Abstract
Transgenerational persistence of parental responses to environmental stimuli has been reported in various organisms, but the underlying mechanisms remain underexplored. In one of these reported examples, we have shown that exposure of fly larvae to G418 antibiotic leads to non-Mendelian inheritance of ectopic induction of certain developmental genes. Here we investigate if this inheritance involves changes in mRNA composition within the early, maternal-stage offspring embryos of exposed flies. Exposure to G418 in F1 modified the maternal RNA levels of many genes in their early (F2) embryos. This includes reduction of maternal Polycomb group genes which persisted in the following generation of embryos (F3). To investigate the functional meaning of this reduction, we compared genetically normal embryos of Polycomb mutant females to normal embryos of normal females. Analysis with two different alleles of Polycomb, Pc1 and Pc3, revealed that maternal reduction in Polycomb gene dosage has a positive influence on the inheritance of induced expression. Together, this shows that exposure to G418 stress reduces the maternal levels of Polycomb in the offspring embryos and this reduction contributes to the inheritance of induced expression.
Key points
Previous work on epigenetic transgenerational phenomena focused on chromatin modifications or small RNAs as potential carriers of non-genetic transgenerational influence.
We describe a hitherto non-appreciated mode of trans-generational influence by which physiological stress in one generation can impact multiple generations of non-exposed offspring.
This mode of transfer involves persistent changes in the composition of maternal RNA in the early offspring embryos.
In particular we show that reduction in maternal Polycomb gene levels have a functional contribution to trans-generational inheritance of induced gene expression.
Our findings extend the mechanistic repertoire of epigenetic inheritance by providing evidence connecting changes in maternal RNA with trans-generational inheritance of induced phenotypes.
Introduction
Recent evidence of transgenerational epigenetic phenomena in various species (Jablonka & Lamb, 1989; Sollars et al. 2003; Cropley et al. 2006; Richards, 2006; Rando & Verstrepen, 2007; Xing et al. 2007; Heijmans et al. 2008; Guerrero-Bosagna et al. 2010; Rechavi et al. 2011; Ashe et al. 2012; Buckley et al. 2012; Daxinger & Whitelaw, 2012; Jablonka, 2012; Lim & Brunet, 2013) suggests that environmental stimuli can induce developmental (Morgan et al. 1999; Anway et al. 2005; Cropley et al. 2006; Seong et al. 2011; Stern et al. 2012) and metabolic changes (Cropley et al. 2006; Carone et al. 2010; Crews et al. 2012) which persist in non-exposed offspring. Yet, the scope, mechanisms and potential implications for offspring health and adaptation are not clear.
Recently, we introduced an experimental model system for confronting the development of the fruit-fly, Drosophila melanogaster, with patterns of toxic G418-mediated stress that are not expected to occur during fly development (Stern et al. 2012). In this system, we supplemented the larval food with G418 and placed a foreign resistance gene fused to GFP (neoGFP) under the regulation of arbitrary, spatio-temporally restricted developmental promoters. This leads to toxic stress in tissues that are exposed to G418 but do not express sufficient levels of neoGFP. Under a wide range of promoter::neoGFP scenarios, this stress led to modified developmental features, including ectopic induction of gene expression and longer time for pupation. We also showed that the ectopic induction of gene expression was assisted by suppression of Polycomb group (PcG) genes in the gut of stressed larvae. This stress-mediated suppression of Polycomb leads to de-repression of developmental regulators and their expression in new domains. Some of these G418-induced phenotypes were inherited by subsequent generations of non-exposed progenies (Stern et al. 2012).
In addition to the direct effect of G418 toxicity on the fly tissues, G418 may also modify the microbiome of the fly. The involvement of the microbiome in the inheritance of one of the induced phenotypes (delay in development) is described elsewhere (Y. Fridmann-Sirkis et al. 2014). Here we focus on molecular changes occurring in the germline of exposed flies and their functional involvement in the inheritance of the stress-induced phenotypes. We show that exposure of larvae in F1 modifies the maternal RNA levels in early-stage (F2) embryos and that reduction in maternal Polycomb dosage supports the inheritance of induced gene expression in the larval foregut, but not the inheritance of the delay in development.
Methods
Drosophila stocks
Drosophila lines hairy-GAL4, Pc3 and Pc1 were obtained from the Bloomington Stock Center. The yw stock was obtained from the laboratory of Dr Eli Arama (Weizmann Institute of Science, Israel). The UAS-neoGFP line was generated as described (Stern et al. 2012). Unless specifically indicated, all F1 experiments were done using lines heterozygous for the GAL4 driver and the UAS-neoGFP transgene.
