Abstract
A challenge in the computational design of enzymes is that multiple properties must be simultaneously optimized -- substrate-binding, transition state stabilization, and product release -- and this has limited the absolute activity of successful designs. Here, we focus on a single critical property of many enzymes: the nucleophilicity of an active site residue that initiates catalysis. We design proteins with idealized serine-containing catalytic triads, and assess their nucleophilicity directly in native biological systems using activity-based organophosphate probes. Crystal structures of the most successful designs show unprecedented agreement with computational models, including extensive hydrogen bonding networks between the catalytic triad (or quartet) residues, and mutagenesis experiments demonstrate that these networks are critical for serine activation and organophosphate-reactivity. Following optimization by yeast-display, the designs react with organophosphate probes at rates comparable to natural serine hydrolases. Co-crystal structures with diisopropyl fluorophosphate bound to the serine nucleophile suggest the designs could provide the basis for a new class of organophosphate captures agents.
Serine hydrolases utilize a conserved nucleophilic serine to hydrolyze ester, amide or thio-ester bonds in proteins and small molecules and constitute one of the largest enzyme families in the human proteome1. The canonical serine-histidine-aspartic acid catalytic triad mechanism has been well-studied: the serine Oγ is the nucleophile, the imidazole ring of the histidine acts as a general acid/base, and the carboxylate of the aspartic acid orients the imidazole ring and neutralizes the charge developed on the histidine in the transition state. In addition to the catalytic triad residues, a highly conserved oxyanion-binding site, commonly referred to as the oxyanion hole, stabilizes the negative charge on the carbonyl oxygen of the tetrahedral intermediate. The peptide backbone NH groups stabilize these high-energy species in most cases2–7. The activated nucleophilic serine can be covalently modified by electrophilic organophosphate compounds resulting in the loss of catalytic activity. This has inspired the development of fluorophosphonates as activity-based probes for serine hydrolases, which enable monitoring of the activity of many serine hydrolases in parallel directly in complex proteomes by the Activity Based Protein Profiling (ABPP) technology8,9.
To make progress in enzyme design, it is useful to focus on critical aspects of catalysis (in this case, the active site nucleophile that initiates catalysis) independent of full multiple turnover reaction cycles whose optimization requires many complex tradeoffs — such reduction in complexity can facilitate the solution of very challenging problems. Recently, we designed esterases with a cysteine-histidine dyad and an oxyanion hole in the active site10. Though these designs contained active site cysteines with heightened nucleophilicity11 and were catalytically active, crystal structures revealed that the active site histidine residues, which were intended to activate the cysteine nucleophiles, were not properly positioned10. Catalytic activity was likely achieved because cysteine, with a pKa around 8, is one of the most intrinsically nucleophilic amino acids and its inherent reactivity can be enhanced without specific interactions with supporting residues in the enzyme’s active site. Compared to lysine, which was used in de novo designed retroaldolases12, and cysteine, serine has a much higher pKa at ~13 and hence its nucleophilicity typically depends on interactions with activating residues such as the histidine and aspartate/glutamate of the catalytic triad. It is therefore methodologically more challenging to design serines with heightened nucleophilicity — success likely depends on accurate designed hydrogen bonding interactions with activating residue(s).
Here we take on the challenge of designing active sites with serines with nucleophilicity approaching that found in the catalytic triads of native hydrolases. De novo designed proteins with activated serine nucleophiles could be useful in their own right as scavengers of organophosphate nerve agents. We use computational methods to design proteins with idealized serine-containing catalytic triads and take advantage of activity-based fluorophosphonate probes13 to screen and assess the serine nucleophilicity of these designs directly in native cellular systems.
Results
Computational design of catalytic triad active sites
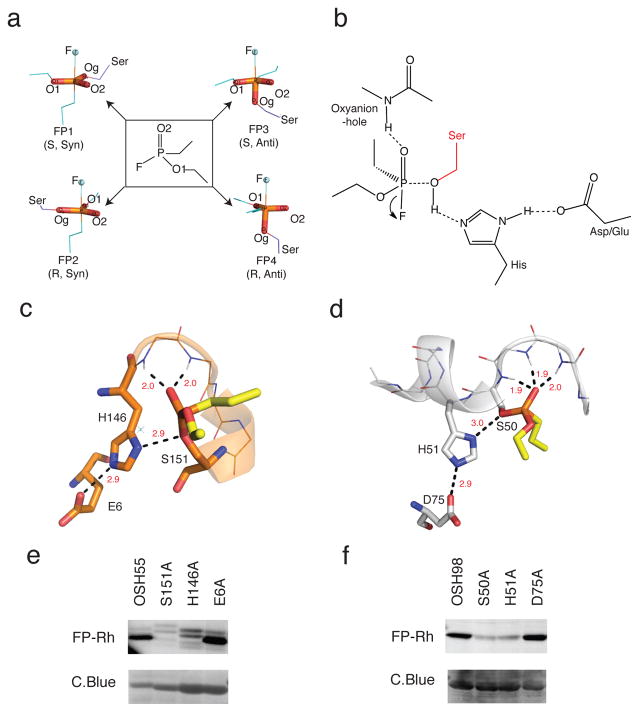
We selected a fluorophosphonate reactive group (Fig. 1a) as the model organophosphate substrate because it provides: 1) a suitably tempered electrophile to test the serine nucleophilicity of protein designs, and 2) a mimic of the tetrahedral transition state that occurs during serine hydrolase catalysis. From a technical perspective, reporter-tagged fluorophosphonates should provide a straightforward way to screen enzyme designs in complex proteomes, as we have shown for other ABPP probes11. Four transition state models corresponding to syn- and anti-attack for two fluorophosphonate isomers (R & S) were generated using bond angles and bond lengths obtained from QM/MM calculations (Fig. 1a and Supplementary Results, Supplementary Table 1)14. Ideal active sites with the transition state model, a serine-histidine-aspartate/glutamate catalytic triad, and a backbone oxyanion hole in orientations optimal for catalysis (Fig. 1b) were built as described in Methods. We used RosettaMatch to search for placements of these “theozymes” in a set of 800 protein scaffolds15, none of which contains an existing catalytic-triad based active site. The residues surrounding the matched sites were then optimized by RosettaDesign16 to stabilize the active site residues and/or to increase affinity for the fluorophosphonate ligand. 85 designs were chosen for experimental characterization based on the accuracy of the active site geometry following unconstrained combinatorial side-chain optimization and the transition state binding energy (See Methods). The overall design workflow is illustrated in Supplementary Fig. 1a and the scaffolds on which the 85 designs are based are listed in Supplementary Table 2.
