49:38–45. doi: 10.1111/j.1365-313X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- Kinsey PT, Sandmeyer SB. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics. 1995;139:81–94. doi: 10.1093/genetics/139.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Connolly CM, Sandmeyer SB. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, Cuppen E. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Koga A, Cheah FS, Hamaguchi S, Yeo GH, Chong SS. Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev Dyn. 2008;237:2466–2474. doi: 10.1002/dvdy.21688. [DOI] [PubMed] [Google Scholar]
- Kokosar J, Kordis D. Genesis and regulatory wiring of retroelement-derived domesticated genes: a phylogenomic perspective. Mol Biol Evol. 2013;30:1015–1031. doi: 10.1093/molbev/mst014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Engelberg-Kulka H. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J Bacteriol. 2008;190:3169–3175. doi: 10.1128/JB.01918-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- Komano T. Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999;33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010;2010:543531. doi: 10.4061/2010/543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 2010;8:2. doi: 10.1186/1741-7007-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz BK, Dworkin M, Dunny GM. Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Int J Med Microbiol. 2006a;296:141–147. doi: 10.1016/j.ijmm.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol. 2006b;62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieghoff-Henning E, Hofmann TG. Role of nuclear bodies in apoptosis signalling. Biochim Biophys Acta. 2008;1783:2185–2194. doi: 10.1016/j.bbamcr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kurosawa A, Saito S, So S, Hashimoto M, Iwabuchi K, Watabe H, Adachi N. DNA ligase IV and artemis act cooperatively to suppress homologous recombination in human cells: implications for DNA double-strand break repair. PLoS One. 2013;8:e72253. doi: 10.1371/journal.pone.0072253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamrani S, Ranquet C, Gama MJ, Nakai H, Shapiro JA, Toussaint A, Maenhaut-Michel G. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol Microbiol. 1999;32:327–343. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Gilbert C, Cordaux R. Cargo capacity of phages and plasmids and other factors influencing horizontal transfers of prokaryote transposable elements. Mob Genet Elements. 2012;2:115–118. doi: 10.4161/mge.20352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SI, Hoffman CS, Levin HL. Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MM, Florizone SM, Taylor TA, Lang AS, Beatty JT. The gene transfer agent of Rhodobacter capsulatus. Adv Exp Med Biol. 2010;675:253–264. doi: 10.1007/978-1-4419-1528-3_14. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao Y, Wang JY. Analysis of Ig gene hypermutation in Ung−/−Polh−/− mice suggests that UNG and A:T mutagenesis pathway target different U:G lesions. Mol Immunol. 2013;53:214–217. doi: 10.1016/j.molimm.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman DA, McClure ML, Bouchard RJ, Laessig TA, Ahmadi FA, Heidenreich KA. Suppression of death receptor signalling in cerebellar Purkinje neurons protects neighboring granule neurons from apoptosis via an insulin-like growth factor I-dependent mechanism. J Biol Chem. 2002;277:24546–24553. doi: 10.1074/jbc.M201098200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2003a;2:479–483. [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003b;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87:579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol. 2010;84:11876–11887. doi: 10.1128/JVI.00955-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Li B, Yu X, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol Biol. 2011a;11:276. doi: 10.1186/1471-2148-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. Widespread endogenization of densoviruses and parvoviruses in animal and human genomes. J Virol. 2011b;85:9863–9876. doi: 10.1128/JVI.00828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes–evidence of exon shuffling? Trends Genet. 2004;20:399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Lloyd AH, Timmis JN. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol. 2011;28:2019–2028. doi: 10.1093/molbev/msr021. [DOI] [PubMed] [Google Scholar]