Quantitative PCR analysis of RNA
About 500–600 adult flies were allowed to lay eggs for 2 h on a 10 cm agar plate. Total RNA was extracted from embryos 0–2 and 8–10 h old. RNA was purified using the RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA) and mRNA was converted to cDNA using a high-capacity reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). Transcript levels were measured using real-time quantitative PCR (qPCR) on a 7900HT Fast Real-Time PCR machine with PerfeCTa SYBR green FastMix, ROX (Quanta Biosciences, Gaithersburg, MD, USA). Specific primers used in this study were: Act5C: 5′-CCCTCGTTCTTGGGAATGG-3′, 5′-CGGTGTTGGCATACAGATCCT-3′; Pc: 5′-AAATCA TCCAAAAGCGCGTTA-3′, 5′-CCGGTTCCCAGGTGTT GTAG-3′; Rfabg: 5′-AAGGGGCCCAGCG TAATGGC-3′, 5′-GCAAGCGTTTTCAGCATTTACAGCA-3′; Aldh: 5′-AGAACTTCGCAGCAGCTGTTG-3′, 5′-TGTTGATAA ATACCCCGGTGTAGA-3′; Ipod: 5′-TCATTCACTA GCCGGGAACAG-3′, 5′-GAAGACCTTCATTTCAGCG TTGA-3; Cralbp: 5′-AGCGTGTGACGTGCTTGAAA-3′, 5′-TCTGCGATTCGTTGGTTCTG-3′; Nplp2: 5′-GGAC GCGAAGAAGGTTGAGGGTC-3′, 5′-GGATGCGGCTGGTGCCTGAA-3′; dnr1: 5′-CAACGAGAGTGCCACGCGCT-3′, 5′-GCGCCAAAGGGGATGCTCTG-3′; TwdIL: 5′-AAAGATGCGCGCCTTCATCGTAATGT-3′, 5′-TGGAGTGTCCCACGGGCTGG-3′; Cpr65Ea: 5′-CAAGCTCCTTCTCGTCGTCGCC-3′, 5′-AGGCGTAGGTGCCGTCCGTA-3′; Mhc: 5′-CGAGCGCGGAAGTTTTGGGC-3′, 5′-CGACTGGCTTCGGCATCTTGCT-3′; obst-E: 5′-TCAATGGCTCTTGGCTCGCCG-3′, 5′-CGGACACAGCTTCTCCACGGG-3′; Osi15: 5′-GCGCTACCTGGAGACCCACG-3′, 5′-CGCTTGCTGCGGGACTCATCC-3′; Ilp4: 5′-CGGTCCAGGGACGCCGAAAG-3′, 5′-TTAGA CGCACTGCTCCGCCTG-3′; hth: 5′-CCTCACCTGCCG ATGCTCGC-3′, 5′-GGCATCACCGCTTGCACTCGT-3′; ci: 5′-CGTCATTCTTGTTGTGGACTAACTTT-3′, 5′-TGGACGCTAAAAACTGCAATTC-3′; Sxl: 5′-ATTTTGATACTGTGACTCCCTGTTCGAC-3′, 5′-CCCAAATCCACGCCCACCACT-3′; Pcl: 5′-CCAGTTGCCCTACCATGCGGATA-3′, 5′-TTCCCGGTTTGCCGCAGTAGC-3′; gdl: 5′-CGGATGCAGGCGGAGAACGAG-3′, 5′-AATGTGGCGCAACTCCTCGGG-3′; knrl: 5′-TCAGACGTGCAAGGTGTGTGGA-3′, 5′-AGA AGGA CTT GCA GCCC TCGC-3′; msl-2: 5′-CGCCAGACTCTTCTGCCCCC-3′, 5′-ATTGAAGCCCTGGTACTCGCCT-3′; E(z): 5′-CAAGGATAATGGCCTGACTGTAAA-3′, 5′- CACTGCGTACTGGTGATGTTCA-3′; trx: 5′-GCCACCGCTATAAAAAGGCG-3′, 5′-TCTTTGCTGACTGCTCAGGG-3′; roX1: 5′-GGCATCCCTCGGAAAGGAAA-3′, 5′-ATG GCGATTCTACGCCTG-3′.
Deep sequencing analysis of RNA (RNA-seq)
Deep sequencing analysis of RNA was performed in the biological services unit of the Weizmann Institute of Science (Rehovot, Israel) using Illumina Genome Analyzer IIx (GA IIx). Sample was prepared using an Illumina mRNA-seq sample prep kit (Illumina, RS-100-0801). Coding and non-coding transcripts were enriched by applying Duplex-Specific Nuclease (DSN) treatment (Zhulidov et al. 2004) which reduces rRNA content. For sequencing we used the following experimental kits and reagents: Standard Cluster Generation Kit (#GD-103-4001, Illumina, San Diego, CA, USA) containing all reagents necessary to load the samples on to the flowcell and to perform the bridge amplification, and the Illumina Sequencing Kit v5 (TruSeq SBS Kit v5 GA (36-cycles), FC-104-5001) which contains the reagents for the sequencing runs. The GA IIx Sequencing Control Software version SCS 2.8 was used to control the sequencer. Sequencing was based on 80 bp unpaired reads.
Reads were mapped to the Ensembl D. melanogaster genome assembly release BDGP5.25.63 (Flicek et al. 2011). Tophat (Trapnell et al. 2009) was used to map the reads to the genome assembly and map splice junctions. The genome assembly FASTA file was indexed using: bowtie–build -o 2. Tophat version 1.3.3 was run using the following flags: ––solexa1.3-quals, –bowtie-n and –no-convert-bam. The GTF annotations from the Ensembl release (BDGP5.25.63) were also supplied to tophat using the -G flag to allow Tophat to utilize known splice junctions. The output of Tophat identifies the loci of each read. Reads were assigned to known transcripts by a custom program. This program reads the Tophat SAM output and the RefSeq gene annotations downloaded from the UCSC genome browser database (http://genome.ucsc.edu/, April 2006 assembly, dm3, BDGP Release 5). The results are imported into matlab (by readRaw.m) and normalized to RPKM (reads per kilobase of exon model per million mapped reads) as follows (Mortazavi et al. 2008):
Here, ‘total reads mapped’ used for the RPKM normalization represents the total number of reads mapped to mRNA genes, so as to avoid including reads mapped to rRNA in the normalization. Similar findings were made when reads were mapped to RefSeq RNA sequences from the April 2006 assembly of the D. melanogaster genome (dm3, BDGP Release 5), downloaded from the UCSC genome browser database (Celniker & Rubin, 2003) using Bowtie, version 0.12.7 (Langmead et al. 2009).