Figure 1. Design strategy.
(a) Transition-state (TS) models. The fluorophosphonate (FP) ligand (ethyl ethylphosphonofluoridate) used in design calculations is in the middle panel flanked by TS models corresponding to syn-and anti- attack on the two isomers (R & S) of the ligand with the leaving atom fluorine (F) in blue-spheres. The serine nucleophile is shown in purple. (b) Theozyme geometry. Geometric parameters were derived from the computed ideal active site geometries and native hydrolase statistics previously reported30. Theozymes with the histidine hydrogen bonding reversed were also considered during RosettaMatch (See Methods). (c.d) Design models of OSH55 (c) and OSH98 (d). Active-site residues of OSH55 and OSH98 are shown in orange and white sticks respectively. The modeled fluorophosphonate ligand is shown in yellow sticks. In both designs, multiple backbone NHs form the oxyanion hole to stabilize the transition state. (e,f) Selective FP-Rh labeling of OSH55 (e) and OSH98 (f) but not the active site serine knockouts. Serine reactivity was assessed by gel-based ABPP (top, in-gel fluorescence; bottom, coomassie staining, see Supplementary Fig. 15 for full-size gel images. The experiments were performed in duplicates with consistent results).
Experimental screening of designs
Genes corresponding to the 85 designs were ordered and expressed in Escherichia coli. A significant fraction (~50%) of the designs did not express as soluble proteins; the requirement that the active site be preformed favored designs with the catalytic triad Asp or Glu at least partially buried, and this may have destabilized many of the proteins. The reactivity of the active site serine in the soluble designs was assessed in cell lysates by gel-based ABPP (Supplementary Fig. 1b) using fluorophosphonate-rhodamine (FP-Rh) and fluorophosphonate-alkyne (FPyne) probes (Supplementary Fig. 2). Two of the designs, OSH55 and OSH98 (Fig. 1c, d) showed modest reactivity with the fluorophosphonate probes. The catalytic triads in the designs involve all non-native residues; for example in OSH55, the catalytic triad residues Ser151, His146 and Glu6 are Asp, Gly and Val respectively in the native scaffold (PDB ID: 2DX6) (Supplementary Fig. 3). Mutagenesis of the designed active site serine and histidine residues to alanine indicated that probe labeling was specific to the designed serines (Fig. 1e, f). OSH55 was chosen for further characterization due to its small size (163 amino acids) and the stability of the starting scaffold. In the design model, OSH55 has two backbone amide protons contributing to the oxyanion hole (Fig. 1c)
Characterization and optimization of OSH55
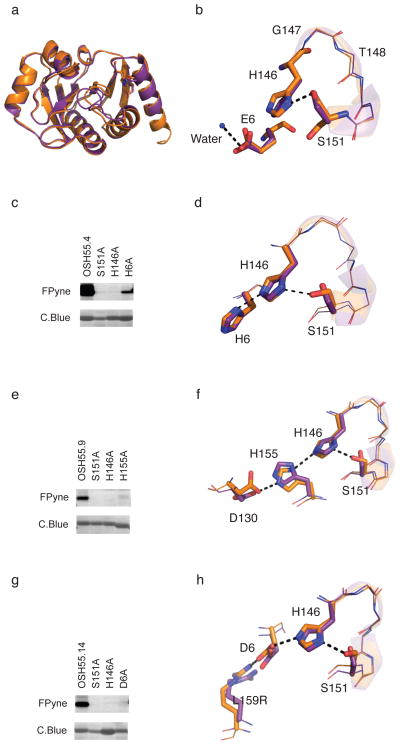
We solved the structure of OSH55 by crystallography at 2.3 Å resolution (PDB ID: 3V45). The design model and crystal structure match closely with an overall backbone RMSD of 0.6 Å (Fig. 2a). The crystal structure conformations of the active site Ser151 and His146 side-chains, and the backbone geometry of the oxyanion hole, composed of the backbone NH of residues Gly147 and Thr148, are nearly identical to those in the design model (Fig. 2b). Glu6, designed to stabilize His146, moves slightly away from the position in the design model and interacts with a nearby water molecule (Fig. 2b). Consistent with this observation, mutation of Glu6 did not affect fluorophosphonate probe labeling (Fig. 1e), suggesting that it does not contribute to activation of the active site serine.
Figure 2. Characterization of OSH55 derived designs.
(a) Structural superposition of design (brown) and crystal structure (purple) of OSH55 with overall backbone RMSD of 0.6 Å. The all-atom RMSD over the binding pocket residues is 0.34 Å. (b) A zoom-in view of the active site of OSH55 showing the orientation of Ser151 and His146 as designed and Glu6 coordinating a water molecule. (c,e,g) Selective fluorophosphonate probe labeling of OSH55.4 (c), OSH55.9 (e) and OSH55.14 (g) but not the active-site knockout mutants. Serine reactivity was assessed by gel-based ABPP (top, in-gel fluorescence; bottom, coomassie staining, see Supplementary Fig. 16 for full-size gel images. The experiments were performed in duplicates with consistent results). There is much less labeling in the catalytic triad knockouts but no decrease in the amount of protein. (d,f,h) Superimposition of the designed catalytic triad active sites (brown) overlaid on the experimentally determined crystal structures (purple) for OSH55.4 (d), OSH55.9 (f) and OSH55.14 (h); the designed hydrogen bond networks are recapitulated with very high accuracy.