- Lo Scrudato M, Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012;8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Lahav G. Cellular conference call: external feedback affects cell-fate decisions. Cell. 2006;124:1128–1130. doi: 10.1016/j.cell.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Long M. Evolution of novel genes. Curr Opin Genet Dev. 2001;11:673–680. doi: 10.1016/s0959-437x(00)00252-5. [DOI] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lu AL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- Lwoff A. The life cycle of a virus. Sci Am. 1954;190:34–37. [Google Scholar]
- Lwoff A. The concept of virus. J Gen Microbiol. 1957;17:239–253. doi: 10.1099/00221287-17-2-239. [DOI] [PubMed] [Google Scholar]
- Lwoff A. Interaction among virus, cell, and organism. Science. 1966;152:1216–1220. doi: 10.1126/science.152.3726.1216. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, Pfaff SL. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G. Mechanism of SOS-induced targeted and untargeted mutagenesis in E. coli. Biochimie. 1985;67:365–369. doi: 10.1016/s0300-9084(85)80082-1. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Caillet-Fauquet P. Genetic control of the UV-induced SOS mutator effect in single- and double-stranded DNA phages. Mutat Res. 1990;230:241–254. doi: 10.1016/0027-5107(90)90062-9. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Janel-Bintz R, Fuchs RP. A umuDC-independent SOS pathway for frameshift mutagenesis. Mol Gen Genet. 1992;235:373–380. doi: 10.1007/BF00279383. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Shapiro JA. The roles of starvation and selective substrates in the emergence of araB–lacZ fusion clones. EMBO J. 1994;13:5229–5239. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal AK, Pandey R, Jha V, Mukerji M. Transcriptome-wide expansion of non-coding regulatory switches: evidence from co-occurrence of Alu exonization, antisense and editing. Nucleic Acids Res. 2013;41:2121–2137. doi: 10.1093/nar/gks1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TJ, Peer CJ, Figg WD. Uncovering the genetic landscape driving castration-resistant prostate cancer. Cancer Biol Ther. 2013;14:399–400. doi: 10.4161/cbt.24426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Fowler KR, Smith GR. Making chromosomes hot for breakage. Cell Cycle. 2013;12:1327–1328. doi: 10.4161/cc.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. Rad54, the motor of homologous recombination. DNA Repair (Amst) 2010;9:286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. A correlation of ring-shaped chromosomes with variegation in Zea mays. Proc Natl Acad Sci U S A. 1932;18:677–681. doi: 10.1073/pnas.18.12.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Nat Acad Sci U S A. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc Nat Acad Sci U S A. 1942;28:458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- McClintock B. Discovery and Characterization of Transposable Elements: The Collected Papers of Barbara McClintock. New York: Garland; 1987. [Google Scholar]
- McDonald JF, Matyunina LV, Wilson S, Jordan IK, Bowen NJ, Miller WJ. LTR retrotransposons and the evolution of eukaryotic enhancers. Genetica. 1997;100:3–13. [PubMed] [Google Scholar]
- McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Rosenberg SM. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr Opin Microbiol. 2001;4:586–594. doi: 10.1016/s1369-5274(00)00255-1. [DOI] [PubMed] [Google Scholar]
- Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Kiff JE, Waterston RH. Germline excision of the transposable element Tc1 in C. elegans. Nucleic Acids Res. 1991;19:5669–5672. doi: 10.1093/nar/19.20.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Mou Z, Kenny AE, Curcio MJ. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics. 2006;172:2157–2167. doi: 10.1534/genetics.105.054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 2012;22:693–703. doi: 10.1101/gr.129460.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Tobler H. Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int J Parasitol. 2000;30:391–399. doi: 10.1016/s0020-7519(99)00199-x. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum Mol Genet. 2007;16(R2):R159–167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo GW, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H. The Trickster in the genome: contribution and control of transposable elements. Genes Cells. 2011;16:827–841. doi: 10.1111/j.1365-2443.2011.01533.x. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39:5813–5825. doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- North JA, Amunugama R, Klajner M, Bruns AN, Poirier MG, Fishel R. ATP-dependent nucleosome unwrapping catalyzed by human RAD51. Nucleic Acids Res. 2013;41:7302–7312. doi: 10.1093/nar/gkt411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O. Chromodomains and LTR retrotransposons in plants. Commun Integr Biol. 2009;2:158–162. doi: 10.4161/cib.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF. RNA-mediated epigenetic programming of genome rearrangements. Annu Rev Genomics Hum Genet. 2011;12:367–389. doi: 10.1146/annurev-genom-082410-101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, Defraia C, Thomas EN, Slotkin RK. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Parks AR, Li Z, Shi Q, Owens RM, Jin MM, Peters JE. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AR, Peters JE. Tn7 elements: engendering diversity from chromosomes to episomes. Plasmid. 2009;61:1–14. doi: 10.1016/j.plasmid.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask AJ, Papenfuss AT, Ager EI, McColl KA, Speed TP, Renfree MB. Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 2009;10:R1. doi: 10.1186/gb-2009-10-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim Pol. 2003;50:891–908. [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Perez-Duran P, Belver L, de Yebenes VG, Delgado P, Pisano DG, Ramiro AR. UNG shapes the specificity of AID-induced somatic hypermutation. J Exp Med. 2012;209:1379–1389. doi: 10.1084/jem.20112253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Proofreading by the epsilon subunit of Escherichia coli DNA polymerase III increases the fidelity of calf thymus DNA polymerase alpha. Proc Natl Acad Sci U S A. 1989;86:3085–3088. doi: 10.1073/pnas.86.9.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 2001;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Polavarapu N, Borodovsky M, McDonald J. Exonization of the LTR transposable elements in human genome. BMC Genomics. 2007;8:291. doi: 10.1186/1471-2164-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher RS, Wilson TE, Doherty AJ. New insights into NHEJ repair processes in prokaryotes. Cell Cycle. 2005;4:675–678. doi: 10.4161/cc.4.5.1676. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Simon MI, Barbour AG. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Prescott DM. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat Rev Genet. 2000;1:191–198. doi: 10.1038/35042057. [DOI] [PubMed] [Google Scholar]
- Rechkunova NI, Lavrik OI. Nucleotide excision repair in higher eukaryotes: mechanism of primary damage recognition in global genome repair. Subcell Biochem. 2010;50:251–277. doi: 10.1007/978-90-481-3471-7_13. [DOI] [PubMed] [Google Scholar]
- Remacle-Bonnet MM, Garrouste FL, Heller S, Andre F, Marvaldi JL, Pommier GJ. Insulin-like growth factor-I protects colon cancer cells from death factor- induced apoptosis by potentiating tumor necrosis factor α-induced mitogen-activated protein kinase and nuclear factor κB signaling pathways. Cancer Res. 2000;60:2007–2017. [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Rho M, Choi JH, Kim S, Lynch M, Tang H. De novo identification of LTR retrotransposons in eukaryotic genomes. BMC Genomics. 2007;8:90. doi: 10.1186/1471-2164-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Micsinai M, Kim JR, Hewitt SL, Souza PP, Trimarchi T, Strino F, Parisi F, Kluger Y, Skok JA. Close proximity to Igh is a contributing factor to AID-mediated translocations. Mol Cell. 2012;47:873–885. doi: 10.1016/j.molcel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Skok JA. The origin of recurrent translocations in recombining lymphocytes: a balance between break frequency and nuclear proximity. Curr Opin Cell Biol. 2013;25:365–371. doi: 10.1016/j.ceb.