Abstract
It is widely accepted that the crosstalk between naive nucleus and maternal factors deposited in the egg cytoplasm before zygotic genome activation is crucial for early development. This crosstalk may also exert some influence on later development. It is interesting to clarify the relative roles of the zygotic genome and the cytoplasmic factors in development. Cross-species nuclear transfer (NT) between two distantly related species provides a unique system to study the relative role and crosstalk between egg cytoplasm and zygotic nucleus in development. In this review, we will summarize the recent progress of cross-species NT, with emphasis on the cross-species NT in fish and the influence of cytoplasmic factors on development. Finally, we conclude that the developmental process and its evolution should be interpreted in a systemic way, rather than in a way that solely focuses on the role of the nuclear genome.
 |
Yong-Hua Sun obtained his bachelor degree from Wuhan University (China) in 1997 and a PhD degree from the University of the Chinese Academy of Sciences in 2002. He is a professor of fish developmental biotechnology at the Institute of Hydrobiology, Chinese Academy of Sciences and the director of the China Zebrafish Resource Center (http://zfish.cn). Professor Sun studied fish developmental biology and fish biotechnology. Zuo-Yan Zhu graduated from Peking University (China) in 1964 and the graduate school of the Chinese Academy of Sciences in 1980. He was elected an Academician of the Chinese Academy of Sciences in 1997 and a member of the Third World Academy of Sciences in 1998. He is currently a professor of fish developmental biology and biotechnology at the Institute of Hydrobiology, Chinese Academy of Sciences, and at Peking University. Professor Zhu is a pioneer in the field of fish transgenic studies.
Introduction: environmental factors in genetics and development
It is widely accepted that the crosstalk between naive nucleus and maternal factors deposited in the egg cytoplasm of vertebrates before zygotic genome activation is crucial for early development (Dosch et al. 2004). This crosstalk may also exert some influence on later developmental characteristics. Therefore, it is interesting to clarify the relative roles of zygotic genome and cytoplasmic factors in development. The direct evidence of maternal control on development comes from the screening of maternal-effect mutants in zebrafish (Dosch et al. 2004; Wagner et al. 2004). Cross-species nuclear transfer (NT) between two distantly related species, which have distinct appearances or phenotypes, provides a unique system to study the relative role and crosstalk between the egg cytoplasm and the zygotic nucleus in development (Fig. 1; Pei et al. 2007). In this review, we will summarize the recent progress of cross-species NT, with emphasis on the influence of cytoplasmic factors on development. We conclude that the developmental process and its evolution should be interpreted in a systemic way, for example, to consider the genome and the environment at different levels, rather than in a way that solely focuses on the role of the nuclear genome.
Figure 1. Diagram of cross-species nuclear transfer (NT).
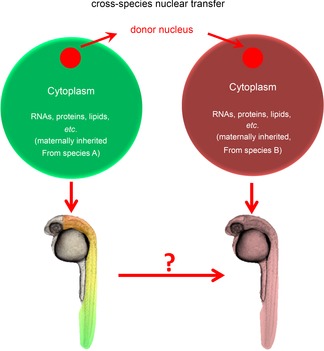
Left: development of a regularly fertilized embryo of species A; right: development of a cross-species NT embryo with a nucleus from species A combined with an enucleated egg from species B. The egg cytoplasm of species A and species B contains different types of maternal factors, such as RNAs, proteins and lipids, etc. After nuclear transfer, the reconstructed embryo that contains the nucleus of species A and the cytoplasm of species B may develop into an animal that does not fully resemble species A.
Cross-species NT in amphibians and mammals
A species, as a basic unit in biological taxonomy, is considered as a group of organisms that can breed naturally and produce fertile offspring. Different species usually have different genomic materials and distinct developmental shapes. NT is defined as transferring the nuclei of donor cells into enucleated oocytes or eggs to generate reconstructed embryos, which may have the ability to develop to term. If NT is done within one species, i.e. donor cells and oocytes (eggs) come from the same species, it is called inter-species NT. Inter-species NT has been used to study developmental plasticity and nuclear reprogramming of the donor nucleus and to generate reprogrammed stem cells from differentiated cells (Gurdon & Wilmut, 2011).
However, if the oocytes and donor cells come from two different species, the NT will be defined as cross-species NT. Cross-species NT was first described in amphibians, within the genera of Rana or Xenopus (Moore, 1960; Gurdon, 1962). In those studies, all the NT embryos showed early developmental arrest, probably due to the incomplete reprogramming of the donor nuclei and/or incompatibility between the nuclei and the egg cytoplasm that contains mitochondria from different species. In mammals, although some reprogramming events such as sperm demethylation occur in cross-species NTs (Beaujean et al. 2004), the NT embryos usually die at the stage when zygotic transcription starts, suggesting that the egg cytoplasmic environment is crucial for the proper development of transferred nuclei. Nevertheless, cross-species NT has succeeded in cloning some endangered mammals, such as the gaur (Lanza et al. 2000), the mouflon (Loi et al. 2001), the African wild cat (Gomez et al. 2004), the sand cat (Gómez et al. 2008) and the coyote (Hwang et al. 2012), by using the oocytes from closely related species. It is commonly reported that the cloned animals are identical to their nuclear donors in genotypes and phenotypes, indicating the significant dominance of the nuclear genome in phenotypic determination.