Since the crystal structure and mutagenesis experiments indicated Glu6 was not interacting with His146 as in the model, we used computational design to identify alternative solutions with His, Asp, or Glu residues positioned to hydrogen bond to and stabilize the desired rotamer conformation of His146 (Supplementary Table 3). The alternative catalytic triad designs were expressed and tested for fluorophosphonate reactivity by ABPP (Supplementary Fig. 4). Three designs were found to have significantly increased reactivity towards the FPyne probe. In OSH55.4, the third catalytic triad residue is His6. Knockout of any of the three catalytic triad residues abolished FPyne labeling, suggesting that, in the Ser151-His146-His6 triad, the nucleophilicity of Ser151 is substantially activated by the supporting His-His dyad (Fig. 2c,d). LC-MS/MS confirmed that the FPyne is covalently bound to Ser151 in OSH55.4 (Supplementary Fig. 5). OSH55.9 also contains a Ser-His-His triad in the designed active site but the third histidine comes from a different position (A155H); mutagenesis of the serine again abolished FPyne labeling (Fig. 2e,f). Ser-His-His triads are less common than Ser-His-Asp triads, but are found in some native serine hydrolases such as herpes virus peptidases3,17. OSH55.14 has a more canonical catalytic triad, generated by replacing His6 with an Asp, and incorporates an additional arginine (L159R) to stabilize the Asp6 side-chain orientation in the catalytic triad. Mutagenesis of the catalytic triad histidine and aspartic acid in addition to the serine nucleophile abolished FPyne labeling (Fig. 2g,h). Thus, like in OSH55.4, both the histidine and the most distal residue in the triad (His in OSH55.4 and OSH55.9, Asp in OSH55.14) contribute to serine activation, suggesting the catalytic triad is functioning as designed.
We solved the structures of the OSH55.4 (PDB ID: 4ETJ), OSH55.9 (PDB ID: 4ETK), and OSH55.14 (PDB ID: 4ESS) to determine if the triads had the designed conformations. In all three cases, the agreement between the crystal structures and design models was quite striking (Fig. 2d,f and h). Most notably, in the crystal structure of OSH55.14 the designed hydrogen-bonding network Ser151-His146-Asp6-Arg159 is almost identical to the design model (Fig. 2h). This is perhaps the most extensive designed hydrogen bonding network that has been structurally confirmed to date, and certainly the most accurate network in a functional protein design.
Insights from structures of inactive designs
To gain insight into the factors that contribute to serine activation, we solved the structures of three designs that expressed at high levels but showed little probe labeling (Supplementary Fig. 6). In the crystal structure of OSH97, the conformations of the designed active site residues (Ser70, His37, Glu320) are very close to those in the design model. However, the oxyanion hole is disrupted by conversion of the loop conformation of Gly113 in the design model into a β-strand in the crystal structure. The lack of activity with a perfect catalytic triad but no oxyanion hole suggests that the oxyanion hole is important for transition state stabilization in the reaction with organophosphates. In the other two inactive designs (OSH26 and OSH49), there are significant backbone conformation changes in loop regions containing the catalytic triad residues, which disrupt the designed sites. Comparison with the active OSH55 series of designs suggests that regular secondary structure elements may be better starting points for building up a new functional site than loop regions, as the energy gap between the designed and undesired alternative conformations will generally be larger.
To further investigate the importance of active site pre-organization, we computed the average Boltzmann weight of the designed configurations of the catalytic triad residues in each of the 85 original designs and several native enzymes (Supplementary Fig. 7). In most of the designs, the predicted occupancies of the designed catalytic triad conformations were relatively low (<5%) (gray shaded). Strikingly, the computed occupancies were significantly higher in both the native enzymes (blue shaded) and the active designs (red shaded). Together with the structures of the inactive designs, this suggests that lack of active site pre-organization may be the primary flaw in many of the inactive designs and that future design efforts should focus on creating pre-organized active sites.
Evolutionary optimization of OSH55.4
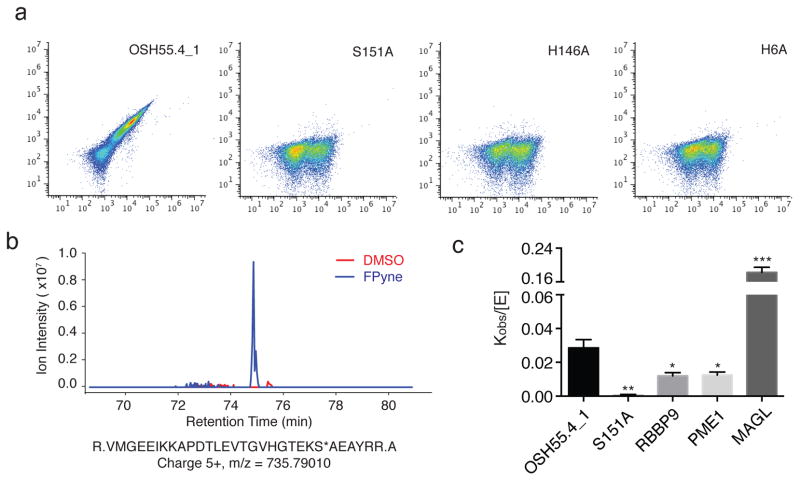
To determine whether the fluorophosphonate reactivity of designs with structurally intact catalytic triads could be further increased, we used yeast display selection18 with a biotinylated fluorophosphonate (FP-biotin) probe (Supplementary Fig. 2). We began with OSH55.4 since it showed the greatest fluorophosphonate reactivity (Supplementary Fig. 4). Libraries were created by randomizing six positions around the designed active site. Four rounds of Fluorescence Assisted Cell Sorting (FACS) enrichment for increased fluorophosphonate reactivity led to two populations of cells with distinct low and high signals (Supplementary Fig. 8). The sequences of 50 clones from the high-reactivity population revealed strong convergence to a single unique sequence -- OSH55.4_1, while sequences from the low-reactivity pool converged to three distinct sequences, OSH55.4_2, OSH55.4_4 and OSH55.4_6. Mutagenesis of each of the three catalytic triad residues of OSH55.4_1 eliminated fluorophosphonate reactivity on yeast, confirming that the designed active site remains functional during evolution (Fig. 3a). Of the six mutations introduced during evolution, four (A112S, G113W, L126H and A155Y) were found to contribute to increased fluorophosphonate reactivity (Supplementary Fig. 9). Unlike the parent OSH55.4 that preferentially reacts with the FPyne probe (likely due to clashes of the bulkier rhodamine and biotin probes with an active site-proximal loop), OSH55.4_1 exhibits strong reactivity with all of the tested fluorophosphonate probes (FPyne, FP-Rh and FP-biotin) (Supplementary Fig. 10). LC-MS/MS confirmed that the site of fluorophosphonate labeling is the designed catalytic residue Ser151 (Fig. 3b). Mutagenesis of the serine eliminated most, but not all, of the FP-Rh labeling (Supplementary Fig. 10). The residual labeling is mostly due to Ser112 (Supplementary Fig. 11). Mutation of the other two catalytic triad residues (H146A or H6A) completely eliminated probe labeling (Supplementary Fig. 11).
