Abstract
This report describes the outcomes of the Data Management Challenges in 3D Electron Microscopy workshop. Key topics discussed include data models, validation and raw-data archiving. The meeting participants agreed that the EMDataBank should take the lead in addressing these issues, and concrete action points were agreed upon that will have a substantial impact on the accessibility of three-dimensional EM data in biology and medicine.
The EMDataBank (http://www.emdatabank.org/)1 is an organization that runs a global deposition and retrieval network for three-dimensional EM (3DEM) maps, molecular models and associated metadata. It consists of three partners, the Protein Data Bank in Europe (PDBe), the Research Collaboratory for Structural Bioinformatics (RCSB PDB) and the National Center for Macromolecular Imaging (NCMI). The EMDataBank manages the Electron Microscopy Data Bank (EMDB)2, the global archive of 3DEM data that now holds over 1,500 maps and thus offers a unique perspective on the state and development of the 3DEM field. The field has experienced rapid growth in recent years, as witnessed by rapidly increasing numbers of publications and 3DEM-derived structures archived in the EMDB. In addition, the number and size of images used to derive maps has been increasing steadily, driven by the quest for higher resolution, and these trends are likely to continue (Fig. 1). A bird’s-eye view on the trends and practices in the 3DEM field can be obtained through the EMstats service3 (http://pdbe.org/emstats/).
Figure 1.
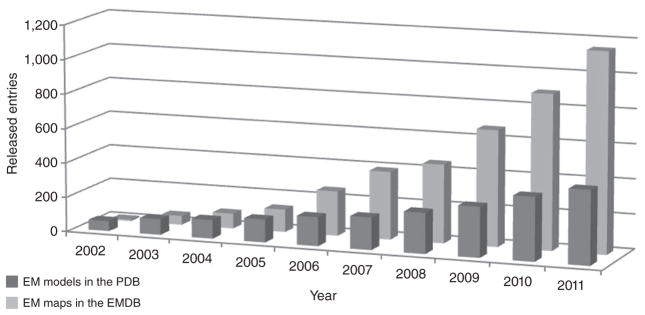
Trends in 3DEM. The cumulative number of released maps in the EMDB and 3DEM-derived models in the PDB are shown as a function of time.
To discuss the growing challenges of storing, sharing, transferring, analyzing, viewing, validating and annotating 3DEM data, PDBe and the Open Microscopy Environment (OME) organized a workshop, Data Management Challenges in 3D Electron Microscopy (DMCEM) at Hinxton Hall, Wellcome Trust Genome Campus, Cambridge, UK on 5 and 6 December 2011.
Participants included experts with established pipelines for 3DEM data collection and processing and thus brought valuable expertise on the data-management challenges facing the field. Several participants have been involved in community-wide initiatives to define standards, conventions and reporting standards for validation in 3DEM, as part of the Electron Microscopy Validation Task Force (EM-VTF)4. Developers of several major EM software packages, including Appion5, Bsoft6, EMAN2 (ref. 7), EMEN2 (http://blake.bcm.edu/emanwiki/EMEN2/), IMOD8, MRC9 and Xmipp10, also participated. M.W. represented two UK-based Collaborative Computational Projects (http://www.ccp.ac.uk/), namely the well-established Collaborative Computational Project Number 4 (CCP4)11 for macromolecular crystallography and the newly formed Collaborative Computational Project for Electron Cryo-microscopy (CCP-EM), and outlined experiences gained with CCP4 and possible synergies with CCP-EM. Three participants from the University of Dundee represented the OME team, which develops scientific image-file-translation libraries and data-management applications12,13. PDBe and OME work together to apply OME resources to 3DEM data in the EMDB, and the workshop was sponsored by a joint Biotechnology and Biological Sciences Research Council (BBSRC) grant that stimulates collaboration between groups with expertise in handling 3DEM and light-microscopy data. Finally, six participants represented the EMDataBank.
The first day of the workshop consisted of presentations of existing data-management solutions and initiatives and talks on 3DEM validation. The second day was devoted to thematic discussions regarding validation, segmentation, standards and formats, and tomography. The goals of the meeting were (i) to obtain feedback on the 3DEM data model currently being developed for EMDB; (ii) to propose concrete measures that the EMDataBank and the Worldwide Protein Data Bank (wwPDB14; http://wwpdb.org) could take to aid implementation of the EM-VTF recommendations4 on how to improve the quality criteria employed in the 3DEM field and to ensure their widespread adoption; and (iii) to discuss possible models for the public archiving of data leading up to the final 3D map.