Cross-species NT in fishes – effect of cytoplasmic factors on development
In fish, a type of relatively primitive vertebrate, cross-species NT could be achieved in quite a few genetically distant species. The art of fish NT was first demonstrated with goldfish and bitterling fish by Tung et al. (1963). Later, cross-species NT was conducted between two different genera, such as a combination of common carp (Cyprinus carpio, genus Cyprinus) nuclei with crucian carp (Carassius auratus, genus Carassius) egg cytoplasm (Tung & Tung, 1980), as well as crucian carp nuclei with common carp egg cytoplasm (Yan et al. 1984), in order to obtain nucleo-cytoplasmic hybrid fish with improved economical traits. In those studies led by Tung, it was found that the vertebral numbers of some NT fish were consistent with those of the egg-providing species, but, unfortunately, no conclusive evidence was provided, and the results have been challenged by the scientific community to a certain extent (Gurdon, 1986; Wakamatsu et al. 2001). Cross-species fish NT was even conducted between members of two different families, such as the goldfish (Carassius auratus, family Cyprinidae, order Cypriniformes) and the loach (Paramisgurnus dabryanus, family Cobitidae, order Cypriniformes), and between two orders, such as the tilapia (Oreochromis nilotica, order Perciformes) and the goldfish, as well as the tilapia and the loach (Yan et al. 1990, 1991). However, there were only suggestions, with no confirming evidence, that the cross-species NT fish actually were nucleo-cytoplasmic hybrids.
In recent years, with the development of transgenic fish (Zhu & Sun, 2000), we were able to generate cross-genus cloned fish by transferring the nuclei of transgenic common carps into the enucleated eggs of goldfish (Carassius auratus) (Sun et al. 2005). By analysing transgene and comparative DNA fingerprint markers, we proved that the nuclear genomes of the cloned fish were exclusively derived from the nuclear donor species, the transgenic common carp, instead of the egg-providing species, the goldfish, whereas the mitochondrial DNA from the donor carp gradually disappeared during the development of NT embryos, and only the mitochondrial DNA from recipient goldfish existed in the NT adults. Therefore, the cross-genus cloned fish is really a type of nucleo-cytoplasmic hybrid, with a nuclear genome from the transgenic donor and egg cytoplasm from the recipient species. All the NT fish were identical with the nucleus-providing common carp regarding exterior phenotypic characteristics, such as long body shape, two pairs of barbels, a normal tail and normal eyes. By contrast, there was almost no visible contribution of distinctive goldfish characteristics, such as spherical body shape, triangular tail and ‘dragon’ eyes (Fig. 2A). Strikingly, somite development and somite number of nuclear transplants were consistent with the recipient species, the goldfish, rather than the nuclear donor species, the common carp. This resulted in a long-lasting effect on the vertebral numbers of the cloned fish, as vertebrae develop from the embryonic somites. The vertebral numbers of the cloned fish belonged within the range of goldfish, which has 28–30 vertebrae, and were distinctly different from those of the common carp, with vertebral numbers of 32–36 (Fig. 2B). This demonstrates that fish egg cytoplasm can not only support the development driven by transplanted nuclei from a distantly related species at the genus scale, but can also significantly modulate development of the nuclear transplants.
Figure 2. Influence of cytoplasmic factors on development of the cloned fish.

A, exterior phenotype of the egg-providing goldfish (left), the common carp (middle) and the cloned fish (right). Note that the NT fish was identical to the nucleus-providing common carp regarding exterior phenotypic characteristics, such as long body shape, two pairs of barbels, normal tail and normal eyes, but there was almost no visible contribution of distinctive goldfish characteristics, such as spherical body shape, triangular tail and dragon eyes; B, X-ray analysis of the egg-providing goldfish (left), the common carp (middle) and the cloned fish (right). Note that the vertebral numbers of the cloned fish belonged to the range of goldfish, which has 28–30 vertebrae, distinctly different from those of the common carp, with vertebral numbers of 32–36.