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM. Evolving responsively: adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Ayliffe MA, Timmis JN. Plastid DNA in the nucleus: new genes for old. Plant Signal Behav. 2012;7:269–272. doi: 10.4161/psb.18762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Krupovic M, Debroas D, Forterre P, Enault F. Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol. 2013;3:130160. doi: 10.1098/rsob.130160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- Rupes I, Webb BA, Mak A, Young PG. G2/M arrest caused by actin disruption is a manifestation of the cell size checkpoint in fission yeast. Mol Biol Cell. 2001;12:3892–3903. doi: 10.1091/mbc.12.12.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Liu Y, Dante RA, Lizarraga LE, Nguyen HN, Brown SW, Klingler JP, Yu J, LaBrant E, Layton TM, Feldman M, Larkins BA. Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc Natl Acad Sci U S A. 2013;110:E1827–1836. doi: 10.1073/pnas.1304903110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisongkorh W, Robert C, La Scola B, Raoult D, Rolain JM. Evidence of transfer by conjugation of type IV secretion system genes between Bartonella species and Rhizobium radiobacter in amoeba. PLoS One. 2010;5:e12666. doi: 10.1371/journal.pone.0012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schlereth K, Beinoraviciute-Kellner R, Zeitlinger MK, Bretz AC, Sauer M, Charles JP, Vogiatzi F, Leich E, Samans B, Eilers M, Kisker C, Rosenwald A, Stiewe T. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2010;38:356–368. doi: 10.1016/j.molcel.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Schmidt T. LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes. Plant Mol Biol. 1999;40:903–910. doi: 10.1023/a:1006212929794. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Brosius J. Exonization of transposed elements: a challenge and opportunity for evolution. Biochimie. 2011;93:1928–1934. doi: 10.1016/j.biochi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Schoemaker JM, Gayda RC, Markovitz A. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J Bacteriol. 1984;158:551–561. doi: 10.1128/jb.158.2.551-561.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld TW, Murugapiran SK, Dodsworth JA, Floyd S, Lodes M, Mead DA, Hedlund BP. Lateral gene transfer of family A DNA polymerases between thermophilic viruses, aquificae, and apicomplexa. Mol Biol Evol. 2013;30:1653–1664. doi: 10.1093/molbev/mst078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hauer M, Gasser SM. Nucleosome remodelers in double-strand break repair. Curr Opin Genet Dev. 2013;23:174–184. doi: 10.1016/j.gde.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol Cell. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- Segurel L, Leffler EM, Przeworski M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 2011;9:e1001211. doi: 10.1371/journal.pbio.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Observations on the formation of clones containing araB–lacZ cistron fusions. Mol Gen Genet. 1984;194:79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- Shapiro JA. A role for the Clp protease in activating Mu-mediated DNA rearrangements. J Bacteriol. 1993;175:2625–2631. doi: 10.1128/jb.175.9.2625-2631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Genome organization, natural genetic engineering and adaptive mutation. Trends Genet. 1997;13:98–104. doi: 10.1016/s0168-9525(97)01058-5. [DOI] [PubMed] [Google Scholar]
- Shapiro JA. Evolution: A View from the 21st Century. Upper Saddle River, NJ: FT Press Science; 2011. [Google Scholar]
- Shapiro JA. How life changes itself: the read-write (RW) genome. Phys Life Rev. 2013;10:287–323. doi: 10.1016/j.plrev.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, Leach D. Action of a transposable element in coding sequence fusions. Genetics. 1990;126:293–299. doi: 10.1093/genetics/126.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Battchikova N, Aro EM, Giglione C, Meinnel T, Glaser F, Pinter RY, Breitbart M, Rohwer F, Beja O. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 2011;5:1178–1190. doi: 10.1038/ismej.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Rio DC. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990;248:1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR. Hotspots of homologous recombination. Experientia. 1994;50:234–241. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- Sonea S. Bacterial viruses, prophages, and plasmids, reconsidered. Ann N Y Acad Sci. 1987;503:251–260. doi: 10.1111/j.1749-6632.1987.tb40612.x. [DOI] [PubMed] [Google Scholar]