Figure 3. Organophosphate reactivity of OSH55.4_1 is comparable to native enzymes.
(a) Flow cytometry analysis of OSH55.4_1 and active site knockouts displayed on yeast following incubation with FP-biotin. Signal along x-axis indicates extent of yeast surface display; signal along y-axis; extent of probe binding. The active site residue knockouts abolish binding to FP-biotin. (b) MS1 chromatographic traces of the OSH55.4_1 active-site peptide adducted with FPyne probe. LC-MS/MS experiments (See Methods) identified a fully tryptic peptide (m/z = 735.79010 and z = 5+) with probe modification on the catalytic Ser151 (denoted with an asterisk). The extracted MS1 chromatographic traces (±15 p.p.m.) showed a distinct peak present only in the FPyne-treated (blue) but not in the DMSO-treated (red) sample. (c) Comparison of rates of FP-Rh labeling of the OSH55.4_1 design to those of a representative set of native serine hydrolases by fluorescence polarization. The kobs/[E] (mean ± s.e.m.) value for OSH55.4_1 is greater than those of RBBP9 and PME1. The S151A knockout mutation reduces the reaction rate to near background level (*p<0.05, **p<0.01, ***p<0.001, t-test, comparison with OSH55.4_1).
Relative FP reactivity of designs and natural hydrolases
We next compared the rate of fluorophosphonate labeling of optimized designs to those of a representative set of natural serine hydrolases using a fluorescence polarization (fluopol) assay19. We tested OSH55.4_1 along with three natural serine hydrolases – retinoblastoma-binding protein 9 (RBBP9, 21.0 kDa), monoacylglycerol lipase (MAGL, 33.2 kDa) and protein phosphatase methylesterase 1 (PME1, 42.3 kDa), all of which contains a canonical Ser-His-Asp triad active site but act on distinct classes of substrates. We monitored the changes in fluorescence polarization over time following the addition of the FP-Rh probe (1 μM) to protein samples ranging from 0.5 to 5 μM (Supplementary Fig. 12a–e). The ratios of the fitted rate constants to enzyme concentration, Kobs/[E] values, are useful measures of serine reactivity. MAGL had the fastest labeling kinetics, perhaps because the probe resembles its natural fatty acylated substrate and hence fits well into the substrate binding site20. The next most reactive protein was OSH55.4_1, which is much smaller (17 kDa) and has a more open active site. It exhibited a notably faster fluorophosphonate labeling rate than PME1 and RBBP9 (Fig. 3c). As expected, the S151A mutant of OSH55.4_1 showed negligible reactivity with FP-Rh in the fluopol assay (Supplementary Fig. 12b). These data suggest that, after evolutionary optimization by yeast display, the fluorophosphonate reactivity of OSH55.4_1 is comparable to those of natural serine hydrolases.
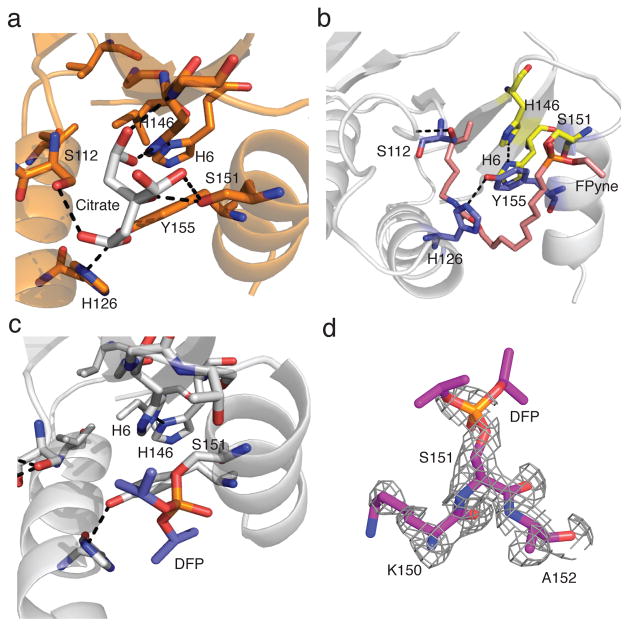
Structural characterization of OSH55.4_1
To investigate how the mutations introduced by yeast display improve fluorophosphonate reactivity, we determined the structure of OSH55.4_1 in both the apo (PDB ID: 4JCA) and FPyne-bound states (PDB ID: 4JLL). The apo structure, with a citrate molecule in the active site from the crystallization solution (Fig. 4a), overlays very well with the design model (data not shown) confirming that the mutations introduced during evolution did not affect the protein fold. Ser112 and Trp113 likely increase the rigidity of a loop near the active site that is flexible in the OSH55.4 structure; Ser112 forms an intra-loop backbone hydrogen bond and Trp113 packs in the core of the protein. His126 and Tyr155 form a hydrogen bond that keeps helices 1 and 2 apart and may provide better access for the ligand (Supplementary Fig. 13a). In the bound structure of OSH55.4_1 with FPyne at 1.6 Å resolution, the entire FPyne probe is visible in the structure and the probe is covalently bound to Ser151 as expected (Fig. 4b). However, because of a slight rotation (~20° around the serine Oγ-phosphorus bond) of the organophosphate relative to the design model, water molecules interact with the organophosphate oxygen (O2 in Fig. 1a) rather than the designed oxyanion hole; this may be a shortcoming of the design or due to a reorientation that occurs following completion of the reaction (the structure is of the end product of the reaction rather than the transition state complex). Water molecules have been suggested to contribute to oxyanion holes in ketosteroid isomerases21. The structure also shows that the alkyl linker of the FPyne probe packs tightly into a hydrophobic groove in the protein (Supplementary Fig. 13b) and this suggests that improved probe binding likely contributes to the enhanced fluorophosphonate reactivity of OSH55.4_1. In order to rule out the possibility that the loss of fluorophosphonate labeling of the H146A mutant was due to disrupted probe binding in the hydrophobic groove, we tested two additional mutants, H146I and H146V that better preserved the hydrophobic properties of the groove. Both of the mutants significantly decreased fluorophosphonate reactivity, consistent with a direct role for His146 in Ser151 activation (Supplementary Fig. 14).