Outcomes
New EM data model
Since the inception of the EMDB archive at the European Bioinformatics Institute (EBI) in 2002 (ref. 2), the 3DEM field has made considerable progress on a number of fronts, including electron tomography, automation and direct electron detectors (Fig. 2). At the same time, the prominence of the archive has grown and so has the need to link it to other relevant bioinformatics resources. Although the EMDB data model2,15 has been updated incrementally to accommodate these changes and address new requirements, it is clear that fundamental changes are required to the data model to keep up with current developments and to allow for future extensions and additions.
Figure 2.
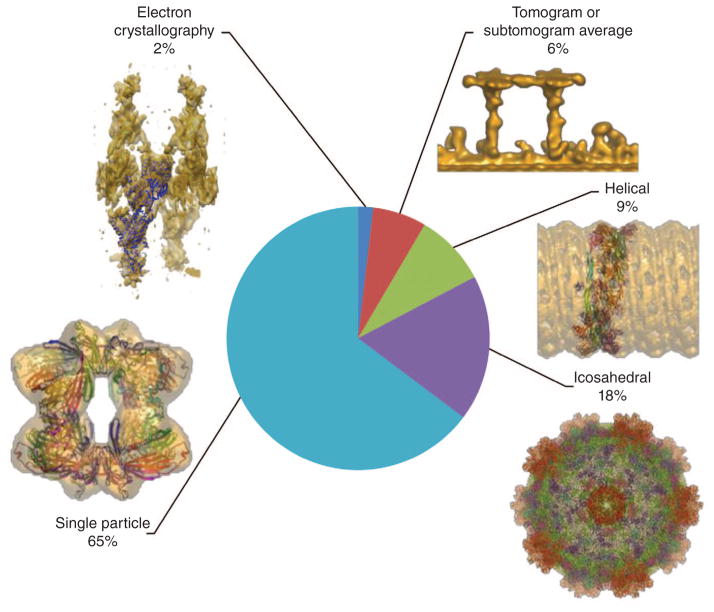
Holdings of the EMDB by 3DEM technique. A representative example of each category is shown: single particle, human αβ-crystallin 24-mer (EMD-1894, PDB 2YGD); icosahedral, cytoplasmic polyhedrosis virus (EMD-5256, PDB 3IZX); helical, GMPPCP-stabilized human dynamin 1 ΔPRD polymer (EMD-1949, PDB 3ZYS); tomogram or subtomogram average, radial spokes from Chlamydomonas reinhardtii flagella (EMD-1941); 2D crystal, pig gastric H+,K+-ATPase with bound BeF and SCH28080 (EMD-1831, PDB 2XZB)24.
The wwPDB and the EMDataBank are jointly developing a new deposition and annotation (D&A) system, which will facilitate the deposition and validation of biomacromolecular structure data (including X-ray crystallography, NMR spectroscopy, 3DEM and any combination of these techniques). The new system will reduce the need for manual annotation and improve the quality, consistency and integrity of the data that are entered into the archive. With an expected life span of at least ten years, the D&A system has prompted the design of a new EMDB data model that will capture the important aspects of the various 3DEM methodologies (such as single-particle analysis, tomography, two-dimensional (2D) crystallography, etc.) and be sufficiently flexible to adapt to changes and as-yet-unforeseen future developments.
The data model is implemented and maintained in an Extensible Markup Language (XML) schema for which there exists a variety of powerful, industry-standard modeling tools. The high level of visual abstraction afforded by these tools allows 3DEM practitioners to modify the schema and to ensure that the model makes scientific sense. For the integration of the model into the D&A system, the model will be translated into the format used internally by the system, the Macromolecular Crystallographic Information File (mmCIF) format extended for the D&A system (PDBx)16. During the meeting, many participants provided feedback on the draft data model. Information about the current version of the data model can be found at http://pdbe.org/emschema/.
Validation
Validation entails the assessment of errors and uncertainties in the results of a 3DEM experiment or their interpretation in terms of a volumetric or atomic model. Validation is essential to provide confidence in the interpretation of the data in a molecular or cellular biological context. There are four essential aspects to single-particle 3DEM validation, namely ensuring the quality of the final map, verifying the claimed resolution, assessing the fit of any models to the map and assessing the quality of the models themselves.
Tilt-pair analysis17,18 has proven to be a valuable tool for establishing the overall quality of a map, and the tilt-pair validation server developed by the Rosenthal group (https://cryoem.nimr.mrc.ac.uk/software/) has made the method generally accessible. To encourage use of this technique, the EMDataBank will collaborate with the Rosenthal group to incorporate support for tilt-pair validation into the new EMDB data model and to migrate the server to the PDBe website in preparation for its eventual integration as a validation tool in the D&A pipeline.