In order to study the cross-species NT in more detail, we established two research models. First, for the study of the early nucleo-cytoplasmic interaction in cross-species NT, we utilized two laboratory fish species from different subfamilies, the zebrafish (Danio rerio) and the rare minnow (Gobiocypris rarus), in order to generate cross-subfamily NT embryos (Pei et al. 2007). We further used suppression subtractive hybridization (SSH) to screen out differentially expressed genes from the forward and reverse subtracted cDNA libraries. After dot blot and real-time PCR analysis, 80 of 500 randomly selected sequences were proven to show transcriptional differences in the cloned embryos. Among them, 45 sequences shared high homology with 28 known zebrafish genes, and 35 sequences were corresponding to 22 novel expressed sequence tags (ESTs). Based on the analysis of gene ontology and literature mining, up- and down-regulated genes in the cross-subfamily cloned embryos were shown to be relevant to transcription and translation initiation, cell cycle regulation, protein binding, etc. Therefore, we concluded that the fish egg cytoplasm can not only support the division and development of nuclei from distantly related species, but can also exert a certain impact on the genetic modulation and cellular homeostasis of the transferred nuclei.
Second, zebrafish and Chinese rare minnow were also utilized to produce mutual crossbred embryos in order to examine the impact of the cytoplasm from different species on a common type of nucleus. Although these two types of crossbred embryos originated from common nuclei of the same genetic materials, diverse developmental capacities were gained due to different cytoplasmic environments from different species (Liu et al. 2008). Using the cDNA amplified fragment length polymorphism (cDNA-AFLP) approach, we compared transcript profiles between the mutual crossbred embryos at two developmental stages (50%- and 90%-epiboly). Three thousand cDNA fragments were generated in four cDNA pools with 64 primer combinations. Compared with ZR (zebrafish ♀ × Chinese rare minnow ♂) embryos, 12 genes were up-regulated and 12 were down-regulated in RZ (Chinese rare minnow ♀ × zebrafish ♂) embryos. The sequences encoded variant proteins which function at different levels of proliferation, growth and development. This strongly suggests that different egg cytoplasms exert completely different impacts on a common nucleus.
Conclusions
Overall, the recent studies of cross-species NT fish experimentally revealed that the nucleus placed in the circumstance of different egg cytoplasm can be strongly influenced by the cytoplasmic factors, at the levels of both genetic regulation and phenotypic determination. On the other hand, introgressive hybridization in animals, especially in fishes, is one of the driving forces of evolution (Smith, 1992; Dowling & DeMarais, 1993), and this type of hybridization certainly includes the crosstalk between nucleus and cytoplasm from different species. Therefore, any type of developmental process and its evolution should be interpreted in a systemic way rather than in a way that solely focuses on the role of the nuclear genome.
Acknowledgments
None declared.
Additional information
Competing interests
The authors have no conflicts of interest to declare.
Funding
This work was supported by the China 973 Project (Grant Nos. 2010CB126306 and 2012CB944504) and the National Science Fund for Excellent Young Scholars of the Natural National Science Foundation of China (NSFC) (Grant No. 31222052) to Y.H.S.
References
- Beaujean N, Taylor JE, McGarry M, Gardner JO, Wilmut I, Loi P, Ptak G, Galli C, Lazzari G, Bird A, Young LE, Meehan RR. The effect of interspecific oocytes on demethylation of sperm DNA. Proc Natl Acad Sci U S A. 2004;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: Mutants from the zebrafish I. Dev Cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Dowling TE, DeMarais BD. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature. 1993;362:444–446. [Google Scholar]
- Gómez MC, Pope CE, Giraldo A, Lyons LA, Harris RF, King AL, Cole A, Godke RA, Dresser BL. Birth of African Wildcat cloned kittens born from domestic cats. Cloning Stem Cells. 2004;6:247–258. doi: 10.1089/clo.2004.6.247. [DOI] [PubMed] [Google Scholar]