Genetic analysis of maternal Polycomb effects
UAS-neoGFP/+;hairy-GAL4/Pc3 or UAS-neoGFP/+;hairy-GAL4/Pc1 larvae were reared with or without 400 mg ml−1 G418. Pupae were isolated to verify collection of virgin females. These females were crossed to UAS-neoGFP/UAS-neoGFP;hairy-GAL4/TM6b,Tb males. Eggs were laid in vials without G418. Only UAS-neoGFP/UAS-neoGFP;hairy-GAL4/TM6b,Tb offspring were selected for analysis. We compared neoGFP expression in offspring of G418-exposed and non-exposed mutant mothers to the expression in offspring of non-mutant mothers.
Statistical analyses
Statistical tests were performed using MATLAB (MathWorks) and SAS software (SAS Institute Inc., Cary, NC, USA). Student's t test (see Figs 3, 5 and 6) and two-way analysis of variance (ANOVA; Fig. 6) were used for evaluating statistical significance of differences in mean values. The ANOVA tested the significance of the interaction between the maternal Pc mutation (either Pc3 or Pc1) and history of exposure to G418. For Fig. 5B, the significance of the Pearson's correlation was numerically calculated using a permutation test, as follows: F2 and F3 fold-changes were randomly shuffled 10,000 times. In each iteration we computed the Pearson's correlation between the two shuffled fold-change vectors. Significance was determined based on the percentage of iterations in which this correlation coefficient was equal to or higher than the correlation in the non-shuffled data (R = 0.4, P < 0.001). Analysis of enrichment of gene ontology annotations in sets of up- and down-regulated genes (Fig. 2) was done using the DAVID web tool with Benjamini correction for multiple hypothesis testing (Huang et al. 2009a,b).
Figure 3. Exposure to G418 in F1 reduces the maternal levels of PcG genes in the F2 embryos.
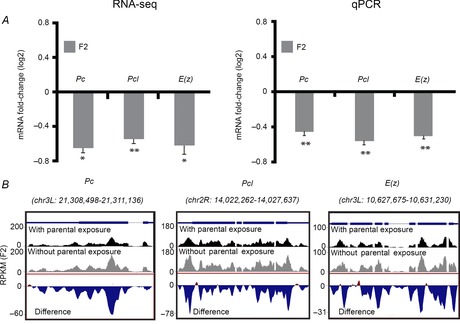
A, left: RNA-seq-based measurements of differences in maternal levels of the PcG genes Pc, Pcl and E(z) in F2 embryos of G418-exposed flies versus embryos of non-exposed flies. Values are mean fold-change ± SEM in two biological replicates. Right: qPCR-based validation of the RNA-seq-based data on the left. Values are mean fold change ± SEM in three biological replicates. *P < 0.05, **P < 0.01 (Student's t test). B, Representative RNA-seq profiles of Pc, Pcl and E(z), indicating reduction in reads throughout these gene loci. Results were normalized and displayed as reads per kilobase of exon model per million mapped reads (RPKM). Bottom track in each panel corresponds to RPKM differences between embryos of G418-exposed versus non-exposed flies.
Figure 5. Genetic reduction in maternal Polycomb dosage enhances the inheritance of induced expression.
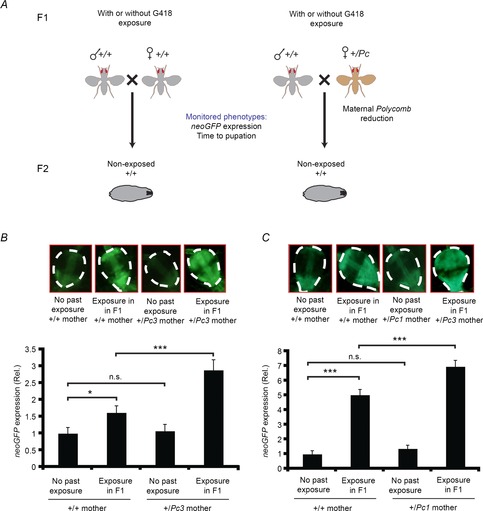
A, a genetic scheme for assessing the impact of changing maternal Polycomb dosage on the inheritance of G418-induced phenotypes (induced expression of neoGFP in the larval proventriculus and delay in pupation time). Differences were analyzed with and without exposure to G418 in F1. B, top: representative proventriculus images of non-exposed progeny produced by wild-type (+/+) or Polycomb heterozygous mutant females (+/Pc3). Shown are examples with and without exposure to G418 in F1. Bottom: statistics of neoGFP expression corresponding to the four categories in the upper panel. Values are mean neoGFP intensity ± SEM in the proventriculi of hairy::neoGFP larval offspring of exposed (n = 76) and non-exposed wild-type flies (n = 45), and exposed (n = 53) and non-exposed flies with +/Pc3 females (n = 34). *P < 0.05, ***P < 0.001 (Student's t test). Significance of the interaction between maternal Pc3 mutation and history of parental exposure to G418 was evaluated by two-way ANOVA (Pinteraction < 0.018). C, Same as B for the Pc1 mutation. Values are mean neoGFP levels ± SEM in the proventriculi of larval offspring of exposed (n = 85) and non-exposed wild-type flies (n = 57), and exposed (n = 86) and non-exposed flies with +/Pc1 females (n = 88). *P < 0.05, ***P < 0.001 (Student's t test). Pinteraction < 0.001 (two-way ANOVA).
Figure 6. Maternal reduction of Polycomb gene dosage does not affect the inheritance of the delay in development.