A.R. suggested that comparing small-angle X-ray scattering (SAXS)19 profiles with simulated SAXS profiles generated from EM maps would be another means of establishing the overall correctness of the map20. It was agreed that the PDBe would investigate the feasibility of adding support for SAXS profiles in the EM data model and of setting up a web service to generate simulated SAXS profiles from EM maps.
Fourier-Shell correlation (FSC)21 is the most commonly used method to estimate the resolution of single-particle maps in EMDB. The shape of the FSC curve depends on the imposed symmetry and mask and whether or not the two 3D reconstructions used were processed from a common reference. The resolution estimated from the curve depends critically on the threshold criterion used. Ideally, an FSC curve is based on two completely independent reconstructions with all relevant variables, including the protocol, symmetry, mask and cutoff, clearly specified. In practice, this means calculating two independent reconstructions, each from separate halves of the recorded data. However, using only half the particles may substantially compromise the achievable resolution. An alternative is to low-pass filter the data to a resolution threshold, say 15 Å, for image processing and to re-introduce the filtered-out information for FSC calculation. Information at spatial frequencies above the resolution threshold will not be affected by reference bias, and the loss of information owing to low-pass filtering would not be expected to have much impact on the accuracy of the alignment and Euler-angle assignment. Another solution, suggested by R.H., is to use any procedure to obtain two reconstructions and calculate the FSC curve (FSCnorm) from them, then randomly scramble the phase information of every particle above a given spatial frequency, rerun the procedure and obtain a new FSC curve (FSCrand). If FSCrand is relatively small compared with FSCnorm (say <25%) in the scrambled spatial-frequency region then one may safely assume that over-fitting is not an issue. However, if it is more significant, for example 50% or higher, then those who have done the test may wish to remove the over-fitting, for example by avoiding the use of high-resolution data in the refinement of particle orientation and position. The success of this more conservative approach should be revealed by the absence of over-fitted noise at high resolution when the test with randomized high-resolution phases is repeated. There are thus a number of protocols that may be followed in calculating the FSC curve that in principle are not tied to specific software packages. The EMDataBank will lead the effort to improve reporting standards for 3DEM validation; as a first step, the new EMDB data model will capture the entire FSC curve and all relevant metadata, including the protocol, symmetry, mask and cutoff.
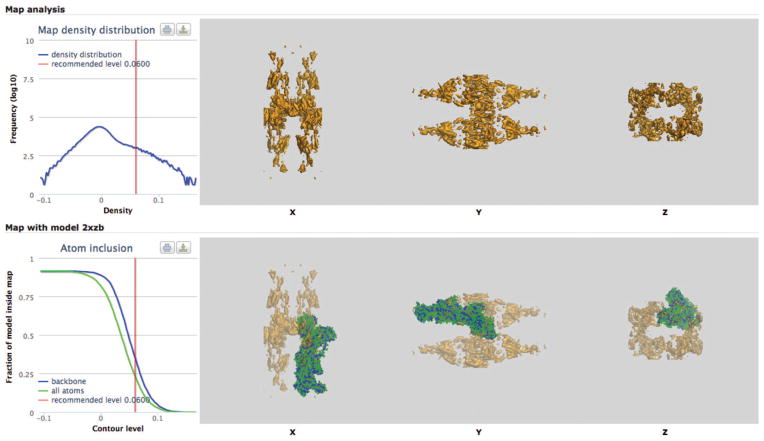
Although the issue of validating the fit of models to maps was not discussed in any detail at this meeting, it was clearly recognized as an area requiring more methods development. Currently, a basic sanity check of the fit of a model to a map is provided by the PDBe ‘Visual analysis’ pages for individual EMDB entries (http://pdbe.org/emd-NNNN/analysis, where NNNN is a four-digit EMDB accession number). These pages present orthogonal surface projections of the map and overlays of the map projections with any fitted models from the PDB (Fig. 3). A map-density-distribution plot and atom-inclusion plots for fitted models are also provided. The overlay of the model fitted to the map shows whether or not the model has been deposited in the same frame as the map, the density distribution reveals whether masking was used (the density spike at zero is often due to masking), and the atom-inclusion plot indicates whether the recommended contour level is reasonable.
Figure 3.
Visual analysis of EMDB entries. Visual analysis page for EMDB entry EMD-1831 (http://pdbe.org/emd-1831/analysis), the structure of pig gastric H+,K+-ATPase with bound BeF and SCH28080 (ref. 24). The top right panel presents orthogonal surface projections of the map. The bottom right panel displays orthogonal surface projections of the map overlaid with the fitted PDB model 2XZB (green, all atoms; blue, backbone only). The chart on the top left shows the histogram of density values, and the one below shows the fraction of model atoms contained in the map as a function of contour level (red line, recommended level).