- Gómez MC, Pope CE, Kutner RH, Ricks DM, Lyons LA, Ruhe M, Dumas C, Lyons J, López M, Dresser BL. Nuclear transfer of sand cat cells into enucleated domestic cat oocytes is affected by cryopreservation of donor cells. Cloning Stem Cells. 2008;10:469–484. doi: 10.1089/clo.2008.0021. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The transplantation of nuclei between two species of Xenopus. Dev Biol. 1962;5:68–83. doi: 10.1016/0012-1606(62)90004-0. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. Nuclear transplantation in eggs and oocytes. J Cell Sci Suppl. 1986;4:287–318. doi: 10.1242/jcs.1986.supplement_4.17. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harb Perspect Biol. 2011;3:a002659. doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Jeong YW, Kim JJ, Lee HJ, Kang M, Park KB, Park JH, Kim YW, Kim WT, Shin T. Successful cloning of coyotes through interspecies somatic cell nuclear transfer using domestic dog oocytes. Reprod Fertil Dev. 2012;25:1142–1148. doi: 10.1071/RD12256. [DOI] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Diaz F, Moraes CT, Farin PW, Farin CE, Hammer CJ, West MD, Damiani P. Cloning of an endangered species (Bos gaurus) using interspecies nuclear transfer. Cloning. 2000;2:79–90. doi: 10.1089/152045500436104. [DOI] [PubMed] [Google Scholar]
- Liu J, Sun YH, Wang YW, Wang N, Pei DS, Wang YP, Hu W, Zhu ZY. Identification of differential transcript profiles between mutual crossbred embryos of zebrafish (Danio rerio) and Chinese rare minnow (Gobiocypris rarus) by cDNA-AFLP. Theriogenology. 2008;70:1525–1535. doi: 10.1016/j.theriogenology.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Loi P, Ptak G, Barboni B, Fulka J, Jr, Cappai P, Clinton M. Genetic rescue of an endangered mammal by cross-species nuclear transfer using post-mortem somatic cells. Nat Biotechnol. 2001;19:962–964. doi: 10.1038/nbt1001-962. [DOI] [PubMed] [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci U S A. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JA. Serial back-transfers of nuclei in experiments involving two species of frogs. Dev Biol. 1960;2:535–550. doi: 10.1016/0012-1606(60)90053-1. [DOI] [PubMed] [Google Scholar]
- Pei DS, Sun YH, Chen SP, Wang YP, Hu W, Zhu ZY. Identification of differentially expressed genes from the cross-subfamily cloned embryos derived from zebrafish nuclei and rare minnow enucleated eggs. Theriogenology. 2007;68:1282–1291. doi: 10.1016/j.theriogenology.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Roche HM, Phillips C, Gibney MJ. The metabolic syndrome: the crossroads of diet and genetics. Proc Nutr Soc. 2005;64:371–377. doi: 10.1079/pns2005445. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Slack JMW. Essential Developmental Biology. 3rd edn. Wiley-Blackwell; 2012. [Google Scholar]
- Smith GR. Introgression in Fishes: Significance for Paleontology, Cladistics, and Evolutionary Rates. Syst Biol. 1992;41:41–57. [Google Scholar]
- Sun YH, Chen SP, Wang YP, Hu W, Zhu ZY. Cytoplasmic impact on cross-genus cloned fish derived from transgenic common carp (Cyprinus carpio) nuclei and goldfish (Carassius auratus) enucleated eggs. Biol Reprod. 2005;72:510–515. doi: 10.1095/biolreprod.104.031302. [DOI] [PubMed] [Google Scholar]
- Tung T, Tung Y. Nuclear transplantation in teleosts. I. Hybrid fish from the nucleus of carp and the cytoplasm of crucian. Scientia (Peking) 1980;14:1244–1245. [Google Scholar]
- Tung T, Wu S, Tung Y, Yan S, Tu M, Lu T. Nuclear transplantation in fish. Chin Sci Bull, Academia Sinica. 1963;7:60–61. (In Chinese.) [Google Scholar]
- Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell. 2004;6:781–790. doi: 10.1016/j.devcel.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Ju B, Pristyaznhyuk I, Niwa K, Ladygina T, Kinoshita M, Araki K, Ozato K. Fertile and diploid nuclear transplants derived from embryonic cells of a small laboratory fish, medaka (Oryzias latipes. Proc Natl Acad Sci U S A. 2001;98:1071–1076. doi: 10.1073/pnas.98.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CY, Wang HP, Zhu ZY, Sun YH. Transcriptional factors Smad1 and Smad9 act redundantly to mediate zebrafish ventral specification downstream of Smad5. J Biol Chem. 2014;289:6604–6618. doi: 10.1074/jbc.M114.549758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Lu D, Du M, Li G, Lin L, Jin G, Wang H, Yang Y, Xia D, Liu A. Nuclear transplantation in teleosts. Hybrid fish from the nucleus of crucian and the cytoplasm of carp. Sci Sin B. 1984;27:1029. [PubMed] [Google Scholar]
- Yan SY, Mao ZR, Yang HY, Tu MA, Li SH, Huang GP, Li GS, Guo L, Jin GQ, He RF, et al. Further investigation on nuclear transplantation in different orders of teleost: the combination of the nucleus of Tilapia (Oreochromis nilotica) and the cytoplasm of Loach (Paramisgurnus dabryanus. Int J Dev Biol. 1991;35:429–435. [PubMed] [Google Scholar]
- Yan SY, Tu M, Yang HY, Mao ZG, Zhao ZY, Fu LJ, Li GS, Huang GP, Li SH, Jin GQ, et al. Developmental incompatibility between cell nucleus and cytoplasm as revealed by nuclear transplantation experiments in teleost of different families and orders. Int J Dev Biol. 1990;34:255–266. [PubMed] [Google Scholar]
- Zhu ZY, Sun YH. Embryonic and genetic manipulation in fish. Cell Res. 2000;10:17–27. doi: 10.1038/sj.cr.7290032. [DOI] [PubMed] [Google Scholar]