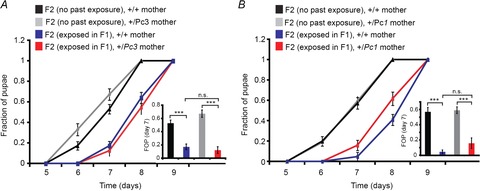
A, growth kinetics of F2 larval offspring (+/+) of wild-type (+/+) or Polycomb mutant females (+/Pc3) with and without exposure to G418 in F1. Values are mean fraction of pupae ± SEM in at least 36 vials pooled from three biological replicates. Inset: statistical analysis of differences between fractions of pupae (FOP) on day 7. B, same as A for the Pc1 mutation. Values are mean fraction of pupae ± SEM in at least 18 vials pooled from two biological replicates. Inset: statistical analysis of differences between fractions of pupae (FOP) on day 7. ***P < 0.001 (Student's t test).
Figure 2. Exposure to G418 in F1 induces widespread changes in the composition of maternal RNA in the early F2 embryos.
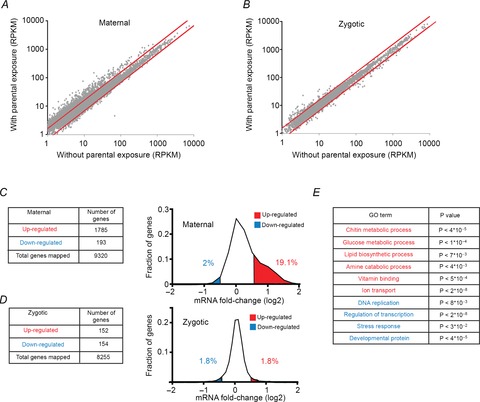
A, scatter plot of RPKM values of maternal RNA (0–2 h AEL) in F2 embryos of exposed versus non-exposed F1 flies. Each dot represents the corresponding transcription level for a particular gene in each case. Red lines indicate a difference of 1.5-fold between RPKM values. RPKM, reads per kilobase of exon model per million mapped reads. B, same as A for RPKM values measured in the zygotic stage (8–10 h AEL). C, number of genes changed (left) and distributions of differences (right) in levels of maternal RNA (0–2 h AEL) in F2 embryos of exposed versus non-exposed flies. Fractions of genes that were up- and down-regulated over 1.5-fold are labelled red and blue, respectively. Values are based on average measurements in two biological replicates. D, same as C for changes observed in the zygotic stage (8–10 h AEL). E, examples of enrichments of specific Gene Ontology (GO) annotations in the groups of genes that were up- (red) and down- (blue) regulated in the maternal stage. P-values, corrected for multiple testing using the DAVID web tool with Benjamini correction (Huang da et al. 2009b; Huang da et al. 2009a).
Data access
RNA-seq data generated in this study have been deposited in the SRA database (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP026270.
Results
Exposure to G418 modifies gene expression in the early offspring embryos
We used the G418 stress paradigm described by Stern et al. (2012) to investigate if this stress modifies the germline and to identify changes that may be involved in the inheritance of induced phenotypes. Specifically, we exposed hairy-GAL4/UAS-neoGFP larvae to a high dosage of G418 in the fly's food. This exposure leads (with high penetrance) to heritable delay in larval development and heritable induction of neoGFP expression in the proventriculus gut region of the larva (Stern et al. 2012). To test if this toxic stress can also modify the offspring embryos, we compared RNA isolated from F2 embryos of F1, G418-exposed and non-exposed flies (Fig. 1). Development of the fly embryo is initially guided by maternal RNA deposited in the egg by the mother. The maternal RNA is degraded within about 2 h after egg laying (AEL), and expression is then based on transcription from the newly synthesized DNA of the embryo (zygotic transcription) (De Renzis et al. 2007; Thomsen et al. 2010). Most of the RNA at this early time window of 2 h is maternal and therefore takes part in two generations (mothers and their embryos). As such, it may serve as a proxy for inherited changes in the germline. We extracted largely maternal (0–2 h AEL) and zygotic RNA (8–10 h AEL) from embryos of previously G418-exposed and non-exposed hairy::neoGFP flies. In each case, we analysed the genome-wide transcriptional profiles of these embryos using RNA-seq (Fig. 2). For some of the genes, we also examined the persistence of changes in the following generation of embryos (F3, Fig. 1). In each generation, we compared embryos with the same genotype, reared in the same environment, but having a different history of environment in F1. As a quality control, we first analysed all pairwise correlations between transcriptional profiles of F2 embryonic samples. As expected, the similarities between transcriptional profiles corresponding to the same embryonic stage were higher than those between samples from different stages. Analysis of replicates further revealed a clear correlation with respect to mRNA fold-change.
Figure 1. Scheme for analyzing the impact of G418 exposure in F1 on gene expression in F2 offspring embryos.