Figure 4. Crystal structures of OSH55.4_1 bound to Diispropyl fluoroposphate (DFP).
(a) Crystal Structure of the apo form of OSH55.4_1 with a citrate molecule (white) from the crystallization buffer cocktail making extensive interactions with active site residues. (b) Crystal structure of OSH55.4_1 covalently modified at Ser151 with FPyne (pink). (c) Crystal structure of OSH55.4_1 covalently modified at Ser151 with DFP (red and blue). (d) Simulated annealing omit map around Ser151 (fo-fc). Density is clearly evident on the serine but the isopropyl groups of DFP are not distinguishable probably due to free rotation about the P-C bond.
A potential application of organophosphate-reactive proteins is the bioremediation and scavenging of toxic organophosphate-based pesticides and nerve agents22–24. To test whether our designs are able to react with organophosphates other than the activity-based fluorophosphonate probes, we solved the structure of OSH55.4_1 in complex with diisopropyl fluorophosphate (DFP), a potent neurotoxin and serine protease inhibitor that mimics some of the structural features of more toxic chemical warfare agents25,26. Even though OSH55.4_1 was neither designed nor evolved to specifically bind DFP, we found that DFP indeed forms a covalent adduct with the active-site Ser151 (Fig. 4c, d) that is very similar to the complex formed by the fluorophosphonate moiety of the FPyne probe (PDB ID: 4JVV).
Discussion
The accuracy and the activity of the de novo designed catalytic triad active sites reported in this paper are significant steps forward for computational enzyme design. The structures of the catalytic triad and quartet designs match the design models much more faithfully than the simpler hydrogen bond networks in previous computational designs12,27. The activation of serine, which has a pKa of ~13, is significantly harder to achieve than the activation of cysteine (pKa ~ 8) and histidine (pKa ~ 7) in previous successful designs. Success in activating the serine nucleophile likely reflects the constraining of the catalytic triad through extensive packing interactions with the rest of the protein; the active site networks in these designs are significantly more buried than in most previous computationally designed enzymes. The control of histidine side chain conformation in the active designs described here contrasts with the lack of control found in the design of His-Cys based esterases10. In addition to burial of the His side chain, correct positioning is likely facilitated by the greater strength of hydrogen bonding between His-Ser compared to His-Cys.
The disrupted catalytic sites in two structures of inactive designs and the success with the catalytic sidechain Boltzmann weight in distinguishing active from inactive designs (Supplementary Fig. 7) together indicate the importance of catalytic triad preorganization for serine activation. The success with the OSH55 series of designs suggests that focusing design efforts on relatively buried sites built primarily on regular secondary structure elements is a good strategy for precisely controlling catalytic sidechain conformation. Surface exposed sites mounted on loop regions are likely to have many alternative conformations with roughly equal energies making precise control over sidechain conformation much harder to achieve.
The most active design in this study, OSH55.4_1, utilizes a Ser-His-His triad which differs from the Ser-His-Asp/Glu triads observed in most natural serine hydrolases. Parallels in nature include herpes virus peptidases with a Ser-His-His triad17, but the contribution of the secondary histidine to catalysis appears to be smaller in these enzymes compared to OSH55.4_1, where mutation of the secondary histidine reduces fluorophosphonate reactivity substantially. Several other variations of the catalytic triad or dyad such as Ser-Glu-Asp, Ser-Ser-Lys, Ser-Lys, and Ser-His are observed in nature, albeit less commonly3. Also unlike most serine esterases, the histidine in the OSH55 series of designs hydrogen bonds to the catalytic serine through the Nδ rather than the Nε. In contrast, the histidine in the structure of an inactive design interacts with the serine via the Nε; evidently the hydrogen bonding orientation of the histidine in the triad is not a major determinant for activation of the serine nucleophile.
Stoichiometric scavenging of organophosphates by injecting human butyrylcholinesterase (BCHE) into the bloodstream of poisoned individual has shown promise in reducing toxicity from nerve agent exposure28. However, due to the high molecular weight of BCHE, large amounts are required for stoichiometric inhibition. For example, 350 mg of human BCHE is required for every 1 mg of cyclosarin29. Another potential disadvantage of BCHE is unwanted hydrolytic cleavage of endogenous esters such as acetylcholine, leading to imbalance of these metabolites in blood. The designed proteins described in this paper have potential advantages over BCHE for organophosphate scavenging in that they are much smaller proteins (17 kDa vs 65 kDa) and are likely to have limited hydrolytic activity against endogenous ester metabolites.
We have shown that computational methods can produce authentic catalytic triads that, with directed evolution, can achieve organophosphate reactivity in levels comparable to those of natural enzymes; starting with properly arrayed active site hydrogen bond networks, improvements in activities could be accomplished relatively quickly. A next challenge is to endow these and similarly designed catalytic triads with substrate recognition and other features required for proficient hydrolytic activity, in particular, the capability to undergo repeated cycles of substrate acylation and deacylation. A separate and more practical challenge is to optimize the current designs to bind specific nerve agents such as Sarin, Tabun or VX for organophosphate scavenging and detection applications. More generally, the current work demonstrates the utility of breaking down the complexity of native enzyme catalysis into simpler subproblems that can be solved independently.
Online Methods
Computational design
Transition state models for syn- and anti- attack of nucleophilic serine to the FP ligand were constructed using data from the previously published results14, and an ensemble of ligand conformers was then generated by Open Eye’s Omega software31. The bond length, angle and dihedrals describing the theozyme geometry were adapted from previous findings30. The geometric parameters used to describe the theozyme are presented in Supplementary Table 1. Some of the constraints were loosened so that the number of initial matches (matched output) could be maximized. Geometric constraints for oxyanion hole were taken from native serine hydrolases as described in Supplementary Table 1. The final theozyme comprised of a conformer library of the substrate in the transition state, the Ser-His-Asp/Glu triad, and one oxyanion-hole contribution from backbone amide protons. The theozyme shown in Fig. 1b is more commonly observed in native proteases but we did not restrict only to this theozyme arrangement during the matching process by RosettaMatch. In the geometric constraints used for Rosetta matching, either Nε or Nδ of histidine is allowed to form an H-bond with Serine-Oγ, and the other unsatisfied N-atom is then allowed to coordinate with either Oε1 or Oε2 of an Asp/Glu. This allows the alternate configuration of the theozyme present in the design to be realized as seen in OSH55. In the structures of an inactive design, OSH97, both the design and crystal structures have the theozyme geometry as shown in Fig. 1b (Supplementary Fig. 6).