- Sonea S. A tentative unifying view of bacteria. Rev Can Biol. 1971;30:239–244. [PubMed] [Google Scholar]
- Sonea S, Mathieu LG. Evolution of the genomic systems of prokaryotes and its momentous consequences. Int Microbiol. 2001;4:67–71. doi: 10.1007/s101230100015. [DOI] [PubMed] [Google Scholar]
- Sonea S, Panisset M. A New Bacteriology. Boston: Jones and Batlett; 1983. [Google Scholar]
- Song J. EMT or apoptosis: a decision for TGF-β. Cell Res. 2007;17:289–290. doi: 10.1038/cr.2007.25. [DOI] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- Stanton TB. Prophage-like gene transfer agents – novel mechanisms of gene exchange for MethanococcusDesulfovibrioBrachyspira, and Rhodobacter species. Anaerobe. 2007;13:43–49. doi: 10.1016/j.anaerobe.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Steiner EM. Fission yeast hotspot sequence motifs are also active in budding yeast. PLoS One. 2012;7:e53090. doi: 10.1371/journal.pone.0053090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M. Pathogen–host interactions in DictyosteliumLegionellaMycobacterium and other pathogens. Semin Cell Dev Biol. 2011;22:70–76. doi: 10.1016/j.semcdb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011;193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD, Smith BT, Godoy VG, Walker GC. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, Ishino F, Renfree MB, Kaneko-Ishino T. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Takeda J, Keng VW, Horie K. Germline mutagenesis mediated by Sleeping Beauty transposon system in mice. Genome Biol. 2007;8(Suppl 1):S14. doi: 10.1186/gb-2007-8-s1-s14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. UmuD’2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Bruenn J. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 2009;7:88. doi: 10.1186/1741-7007-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Scott MI, Holland AJ. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Tentner AR, Lee MJ, Ostheimer GJ, Samson LD, Lauffenburger DA, Yaffe MB. Combined experimental and computational analysis of DNA damage signaling reveals context-dependent roles for Erk in apoptosis and G1/S arrest after genotoxic stress. Mol Syst Biol. 2012;8:568. doi: 10.1038/msb.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CA, Paquola AC, Muotri AR. LINE-1 retrotransposition in the nervous system. Annu Rev Cell Dev Biol. 2012;28:555–573. doi: 10.1146/annurev-cellbio-101011-155822. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, Alba MM. Emergence of novel domains in proteins. BMC Evol Biol. 2013;13:47. doi: 10.1186/1471-2148-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, Castelo R, Bellora N, Alba MM. Evolution of primate orphan proteins. Biochem Soc Trans. 2009;37:778–782. doi: 10.1042/BST0370778. [DOI] [PubMed] [Google Scholar]
- Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter. J Steroid Biochem Mol Biol. 1992;43:365–378. doi: 10.1016/0960-0760(92)90071-p. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- Varetti G, Musacchio A. The spindle assembly checkpoint. Curr Biol. 2008;18:R591–595. doi: 10.1016/j.cub.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Walker GC. Understanding the complexity of an organism's responses to DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:1–10. doi: 10.1101/sqb.2000.65.1. [DOI] [PubMed] [Google Scholar]
- Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MC, Wilson MD, Barbosa-Morais NL, Schmidt D, Stark R, Pan Q, Schwalie PC, Menon S, Lukk M, Watt S, Thybert D, Kutter C, Kirschner K, Flicek P, Blencowe BJ, Odom DT. Latent regulatory potential of human-specific repetitive elements. Mol Cell. 2013;49:262–272. doi: 10.1016/j.molcel.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weigle JJ. Induction of mutations in a bacterial virus. Proc Natl Acad Sci U S A. 1953;39:628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle JJ, Bertani G. Variations des bacteriophages conditionnees par les bacteries hotes [Variations of bacteriophage conditioned by host bacteria.] Ann Inst Pasteur (Paris) 1953;84:175–179. [PubMed] [Google Scholar]