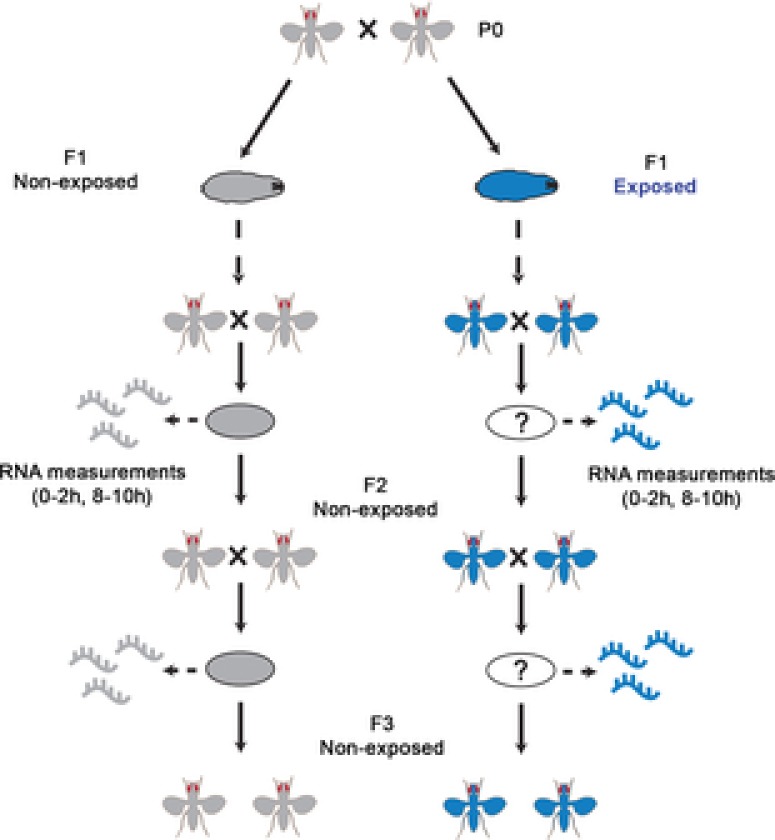
RNA was collected from non-exposed offspring embryos (F2, F3) of G418-exposed and non-exposed F1 hairy::neoGFP flies. Transcriptional profiles corresponding to two developmental intervals (0–2 and 8–10 h AEL) were analysed. Profiles in decedents of G418-exposed flies were compared to profiles of embryos from the same generation, but without past exposure in F1.
Comparison of embryos of exposed and non-exposed flies identified many up- and down-regulated genes as determined by all or most exonic reads of the genes (Fig. 2A–D). Maternal-stage (0–2 h) embryos exhibited a considerably larger extent of gene expression changes than zygotic-stage (8–10 h) embryos (Fig. 2A and C vs. Fig. 2B and D). The maternal changes also exhibited a strong preference for up-regulation of transcripts in embryos of exposed flies; 1785 maternal genes were induced above 1.5-fold versus only 193 genes that were repressed to the same extent (Fig. 2C). On the other hand, the zygotic differences were limited to 152 induced genes and 154 repressed genes (Fig. 2D). Gene Ontology (GO) analysis of the changes in maternal RNA revealed statistically significant functional enrichments within sets of down-regulated and up-regulated transcripts (Fig. 2E and Supplementary Table S1). In particular, the set of down-regulated transcripts was enriched with developmental genes and up-regulated transcripts were enriched with metabolic genes. Thus, exposure to G418 led to widespread changes in the levels of metabolic and developmental genes in early-stage embryos of exposed flies compared to embryos of non-exposed flies. These early changes may influence later-stage developmental and metabolic modifications.
Maternal transcripts of PcG genes are reduced in F2 and F3 embryonic offspring of G418-exposed versus non-exposed flies
The maternal transcripts that were down-regulated in F2 embryos were particularly enriched with regulatory functions. Among these down-regulated transcripts, we identified PcG genes known to maintain an epigenetically repressed state of the chromatin in somatic tissues: Polycomb (Pc), Polycomblike (Pcl) and Enhancer of Zetse (E(z)) (Fig. 3A, left). Pcl and E(z) are members of Polycomb Repressive Complex 2 (PRC2) and E(z) is responsible for the trimethylation of H3K27me3. The Pc gene, in turn, is a PRC1 member with a chromodomain, which recognizes the H3K27me3 mark (Schuettengruber et al. 2007). Higher resolution analysis of all the reads along each of these three genes revealed consistent reduction throughout almost all the genomic regions of the genes (Fig. 3B). This reduction in maternally deposited transcripts of PcG genes was further validated using qPCR (Fig. 3A, right). The changes in PcG genes are particularly noteworthy because we have previously shown that down-regulation of PcG genes in the gut of G418-exposed flies contributes to the induction of neoGFP expression in this region (Stern et al. 2012).
Changes which contribute to the transgenerational inheritance are expected to persist in subsequent generations (>F2) of non-exposed embryos. We therefore checked if the reduction in PcG genes persists in the F3 generation. F2 embryos of exposed and non-exposed flies were developed to adulthood without G418 and these F2 flies were allowed to lay eggs for 2 h (0–2 h AEL). We used qPCR to examine the persistence of changes in these F3 embryos (two generations after exposure to G418). Analysis of maternal RNA revealed lower levels of Pc, Pcl and E(z) in F3 embryos that descended from flies that were exposed in F1 compared to the case of no exposure in F1 (Fig. 4A). These results indicate that, in addition to the suppressive effect of G418 on PcG gene expression in the exposed F1 larval gut (Stern et al. 2012), exposure to G418 leads to persistent reduction of maternal PcG RNA in subsequent generations.
Figure 4. Changes in maternal RNA in F2 embryos of G418-exposed flies tend to persist in the following generation (F3).
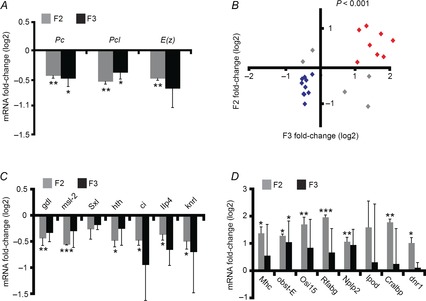
A, qPCR-based measurements of RNA of the PcG genes Pc, Pcl and E(z) in 0–2 h embryos of the F2 and F3 generations. Shown are relative levels in decedents of G418-exposed versus non-exposed F1 flies. Values are mean fold-change ± SEM based on three biological replicates. B, correlation between measurements in the F2 and F3 generations. Shown are fold-change qPCR-based measurements of maternal RNA corresponding to 23 genes. Fold-change refers to the difference between decedents of G418-exposed and non-exposed F1 flies. Genes that were up- and down-regulated in both generations are highlighted in red and blue, respectively. Significance of the positive correlation (P < 10−3) was determined by a random permutation test (see Methods). C, fold-change of maternal RNA levels for the blue highlighted genes in B. Values are mean fold-change ± SEM based on three biological replicates. D, as in C for red highlighted genes. *P < 0.05, **P < 0.01, ***P < 0.001 (Student's t test).