Positions in protein scaffolds where the theozymes could be realized were identified using the RosettaMatch algorithm15. Catalytic interactions (imposed by theozyme geometry) were optimized by three rounds of sequence design and gradient-based energy minimization of the matches. During this minimization, restraints were added to the energy function to favor the desired theozyme geometry. Finally, the designed structures were repacked without the catalytic restraints. The designs to be tested experimentally were obtained by filtering for the following criteria: (1) pre-organization of active site – RMSD between idealized theozyme geometry and active site in the final structures is less than 3Å across all the residues (2) a ligand binding energy is less than −4.0 Rosetta energy units (REU) and (3) there are no more than two unsatisfied, buried polar ligand atoms. Out of ~380 designs that passed these filters, 100 top scoring designs were selected of which 85 designs with unique scaffold (Supplementary Table 2) were chosen for experimental characterization. For the second-round of optimization, crystal structure of OSH55 (PDB ID: 3V45) was used as a starting model. Scaffold residues that are within 15 Å of the active site residues were considered as design shell residues. Ser151 and His146 were not allowed to change during this round of design. Designs were filtered for additional interactions with His146, thereby stabilizing its designed rotamer. Boltzmann rotamer probability distribution was calculated as previously described32. It must be noted that in our original design calculation, we did require that the desired active-site conformation be the lowest energy state, but did not impose a large energy gap between this (catalytically-competent) and other conformations to bias it to be highly populated.
Screening designs by gel-based ABPP
FP-Rh, FP-biotin, FPyne probes and alkyne-azide were previously synthesized in house with >99% purify and aliquots from the more concentrated lab stocks were diluted to be used in the current study. Genes corresponding to the 85 designs were ordered from Genescript, USA. Genes in pet29b vector were then expressed in E.coli BL21 cells in a 2 mL culture. One of the main advantages of ABPP is that the reactivity of each design could be assessed directly from cell lysate in parallel without laborious task of purification of a large number of proteins9. In a typical batch of screening, E.coli cells expressing the designed proteins were pelleted down and lysed in 0.5 mL of ice-cold PBS buffer by sonication. Soluble lysates were obtained by spinning at 14,000 rpm in a desktop centrifuge for 30 min at 4°C and protein concentrations were adjusted to 0.5 mg/mL. 50 μL of each lysate was labeled with 1 μM of FP-Rh, FP-biotin or FPyne probes for 1 h at room temperature (1 μL of 50 μM stock in DMSO). When the FPyne probe was used, copper-catalyzed alkyne-azide cycloaddition (“click chemistry”)33 was performed to conjugate a fluorescent reporter tag to the probe-labelled proteins by the addition of 50 μM of azide-rhodamine (1 μL of a 2.5 mM stock in DMSO), 1 mM TCEP (1 μL of a fresh 50 mM stock in water), 100 μM ligand TBTA (3 μL of a 1.7mM stock in DMSO:t-Butanol 1:4) and 1 mM CuSO4 (1 μL of a 50mM stock in water). Samples were allowed to react at room temperature for 1 h before 2x gel loading buffer was added to quench the probe labelling/click-chemistry reaction. 12.5 μg of each lysate was separated by an in-house 10% SDS-PAGE long gel and in-gel fluorescence was visualized using a Hitachi FMBio II flatbed laser-induced fluorescence scanner (MiraiBio, Alameda, CA). For the FP-biotin labelled samples, immunoblotting was performed with IRDye Streptavidin (1:10,000) and scanned by an Odyssey imaging system (LI-COR). The gels were stained afterwards with coomassie blue to assess the expression and relative abundance of each design. About 50% of the designs did not overexpress in soluble fraction based on visual examination of the coomassie stained gel. For those designs that did show overexpression, their FP reactivity were ranked based on the fluorescence intensity normalized by the intensity from coomassie staining and designs exhibiting significant probe labeling (“hits”) were chosen for further characterization. An example of fluorescence and coomassie blue stained gel images are shown in Supplementary Fig. 1b and FP probe structures are shown in Supplementary Fig. 2.
For the identified hits, knock-out mutants of the active site residues were made using the Multi-site Lightning kit (Startgene). After their sequences were confirmed, the mutants were subjected to the ABPP screening along with their wild-type design to confirm the specific probe labeling on the designed active-site serine.
Protein expression and purification
Positive designs identified by ABPP were grown in 1 L of LB media at 37° C until induction (O.D~0.6) and the cells were switched to 22 °C to continue growing overnight. The harvested cell pellets were re-suspended in the lysis buffer, which contains 1X PBS buffer, 300 mM NaCl, and 1 mM TCEP. No protease inhibitors were added in order to keep the designed active sites free from pre-inhibition. 1 mg/mL lysozyme was added right prior to the sonication step. After the lysate was centrifuged at 4000 g for 20 min, the soluble fraction was applied to 5 mL of TALON resin (Clontech) and then washed with 20mM imidazole three times. The protein bound to the resin was eluted with 20 ml of the lysis buffer supplemented with 250 mM imidazole and the eluted fraction was then further purified by gel–filtration (HiLoad 26/60 Superdex 75) chromatography. The purified proteins were dialyzed overnight against the dialysis buffer containing 1X PBS, 100 mM NaCl and 1mM TCEP overnight and its purity was confirmed by SDS-PAGE. Aliquots of the purified protein were flash frozen in liquid nitrogen and stored in −80 °C for further use.