Archiving of raw 3DEM data
Some members of the 3DEM community are in favor of public archiving of raw data associated with a reconstructed map, for example the raw 2D image data or all the intermediate steps, files and parameters to achieve a full ‘electronic notebook’ of a 3DEM experiment. These data could be used to validate the final map, and software developers could use them to test new algorithms. Moreover, deposited raw data could be reprocessed in the future, for example to a higher resolution, by using improved image-processing techniques or (especially in the case of tomography) a different focus compared to the original deposition. Some meeting participants were positively inclined toward the idea of raw-data storage. However, several of the participants were more cautious, on the basis of their own experiences with local archives. They suggest that the challenge of archiving raw data on a global scale—in terms of storage requirements, the logistics of moving data around and especially the cost and effort of manual annotation—could prove to be prohibitively expensive. In tomography, there are projects that generate 100 gigabytes–1 terabyte of both raw data and final reconstructions. J.R.S. suggested that the 3DEM community might be interested in how the light-microscopy community has addressed similar problems, for example by using the Journal of Cell Biology (JCB) DataViewer22.
There was a consensus that routine deposition of raw data to the EMDB is premature but that it would be useful to set up a test-image database of particle images used in single-particle processing and tilt series used in tomography. The PDBe and the OME will use OMERO12 to set up such a database. The database will provide test data for development work and enable investigation of the many issues surrounding raw-data archiving highlighted above. Several meeting participants, including B.C. and G.J.J., agreed to contribute data to this resource.
Segmentation
Segmentation is the process of dividing a map into regions that may or may not overlap and to which, ideally, biologically relevant identifications or annotations can be assigned. The new EMDB data model will improve the handling of segmentations and annotations. An overview presented by S.J.L. at the meeting showed that no single method for representing map segmentations was superior in all respects, and comparable levels of compression could be achieved by all binary segmentation representations. Some methods can handle overlapping regions, and some require a separate file for each segment, whereas others can store all segments in one file, and only a few support non-binary segmentation (usually at the expense of storage efficiency). In the face of a wide variety of storage options, participants agreed that the PDBe will draft a specification as the basis for communitywide discussion involving all software developers, the EMDataBank, and others.
Tomography
Although single-particle reconstruction relies on averaging information from a large number of molecules, electron tomography, in which a series of images is collected from a specimen region at different tilt angles, can be used to obtain 3D reconstructions of individual macromolecules and to study the 3D organization of macromolecular complexes and organelles in their native environment in the cell. 3D single-particle computational methods can also be applied in the context of a tomogram to obtain subtomogram averages at a higher resolution than the tomogram itself.
The number of tomograms deposited in the EMDB is much lower than the number published; in a survey by the PDBe of tomography-related journal publications for the period 2006–2010, only 14% were found to have associated depositions to the EMDB archive. This is because of a lack of consensus in the tomography community as to the need for deposition and the current EMDB data model that inadequately describes the idiosyncrasies of the technique, for example failing to distinguish between tomograms and subtomogram averages. Although the latter issue is addressed in the new EMDB data model, the former is not so easily resolved. The EMDataBank will engage the tomography community on this issue by organizing thematic discussion sessions at 3DEM-related meetings such as the 3DEM Gordon Research Conference and the International Congress on Electron Tomography.
The issue of whether cellular tomograms should be archived in the EMDB was also discussed. It is difficult to define a sharp threshold (for example, based on size or complexity) to distinguish between macromolecular and cellular tomographic reconstructions. Therefore, it was recommended that no such distinction be made at this time and that the EMDB should continue to accommodate both types of reconstructions. This means that the new EMDB data model will need to handle annotations relevant for both types of tomography. For instance, a molecular description of a sample may not be possible or make sense in cellular tomography, and specimen-preparation techniques differ.