To test the persistence of changes in the expression of other genes, we analysed the F3 maternal levels of 23 developmental and metabolic genes that we have initially chosen for validating the RNA-seq data. For these genes, we evaluated the expression difference by qPCR and examined the persistence of the changes in the F3 embryos. We found a significant correlation between the changes in F2 and F3, with 18 out of the 23 genes (78%) exhibiting correlated changes (P < 10−3; Fig. 4B). Ten genes that were down-regulated in F2 were also down-regulated in the F3 generation following G418 exposure in F1 (Fig. 4C). Eight genes that were up-regulated in the F2 generation were also up-regulated in the F3 generation (Fig. 4D). Despite the statistically significant consistency between the two generations with respect to the overall direction of change, the changes in F3 were much more variable, suggesting gradual reversal of changes in the absence of further exposure to the stress.
Reduction in maternal gene dosage of Polycomb contributes to the inheritance of induced neoGFP expression in the proventriculus
Next, we sought to investigate whether the reduction in maternal Polycomb RNA is causally involved in the inheritance of induced responses to G418. Polycomb mutant mothers have been shown to exhibit maternal effects in their progeny (Denell, 1982). We therefore used Polycomb heterozygous mutants to test if the reduction in maternal Polycomb gene dosage influences inheritance in the F2 generation (Fig. 5A). We crossed hairy::neoGFP females carrying a Polycomb loss-of-function allele (Pc3 or Pc1) to non-mutant hairy::neoGFP males and analysed the 50% of the progeny that did not genetically inherit the Polycomb mutation. We compared larval progenies of Polycomb mutant mothers to larvae of the same genotype which were produced by non-mutant males and females. This comparison was performed with or without G418 exposure of both parents in F1 (Fig. 5A).
Analysis of neoGFP expression in the proventriculli of third-instar hairy::neoGFP, F2 larvae revealed significantly higher levels in offspring of exposed Pc3 females compared to offspring of exposed females with normal Polycomb gene dosage (Fig. 5B). Similar findings were observed using the Pc1 mutant line (Fig. 5C), indicating that the difference in the progeny indeed reflects the reduction in maternal Polycomb gene dosage and not potentially other line-specific factors. Notably, the observed difference between the progeny of Polycomb mutant and normal mothers depended on past exposure to G418 and was not observed without the exposure in F1 (Fig. 5B and C). This suggests that the phenotypic difference between these progeny does not reflect potential genetic variations between them.
In contrast to the effect of maternal Polycomb gene dosage on the inheritance of neoGFP induction, the reduction in Polycomb had no effect on the inheritance of delayed development; we found no clear difference in pupation time between offspring of exposed Pc3 or Pc1 females and offspring of exposed females with normal Polycomb gene dosage (Fig. 6A and B). This lack of difference in pupation time suggests strongly that the mechanisms underlying the inheritance of induced expression do not fully overlap with the mechanisms responsible for the inheritance of the delay in development. Additionally, our data indicate that the stress-induced suppression of maternal Polycomb contributes to the transgenerational inheritance of induced neoGFP expression in the foregut, but not to the inheritance of delayed development.
Discussion
We have previously shown that exposure of fly larvae to G418 toxicity induces multiple phenotypes that are inherited across several generations of non-exposed offspring (Stern et al. 2012). We have also shown that some of the phenotypes were induced by down-regulation of PcG genes in the gut of stressed larvae (Stern et al. 2012). Here, we investigated molecular events associated with the inheritance of one of the induced phenotypes, namely induced expression of neoGFP under regulation of the hairy promoter. As a starting point for this investigation we analysed embryos containing mostly maternal RNA (0–2 h AEL), aiming to evaluate the possibility of transgenerational influence by differential loading of maternal transcripts into the embryos. While the RNA in the 0–2 h interval is dominated by maternal RNA, it also includes lower levels of zygotic transcripts (Thomsen et al. 2010). Still, the difference in the RNA content at the mostly maternal (0–2 h) stage was substantially larger than the difference observed in the purely zygotic (8–10 h stage). This suggests that the extent of change by differential deposition of maternal transcripts following the G418 stress exceeds the extent of change by differential expression of zygotic genes.
Among the changes in the 0–2 h interval we identified reduction in RNA levels of PcG genes. This shows that, in addition to the effect of G418 on the levels of PcG genes in the gut, G418 also reduces the levels of PcG transcripts in the early-stage embryos of exposed flies. Unlike the down-regulation of PcG gene levels in the gut (Stern et al. 2012), the reduction of PcG genes in the early embryo persisted for at least another generation without exposure to G418. Independent analysis with Polycomb mutant lines further indicated that reduction of maternal Polycomb gene dosage enhances the inheritance of induced neoGFP expression in the larval foregut. These findings show that the effect of G418 on the levels of PcG genes is not limited to the foregut and that reduction of PcG gene levels in different tissues leads to related but nonetheless distinct effects. Specifically, the down-regulation of PcG genes in the gut promotes the induction of expression (Stern et al. 2012), while the reduction of Polycomb levels in the early embryos contributes to the inheritance of this induction.