LC-MS/MS assay to identify serine-FP adducts
Each of the purified protein designs (20 μM, 50 μL total volume) was incubated with DMSO or FPyne (50 μM) for 60 min at room temperature. The labeled protein samples were then prepared according to the in-solution trypsin digestion protocol published previously34. Briefly, the proteins were denatured in 6 M Urea (add 150 μL of 8 M Urea in PBS), reduced by 10 mM of Dithiothreitol for 30 min at room temperature (add 10 μL of 200 mM stock in water), and alkylated by 20 mM Iodoacetamide for 30 min at room temperature in dark (add 10 μL of 400 mM stock in water). The samples were diluted with ammonium bicarbonate (25 mM, 400 μL) to 2 M Urea and subjected to trypsin digestion (Promega; 4 μL of 0.5 μg/μL) overnight at 37 °C in the presence of 2 mM CaCl2. Digested peptide samples were desalted using the MacroSpin columns (The Nest Group, Inc.), concentrated and re-suspended in 20 μL of Buffer A (95% water, 5% acetonitrile, 0.1% formic acid). A 10μL aliquot was pressure-loaded onto a 100 μm (inner diameter) fused silica capillary column (Agilent) with a 5 μm tip that contained 10 cm C18 resin (Aqua 5 μm, Phenomenex). LC-MS/MS analysis was performed on an LTQ-Orbitrap mass spectrometer (Thermo Scientific) coupled to an Agilent 1100 series HPLC. Peptides were eluted from the column using a 125 min gradient of 5–100% Buffer B (20% water, 80% acetonitrile, 0.1% formic acid). The flow rate through the column was 0.25 μL/min and the spray voltage was 2.5 kV. The LTQ was operated in data-dependent scanning mode, with one full MS scan (400–1600 m/z) followed by MS/MS scans of the seven most abundant ions with dynamic exclusion enabled. The MS data was analyzed by SEQUEST35 using a Uniprot E.coli sequence database (as of 07/20/2010) supplemented with the FASTA sequences of the designed proteins. A differential modification on serine of 371.22255 Da was defined in the search and after the adducted peptides were identified, the corresponding MS1 chromatographic traces were extracted with ± 15 ppm accuracy from both the FPyne- and DMSO-treated samples and shown in Fig. 3b and Supplementary Fig. 5.
Yeast display selection
Libraries of OSH55.4 were constructed by randomizing 6 positions around the active site (112,113,126,130,155,159) using NNK oligos and then assembled to make the full-length sequence. DNA libraries and linearized PETCON vectors were then transformed into EBY100 cells using the standard yeast display protocol18. The complexity of library was determined to be ~ 106. Cells were re-suspended in SDCAA media (107 cells/mL) and grown at 30 °C overnight. Cells were then centrifuged and induced at 22 °C in SGCAA media for 24–48 h. Cells were labeled with 2 μM of FP-biotin in PBSF buffer, washed with PBSF, secondary labeled with SAPE (Invitrogen) and anti-cmyc FITC (Miltenyi Biotech), and sorted by fluorescent gates (BD Influx sorter). Four rounds of sorting were carried out after which the sequences converged to unique sequence (Supplementary Fig. 8).
Fluorescence polarization (Fluopol) assays
The fluopol assay was performed in a 384-well format based on a modified method published previously19. Briefly, purified OSH55.4_1 (wild-type and the active-site serine knockout mutant) as well as native serine hydrolases RBBP9, PME1 and MAGL were diluted in assay buffer containing 50 mM Hepes, pH 7.5, 150 mM NaCl and 0.01% Pluronic F-127 (Invitrogen) to a series of concentrations ranging from 0.1 to 5 μM. 10 μL of each enzyme or blank assay buffer was added to each well and 1.1 μL of FP-Rh probe (10 μM stock in assay buffer and 1.0 μM final concentration) was added to all wells by an automatic sample dispenser. The plates were read on an Envision plate reader (Perkin Elmer) to measure fluorescence polarization signals at 3 min interval for 45 min or longer. Each condition was represented by 8 replicate wells of which the averaged fluopol signal (± s.d.) was plotted in Supplementary Fig. 12a–e. The time-dependent fluopol signals for each enzyme at each concentration were fitted into a one-phase association model Y=Y0 + (Plateau−Y0)*(1−exp(−kobs*X)) in Prism 6 (GraphPad Software) to obtain Y0, Plateau and the observed rate constant kobs. The fitted kobs value was normalized by the enzyme concentration [E] and the mean ± s.e.m. of kobs/[E] for each enzyme was reported in Fig. 3c.
Structure Determination
Protein crystallization and data collection were carried out in Northeast Structural Genomics Consortium (NESG). Proteins were shipped to NESG where they were assigned with NESG target identifiers (Supplementary Table 4). The proteins were subjected to another round of quality control that includes light scattering experiments to confirm the mono-dispersity of samples and MALDI-TOF analysis to confirm the molecular mass. pET expression vectors for these proteins have been deposited in the PSI Materials Repository (http://psimr.asu.edu/). FP-alkyne complex of OSH55.4_1 was made by incubating 2-fold molar excess of FP-alkyne with OSH55.4_1 for 1 h and the unreacted FPyne probe was removed by gel-filtration. In the case of the DFP complex, protocol will be made available upon request per biosafety requirement.
Crystallization and optimization of crystals
Initial crystallization conditions for the proteins were found by high throughput robotic screening of 1536 different conditions at the Hauptmann Woodward Institute, Buffalo, NY36. The hits were then used for laboratory optimization to grow crystals suitable for X-ray analysis. Crystals were obtained by the method of either microbatch under oil or vapor diffusion sitting drop. A 1–3 μL protein solution drop (8–10 mg/mL protein in 10 mM Tris, pH 7.4, 100 mM sodium chloride, 0.02% sodium azide and 5 mM dithiothreitol) was mixed with 1 μL of the crystallization cocktail solution and the mixture was incubated at 4 °C for several days to allow crystal formation. The crystals were cryo-protected using 15%–20% glycerol, ethylene glycol or N-paratone. Detailed crystallization conditions for each structure are available in Supplementary Table 4.