- Weinert T, Hartwell L. Control of G2 delay by the rad9 gene of Saccharomyces cerevisiae. J Cell Sci Suppl. 1989;12:145–148. doi: 10.1242/jcs.1989.supplement_12.12. [DOI] [PubMed] [Google Scholar]
- Weinert TA. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat Res. 1992;132:141–143. [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene- breaking: a new paradigm for human retrotransposon- mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker JW, McConkey GA, Westhead DR. The transferome of metabolic genes explored: analysis of the horizontal transfer of enzyme encoding genes in unicellular eukaryotes. Genome Biol. 2009;10:R36. doi: 10.1186/gb-2009-10-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S, Nagel G, Eshkind L, Neurath MF, Samson LD, Kaina B. Both base excision repair and O6-methylguanine-DNA methyltransferase protect against methylation-induced colon carcinogenesis. Carcinogenesis. 2010;31:2111–2117. doi: 10.1093/carcin/bgq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dye F, Vial L. Phase and antigenic variation mediated by genome modifications. Antonie Van Leeuwenhoek. 2008;94:493–515. doi: 10.1007/s10482-008-9267-6. [DOI] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 2013;5:439–455. doi: 10.1093/gbe/evt009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM. Ultraviolet mutagenesis in bacteria: the inducible nature of error-prone repair. An Acad Bras Cienc. 1973;45(Suppl):185–191. [PubMed] [Google Scholar]
- Witkin EM. Relationships among repair, mutagenesis, and survival: overview. Basic Life Sci. 1975;5A:347–353. doi: 10.1007/978-1-4684-2895-7_47. [DOI] [PubMed] [Google Scholar]
- Witkin EM. RecA protein in the SOS response: milestones and mysteries. Biochimie. 1991;73:133–141. doi: 10.1016/0300-9084(91)90196-8. [DOI] [PubMed] [Google Scholar]
- Woehle C, Dagan T, Martin WF, Gould SB. Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol Evol. 2011;3:1220–1230. doi: 10.1093/gbe/evr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- Wollman EL, Jacob F. Sexuality and the Genetics of Bacteria. New York: Academic Press; 1961. [Google Scholar]
- Wu D, Zhang C, Shen Y, Nephew KP, Wang Q. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol Metab. 2011;22:474–480. doi: 10.1016/j.tem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Bradley RK, Wurdack KJ, Wong KM, Sugumaran M, Bomblies K, Rest JS, Davis CC. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Nunoshiba T, Shimizu M, Gruz P, Kamiya H, Harashima H, Nohmi T. Involvement of Y-family DNA polymerases in mutagenesis caused by oxidized nucleotides in Escherichia coli. J Bacteriol. 2006;188:4992–4995. doi: 10.1128/JB.00281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Ohta K, Yamada T. Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 2013;41:3504–3517. doi: 10.1093/nar/gkt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XM. Signal transduction mediated by Bid, a pro-death Bcl-2 family proteins, connects the death receptor and mitochondria apoptosis pathways. Cell Res. 2000;10:161–167. doi: 10.1038/sj.cr.7290045. [DOI] [PubMed] [Google Scholar]
- Youngson NA, Kocialkowski S, Peel N, Ferguson-Smith AC. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J Mol Evol. 2005;61:481–490. doi: 10.1007/s00239-004-0332-0. [DOI] [PubMed] [Google Scholar]
- Yousuf FA, Siddiqui R, Khan NA. Acanthamoeba castellanii of the T4 genotype is a potential environmental host for Enterobacter aerogenes and Aeromonas hydrophila. Parasit Vectors. 2013;6:169. doi: 10.1186/1756-3305-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Colson P, Raoult D, Koonin EV. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Penzkofer T, Schultz J, Dandekar T, Badge R, Vingron M. Exonization of active mouse L1s: a driver of transcriptome evolution? BMC Genomics. 2007;8:392. doi: 10.1186/1471-2164-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang K, Budinoff C, Buchan A, Lang A, Jiao N, Chen F. Gene transfer agent (GTA) genes reveal diverse and dynamic Roseobacter and Rhodobacter populations in the Chesapeake Bay. ISME J. 2009;3:364–373. doi: 10.1038/ismej.2008.115. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 2000;23:11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]