Polycomb and other regulators have been previously implicated in transgenerational epigenetic phenomena (Cavalli & Paro, 1998; Katz et al. 2009; Greer et al. 2011; Seong et al. 2011). Additional mechanisms that have been shown to mediate transgenerational phenomena in various organisms include DNA methylation (Cubas et al. 1999; Morgan et al. 1999; Rakyan et al. 2002; Lane et al. 2003; Xing et al. 2007; Dunn & Bale, 2009; Franklin et al. 2010; Ng et al. 2010), histone modifications (Hammoud et al. 2009; Katz et al. 2009; Brykczynska et al. 2010; Carone et al. 2010; Furuhashi et al. 2010; Rechtsteiner et al. 2010; Arico et al. 2011; Burton et al. 2011; Greer et al. 2011; Buckley et al. 2012; Gu et al. 2012; Shirayama et al. 2012) and non-coding RNAs such as piRNA (Ashe et al. 2012; Bagijn et al. 2012; de Vanssay et al. 2012; Grentzinger et al. 2012; Lee et al. 2012; Shirayama et al. 2012), miRNA (Rassoulzadegan et al. 2006; Wagner et al. 2008; Grandjean et al. 2009; Carone et al. 2010; Morgan & Bale, 2011; Halfmann et al. 2012) and siRNA (Grishok et al. 2000; Vastenhouw et al. 2006; Alcazar et al. 2008; Burton et al. 2011; Rechavi et al. 2011; Buckley et al. 2012; Gu et al. 2012). Here we extend this repertoire by providing evidence connecting changes in maternal RNA levels of Polycomb with transgenerational inheritance of environmentally induced gene expression in flies.
Our results indicate that reduction in Polycomb levels at a stage preceding the bulk of zygotic transcription affects the expression of genes at a much later stage (post-embryonic, third-instar larva). Although the main function of Polycomb occurs during zygotic transcription (Nègre et al. 2006), maternal effects of PcG genes (Struhl, 1981; Denell, 1982; Haynie, 1983; Phillips & Shearn, 1990; Martin & Adler, 1993) and trithorax have also been described (Ingham & Whittle, 1980). In particular, the Pc3 allele included in this study was shown to have a stronger maternal effect than other Polycomb mutant alleles (Denell, 1982). This suggests that the impact of the maternal effect might depend on the degree of reduction in Polycomb function. Our results are consistent with this suggestion. We found that the maternal reduction of Polycomb dosage was consequential for the inheritance of neoGFP expression in the offspring only when the parents were exposed to G418. In this case, the reduction in maternal PcG gene levels combines with the genetic reduction in Pc gene dosage, resulting in increased shortage in maternal Polycomb function. Yet, the lack of an observed maternal effect without a history of exposure to G418 suggests that additional, stress-dependent factors are also required to support the maternal effect of Polycomb on the inheritance. Together, the evidence suggests that parental environment may influence the development of offspring by modifying the maternal RNA levels of key epigenetic regulators. This evidence for functional involvement of maternal RNA does not exclude the possibility of additional contribution of stress-induced modifications in the male, which could persist in the offspring by means that are different from maternal deposition of RNA.
In addition to the change in PcG genes, we observed broad changes in the milieu of maternal RNAs in progenies of flies that were exposed to the toxic stress. The affected genes are involved in a wide range of biological processes, including developmental, metabolic and stress responses. This indicates that, in addition to the effect of G418 toxicity on somatic tissues, it has a broad influence on the germline. This influence might reflect direct exposure of the germline to G418 and/or indirect interactions with somatic tissues that have been exposed to G418. The persistence of some of the changes following another generation (i.e. in F3) indicates that at least some of the modifications in the germline are not caused by direct exposure to G418. This probably reflects mechanisms which preserve the altered state of development in the F2 generation. Further studies are required to determine if this persistence can be enhanced by successive exposure to the stress, and if the inheritance is mediated solely by changes in maternal RNA or involves additional epigenetic, metabolic and other types of changes that are not completely reset between generations.
Involvement of additional mechanisms of inheritance is indeed supported by the apparent lack of influence of Polycomb on the inheritance of the delay in development. While the reduction in maternal Polycomb gene dosage contributed to the induced expression in the gut of the offspring larvae (Fig. 5), it had no clear effect on the rate of development of these larvae (Fig. 6). This suggests that inheritance of the delay is mediated by other mechanisms. In line with this suggestion, we have recently uncovered involvement of the commensal microbiome in the inheritance of this delay in development (Y. Fridmann-Sirkis et al. 2014). Specifically, we have found that in addition to the effect of G418 on the host tissue, it also depletes Acetobacter species from the gut. This depletion was itself heritable but it did not change the levels of Polycomb in the gut and it did not induce the expression of the resistance gene (Y. Fridmann-Sirkis et al. 2014). Nonetheless, the depletion of Acetobacter species in F1 led to a considerable delay in larval development in F2. We showed that this transgenerational effect of bacterial depletion was responsible for the inheritance of the delay in development following exposure to G418 in F1 (Y. Fridmann-Sirkis et al. 2014). Together, the current work and the study of Y. Fridmann-Sirkis et al. portray two separate modes of transgenerational influence, each contributing to the inheritance of a different phenotype, namely: the reduction of maternal levels of Polycomb has a positive effect on the inheritance of induced expression while the disruption of the microbiome is responsible for the inheritance of the delay in larval development.