X-ray data collection, processing, and structure determination
Single anomalous dispersion (SAD) x-ray data were collected at the wavelength 0.979Å from single crystals under temperature 100K on beamline X4A or X4C at the National Synchrotron Light Source at Brookhaven National Laboratory. The diffraction images were processed with HKL2000 and scaled with SCALEPACK37. Structures were solved by molecular replacement (MR) using BALBES38 with the structure of a conserved hypothetical protein, TTHA0132 from Thermus thermophilus HB8 (PDB id: 2DX6) as the search model. The MR models were refined with PHENIX39 and then manually corrected with COOT40 by analyzing 2Fo-Fc and Fo-Fc electron density maps. Each of the current and final models was refined with translation, libration, and screw-rotation (TLS) displacement of a pseudo-rigid body41. For the complex structures of OSH55.4_1 with FP-alkyne and DFP, the program eLBOW (http://www.phenix-online.org/documentation/elbow.htm) was used to generate crystallographic input files for model refinement and correction by COOT. Data collection and refinement statistics are reported in Supplementary Table 5.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4JVV, 4JLL, 4JCA, 4ESS, 4ETJ, 4ETK, 3V45, 3TP4, 4F2V and 4DRT.
Supplementary Material
Acknowledgments
We thank K. Masuda and D. Milliken for helping with ABPP screening, R. Xiao and G. Kornhaber for experimental support with sample preparation and structure determination, and the Rosen Lab for sharing instrumentation for performing fluopol experiments. This work was supported in part by the Defense Threat Reduction Agency (DTRA) (D.B.), the National Institute on Drug Abuse grant DA033670 (B.F.C.) and by a grant from the National Institute of General Medical Sciences Protein Structure Initiative (PSI), U54-GM094597 (J.H.). Fellowship support from Sir Henry Wellcome Postdoctoral Fellowship (S.R), NIH/NIEHS K99/R00 Pathways to Independence Postdoctoral Award 1K99ES020851-01 (C.W.) and Helen Hay Whitney Fellowship (M.L.M) are gratefully acknowledged.
Footnotes
Author Contributions S.R., C.W., B.F.C. and D.B. conceived the project. S.R and F.R. performed the computational design, S.R expressed and purified the designed proteins. S.R. and K.Y. performed the yeast display experiments. C.W. performed the ABPP screening and mass spec experiments. C.W. and M.L.M. performed the fluopol experiments. A.P.K., A.E.M. S.L. J.S., M.S. and J.F.H. performed the crystallography experiments. S.R. C.W., B.F.C. and D.B. analyzed data and wrote the manuscript.
Competing financial interests: The authors declare no financial competing interests.
References
- 1.Botos I, Wlodawer A. The expanding diversity of serine hydrolases. Curr Opin Struct Biol. 2007;17:683–690. doi: 10.1016/j.sbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 3.Ekici OD, Paetzel M, Dalbey RE. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 5.Corey DR, Craik CS. An Investigation into the Minimum Requirements for Peptide Hydrolysis by Mutation of the Catalytic Traid of Trypsin. J Am Chem Soc. 1992;114:1784–1790. [Google Scholar]
- 6.Corey DR, McGrath EM, Vasquez JR, Fletterick RJ, Craik CS. An Alternate Geometry for the Catalytic Triad of Serine Proteases. J Am Chem Soc. 1992;114:4905–4907. [Google Scholar]
- 7.Carter P, Wells JA. Engineering enzyme specificity by “substrate-assisted catalysis”. Science. 1987;237:394–399. doi: 10.1126/science.3299704. [DOI] [PubMed] [Google Scholar]
- 8.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 10.Richter F, et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J Am Chem Soc. 2012;134:16197–16206. doi: 10.1021/ja3037367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwasnieski O, Verdier L, Malacria M, Derat E. Fixation of the two Tabun isomers in acetylcholinesterase: a QM/MM study. J Phys Chem B. 2009;113:10001–10007. doi: 10.1021/jp903843s. [DOI] [PubMed] [Google Scholar]
- 15.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De novo enzyme design using Rosetta3. PLoS One. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh HS, et al. Three-dimensional structure of human cytomegalovirus protease. Nature. 1996;383:279–282. doi: 10.1038/383279a0. [DOI] [PubMed] [Google Scholar]
- 18.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 19.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labar G, et al. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 21.Schwans JP, Sunden F, Gonzalez A, Tsai Y, Herschlag D. Evaluating the catalytic contribution from the oxyanion hole in ketosteroid isomerase. J Am Chem Soc. 2011;133:20052–20055. doi: 10.1021/ja208050t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai PC, et al. Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents. Biochemistry. 2012;51:6463–6475. doi: 10.1021/bi300811t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer S, Arad A, Margalit R. Lipsome-formulated enzymes for organophosphate scavenging: butyrylcholinesterase and Demeton-S. Arch Biochem Biophys. 2005;434:108–115. doi: 10.1016/j.abb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 24.diTargiani RC, Chandrasekaran L, Belinskaya T, Saxena A. In search of a catalytic bioscavenger for the prophylaxis of nerve agent toxicity. Chem Biol Interact. 2010;187:349–354. doi: 10.1016/j.cbi.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Tsay OG, Atwood DA, Churchill DG. Destruction and detection of chemical warfare agents. Chemical reviews. 2011;111:5345–5403. doi: 10.1021/cr100193y. [DOI] [PubMed] [Google Scholar]
- 26.Pacsial-Ong EJ, Aguilar ZP. Chemical warfare agent detection: a review of current trends and future perspective. Front Biosci (Schol Ed) 2013;5:516–543. doi: 10.2741/s387. [DOI] [PubMed] [Google Scholar]
- 27.Rothlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 28.Lenz DE, et al. Stoichiometric and catalytic scavengers as protection against nerve agent toxicity: a mini review. Toxicology. 2007;233:31–39. doi: 10.1016/j.tox.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 29.Raushel FM. Chemical biology: Catalytic detoxification. Nature. 2011;469:310–311. doi: 10.1038/469310a. [DOI] [PubMed] [Google Scholar]
- 30.Smith AJ, et al. Structural reorganization and preorganization in enzyme active sites: comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J Am Chem Soc. 2008;130:15361–15373. doi: 10.1021/ja803213p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bostrom J, Greenwood JR, Gottfries J. Assessing the performance of OMEGA with respect to retrieving bioactive conformations. J Mol Graph Model. 2003;21:449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 32.Fleishman SJ, Khare SD, Koga N, Baker D. Restricted sidechain plasticity in the structures of native proteins and complexes. Protein Sci. 2011;20:753–757. doi: 10.1002/pro.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angewandte Chemie. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Bachovchin DA, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc Natl Acad Sci U S A. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 36.Luft JR, et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 37.Zbyszek Otwinowski WM. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.