Standards and conventions
Community-wide standards and conventions are important for the exchange of data between software packages and the deposition of data in public archives. There was a community-wide effort in 2004 to define such conventions (http://rcsb-cryo-em-development.rutgers.edu/), and although some packages have adopted these to various degrees, many of the major ones have not. In many cases, developers have legitimate reasons for adopting proprietary formats; having already implemented a lot of routines using these formats, they find little to be gained by making a change. As highlighted by J.R.S., the experience in the light-microscopy field has been that it is not sufficient to simply define standards and conventions and that one also has to provide the community with the necessary software libraries to allow data conversion to and from the convention standard13. The OME has done so with the BioFormats library (http://www.openmicroscopy.org/site/support/bio-formats/), which now also supports several 3DEM formats, including Spider (http://www.wadsworth.org/spider_doc/spider/docs/image_doc.html), Imagic (http://www.imag-escience.de/formats.html) and MRC (http://www2.mrc-lmb.cam.ac.uk/image2000.html). Also, as J.B.H. argued, validation tools to examine map parameters, to assess map symmetry and to check orientation conversions are essential to ensure accurate translation of data formats. The EMDataBank will set up a portal to provide access to such tools and will promote the development of new tools.
J.B.H. proposed a new map format that would be able to deal with up to five-dimensional data (channels, x, y, z and volumes) and would not be limited by the legacy issues that plague other formats (often derived from the CCP4 map format (http://www.ccp4.ac.uk/html/maplib.html#description) used in macro-molecular crystallography). However, some participants argued that compatibility with CCP4 was a requirement. M.W. suggested that the 3DEM community should not be hindered by the requirements of the X-ray community in defining its formats. This is partly because crystallographers increasingly store map coefficients and compute maps on the fly, which reduces the importance of maps and map formats. No consensus was reached on this issue, but the recent CCP-EM initiative, with its close ties to CCP4 and the Collaborative Computing Project for NMR (CCPN)23, provides an opportunity to develop a format acceptable to 3DEM developers and supported by relevant crystallography software.
From the perspective of the EMDataBank and the wwPDB, there is an urgent need to resolve issues around standards and conventions in order to incorporate 3DEM validation methods into the D&A pipeline. It was generally agreed that the participants would adapt their software and practices to standards proposed and adopted by the EMDataBank and that therefore EMDataBank should lead this initiative.
Conclusions
The PDBe will investigate the use of SAXS data for 3DEM map validation.
The EMDataBank will lead the effort to improve reporting standards for 3DEM validation and begin by providing a comprehensive description of the FSC method in the new EMDB data model.
The PDBe and OME will use OMERO to set up a test-image database for 3DEM.
The EMDataBank will lead the effort to develop a segmentation file format with the 3DEM community.
The EMDataBank will engage with the electron-tomography community to resolve issues surrounding the deposition of tomographic data to the EMDB archive.
The EMDataBank will set up a portal to provide access to validation tools and will promote the development of new ones.
The EMDataBank will lead the effort to define and promote standards and conventions for 3DEM.
Acknowledgments
We thank P. Haslam for help with the manuscript. This workshop and the OMERO-EMDB project are supported by the BBSRC (BB/G022577). The EMDataBank is funded by the US National Institutes of Health National Institute of General Medical Sciences (R01GM079429). The work on the EMDB at the PDBe also profits from funding by European Molecular Biology Laboratory–EBI and the Wellcome Trust (088944).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available at http://www.nature.com/doifinder/10.1038/nsmb.2426.
References
- 1.Lawson CL, et al. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tagari M, Newman R, Chagoyen M, Carazo JM, Henrick K. Trends Biochem Sci. 2002;27:589. doi: 10.1016/s0968-0004(02)02176-x. [DOI] [PubMed] [Google Scholar]
- 3.Velankar S, et al. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson R, et al. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander GC, et al. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heymann JB, Belnap DM. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Tang G, et al. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Kremer JR, Mastronarde DN, McIntosh JR. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 9.Crowther RA, Henderson R, Smith JM. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 10.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winn MD, et al. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan C, et al. Nat Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linkert M, et al. J Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H, Henrick K, Nakamura H. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 15.Henrick K, Newman R, Tagari M, Chagoyen M. J Struct Biol. 2003;144:228–237. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald PMD, et al. International Tables for Crystallography G. In: Hall SR, McMahon B, editors. Definition and exchange of crystallographic data. Springer; 2005. pp. 295–443. [Google Scholar]
- 17.Henderson R, et al. J Mol Biol. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenthal PB, Henderson R. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Mertens HD, Svergun DI. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Berry R, et al. Proc Natl Acad Sci USA. 2009;106:8561–8566. doi: 10.1073/pnas.0812178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harauz G, van Heel M. Optik (Stuttg) 1986;73:146–156. [Google Scholar]
- 22.Hill E. J Cell Biol. 2008;183:969–970. [Google Scholar]
- 23.Vranken WF, et al. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 24.Abe K, Tani K, Fujiyoshi Y. Nat Commun. 2011;2:155. doi: 10.1038/ncomms1154. [DOI] [PMC free article] [PubMed] [Google Scholar]