Having more than one mechanism of non-Mendelian inheritance in a single setting of stress suggests that the inheritance of environmentally induced phenotypes is probably more prevalent than often assumed. Although we identified involvement of maternal RNA and commensal bacteria in transgenerational inheritance of induced responses in flies, these modes of transgenerational influence may be broadly relevant to many other animals. Indeed, early development of many animal species, including in mammals, is guided and influenced by RNA and other cytoplasmic determinants deposited by the mother prior to the onset of zygotic transcription (Telford et al. 1990; Tadros & Lipshitz, 2009). How this deposition is affected by maternal physiology and history of environmental exposures is largely unknown. It is also unknown if this influence can be used to prepare the organism for an anticipated change in the environment and whether these maternal influences can accumulate and contribute to offspring adaption over time in the altered environment. Similar considerations apply to the potential involvement of the commensal microbiome, which is an integral part of the physiology and development of many if not all organisms, including all mammals. These host-intrinsic (e.g. epigenetic) and extrinsic (i.e. microbiome-mediated) examples offer substantial infrastructure for influence that may extend well beyond one generation, thus providing a hitherto under-appreciated bridge between physiological responses and the much larger time scales of genetic adaptation. Deciphering how the environment influences physiology on time scales of multiple generations and whether this might feedback on the host genome may add another dimension to our understanding of gene regulation, inheritance, adaptation and evolution.
Acknowledgments
We thank the biological services unit of the Weizmann Institute of Science for the hard work and effort they have put into this project.
Glossary
- AEL
after egg laying
- ANOVA
analysis of variance
- GO
Gene Ontology
- PcG
Polycomb group
- RPKM
reads per kilobase of exon model per million mapped reads
Additional information
Competing interests
None declared.
Author contributions
Study concept and design: S.S., S.O. and S.Y. Acquisition of data: S.S., S.O., M.E., G.M. and Z.I. Analysis and interpretation of data: S.S., S.O., M.E. and S.Y. Drafting of manuscript: S.S., S.O. and S.Y. All authors approved the final version of the manuscript. All experiments were performed in the Department of Biological Chemistry, Weizmann Institute of Science, Rehovot, Israel.
Funding
This work was supported by the John Templeton Foundation (grant ID: #40663), the Minerva-Weizmann Program, and The Israel Science Foundation (grant No. 1860/13). Y.S. is Incumbent of the Daniel E. Koshland Sr. Career Development Chair at the Weizmann Institute. S.S. was supported by the Clore fellowship and the ‘Kahn Family Research Center on Systems Biology of the Human Cell’ award.
Supporting Information
The following supporting information is available in the online version of this article.
Table S1. Enrichments of Gene Ontology (GO) annotations in the group of genes which changed in the maternal stage (0–2 h AEL) following parental exposure to G418.
References
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico JK, Katz DJ, van der Vlag J, Kelly WG. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 2011;7:e1001391. doi: 10.1371/journal.pgen.1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Rubin GM. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci U S A. 2012;109:9143–9148. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropley JE, Suter CM, Beckman KB, Martin DIK. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc Natl Acad Sci U S A. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bougé A-L, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Denell RE. Homoeosis in Drosophila – evidence for a maternal effect of the Polycomb locus. Dev Genet. 1982;3:103–113. [Google Scholar]
- Dunn GA, Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150:4999–5009. doi: 10.1210/en.2009-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Larsson P, Longden I, McLaren W, Overduin B, Pritchard B, Riat HS, Rios D, Ritchie GR, Ruffier M, Schuster M, Sobral D, Spudich G, Tang YA, Trevanion S, Vandrovcova J, Vilella AJ, White S, Wilder SP, Zadissa A, Zamora J, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernández-Suarez XM, Herrero J, Hubbard TJ, Parker A, Proctor G, Vogel J, Searle SM. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Stern S, Elgart M, Galili M, Zeisel A, Shental N, Soen Y. Delayed development induced by toxicity to the host can be inherited by a bacterial-dependent, transgenerational effect. Front Genet. 2014;5 doi: 10.3389/fgene.2014.00027. doi: 10.3389/fgene.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi H, Takasaki T, Rechtsteiner A, Li T, Kimura H, Checchi PM, Strome S, Kelly WG. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, Chambeyron S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–1888. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang SH, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie JL. The maternal and zygotic roles of the gene Polycomb in embryonic determination in Drosophila melanogaster. Dev Biol. 1983;100:399–411. doi: 10.1016/0012-1606(83)90234-8. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingham P, Whittle R. Trithorax: a new homoeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet. 1980;179:607–614. [Google Scholar]
- Jablonka E. Epigenetic inheritance and plasticity: the responsive germline. Prog Biophys Mol Biol. 2012;111:99–107. doi: 10.1016/j.pbiomolbio.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ. The inheritance of acquired epigenetic variations. J Theor Biol. 1989;139:69–83. doi: 10.1016/s0022-5193(89)80058-x. [DOI] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JP, Brunet A. Bridging the transgenerational gap with epigenetic memory. Trends Genet. 2013;29:176–186. doi: 10.1016/j.tig.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EC, Adler PN. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development. 1993;117:641–655. doi: 10.1242/dev.117.2.641. [DOI] [PubMed] [Google Scholar]
- Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31:11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nègre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S-F, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Phillips MD, Shearn A. Mutations in polycombeotic, a Drosophila polycomb-group gene, cause a wide-range of maternal and zygotic phenotypes. Genetics. 1990;125:91–101. doi: 10.1093/genetics/125.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang WC, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation–revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Seong K-H, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- Stern S, Fridmann-Sirkis Y, Braun E, Soen Y. Epigenetically heritable alteration of fly development in response to toxic challenge. Cell Rep. 2012;1:528–542. doi: 10.1016/j.celrep.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11:R93. doi: 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882–882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Xing Y, Shi S, Le L, Lee CA, Silver-Morse L, Li WX. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 2007;3:1598–1606. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37–e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


